
The meeting on Translational Control and Non-Coding RNA took place between 8 and 12 November 2006, in Nove Hrady, the Czech Republic, and was organized by M. Pospisek and L. Valasek.
Introduction
Many pathways that regulate gene expression target the complex process of protein synthesis and in particular its initiation stage. During the past 40 years or so, all the initiation factors required for the assembly of translationally competent ribosomes have been identified and their principal roles determined. Recently, there has been significant progress in determining the structure of the components of the translation apparatus: from crystal structures of prokaryotic ribosomes, to crystal and nuclear magnetic resonance structures of factors, and cryo-electron microscopy reconstructions of functional ribosomal complexes. These advances now allow the molecular mechanisms of the stages involved in initiation to be investigated in detail. Concurrently, diverse studies have revealed that translational control regulates processes ranging from development to learning. This meeting highlighted recent advances about canonical and internal ribosome entry site (IRES)-mediated mechanisms of eukaryotic initiation, and the control of translation during development in response to stress and by mechanisms ranging from signal transduction pathways to regulation by noncoding RNAs and RNA-binding proteins.
Translation initiation mechanisms
Canonical 5′-end-dependent eukaryotic translation initiation requires at least 11 initiation factors (eIFs) and occurs in two stages: formation of the 48S initiation complex at the AUG codon of messenger RNA (mRNA), and its joining with a 60S ribosomal subunit (Fig 1; Hinnebusch et al, 2007; Pestova et al, 2007). eIF1 has a crucial role in the selection of initiation codons, allowing scanning 43S complexes to discriminate against codon–anticodon mismatches and preventing premature eIF5-induced hydrolysis of eIF2-bound GTP and Pi release. According to a recent model, eIF1—in cooperation with eIF1A—promotes a scanning-competent ‘open' conformation of the 43S complex. The establishment of codon–anticodon base-pairing leads to tightening of the eIF1A–40S interaction and subsequent eIF1 dissociation, which switches the complex to a ‘closed' conformation and relieves repression of hydrolysis of eIF2-bound GTP and Pi release. In his opening keynote lecture, A. Hinnebusch (Bethesda, MD, USA) presented work in collaboration with J. Lorsch (Baltimore, MD, USA), which provides more evidence that dissociation of eIF1 from the 40S subunit is required to arrest the scanning 43S complex at the AUG codon and to release Pi from eIF2. Hinnebusch showed that the Sui− phenotype—enhanced initiation on UUG codons—of yeast eIF1 mutants correlated with a reduced affinity of eIF1 for 40S subunits in vitro and, importantly, with increased rates of mutant eIF1 dissociation and Pi release from in vitro-assembled 48S complexes. The eIF1A carboxy-terminal domain (CTD) and its amino-terminal domain (NTD) had opposite effects on scanning and start codon selection: CTD mutations yielded a Sui− phenotype, whereas NTD mutations led to hyperaccuracy. Consistent with the current model, these defects could be accounted for by altered rates of eIF1A dissociation from 48S complexes: eIF1A CTD mutations stabilized the binding of eIF1A to 48S complexes assembled in vitro on UUG triplets, whereas NTD mutations had the opposite effect and accelerated eIF1 release from 40S subunits. Importantly, the hyperaccurate eIF1A mutant also decreased eIF1 dissociation from 48S complexes.
Figure 1.
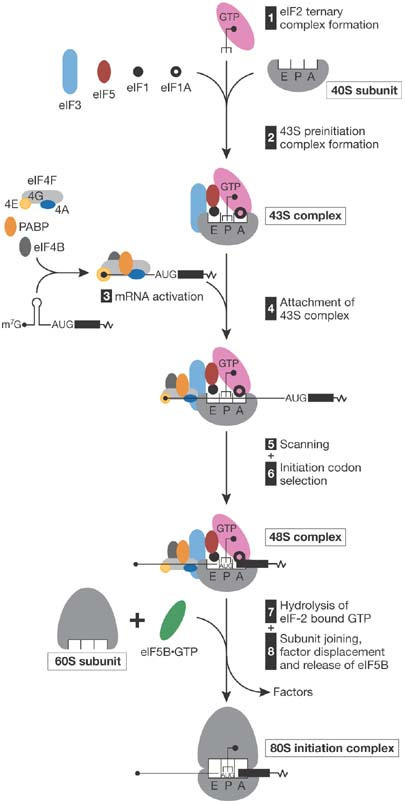
A simplified model of the mechanism of cap-dependent initiation. eIF2·GTP/Met-tRNAiMet ternary complex, eIFs 3, 1, 1A, 5 and a 40S subunit form a 43S pre-initiation complex, which initially binds to the 5′-proximal region of mRNA. The secondary structure of the mRNA is unwound in an ATP-dependent manner by the cooperative action of eIFs 4A, 4B, 4F and poly(A) binding protein (PABP). This complex then scans downstream to the initiation codon, where it forms a 48S initiation complex with an established codon–anticodon interaction. Joining of a 60S subunit is mediated by eIF5, which induces hydrolysis of eIF2-bound GTP and partial dissociation of eIF2-GDP from a 40S subunit, and eIF5B, which promotes subunit joining and dissociation of other factors. For simplicity, PABP bound to the 3′-poly(A) tail and recycling of eIF2·GDP by eIF2B are not shown. Met, methionine.
The multisubunit factor, eIF3, has crucial roles in ribosomal subunit anti-association, 43S complex formation and scanning. A composite cryo-electron microscopy reconstruction of human eIF3–40S complexes showed that eIF3 binds primarily to the solvent side of the 40S subunit, but the positions of individual subunits have not been assigned. The CTD of yeast eIF3a binds to helices 16–18 of 18S rRNA, and eIF3a–NTD and eIF3c bind to ribosomal proteins (rp) S10A and rpS0A (on the back of the 40S subunit), which is consistent with yeast eIF3 also binding to the solvent side of the 40S subunit. E. Rutkai (Prague, Czech Republic) identified several yeast eIF3–40S interactions involving eIF3j (HCR1) and rpS2, the HCR1-like domain of eIF3a and rpS10, and eIF3c–CTD and rpS33/Asc1. The last interaction suggests that the part of eIF3 that was previously modelled to stretch towards the left foot of the 40S subunit is instead probably located near the mRNA exit channel. B. Szamecz (Prague, Czech Republic) presented data supporting the important role of eIF3a–CTD, which interacts with rpS0A, in 43S complex formation, reinitiation after translation of open reading frame 1 (ORF1) of the general control non-derepressible 4 (GCN4) mRNA and in maintaining the 60S/40S ratio. Considering the role of rpS0A in 40S subunit maturation, this suggests that the eIF3a–CTD/rpS0A contact is also important for processes other than initiation.
Attachment of 43S complexes to mRNA is thought to require eIF4F, which comprises eIF4E (cap-binding subunit), eIF4A (RNA helicase) and eIF4G, which binds to eIF4E, eIF4A and eIF3. F. Ramirez-Valle (New York, NY, USA) reported that, surprisingly, RNA interference-mediated knockdown of eIF4GI and eIF4GII isoforms reduced total protein synthesis by only about 20%, which is similar to the effect of silencing p97, an eIF4G family member. Conversely, silencing eIF4GI and p97 reduced translation by more than 60%. These data reveal unexpected complexity in eukaryotic initiation mechanisms and suggest that eIF4GI and p97 might preferentially promote translation of different mRNAs, although partly redundant roles of eIF4G and p97 in initiation cannot be excluded. Consistently, most mRNAs were unaffected by eIF4GI knockdown, but mRNAs that encode factors involved in growth and metabolism were depleted from polysomes and mRNAs that encode catabolic pathway factors were enriched. The effects of eIF4GI silencing resemble the effects of silencing mammalian target of rapamycin (mTOR), which integrates signals from growth factors and nutrient availability to regulate cell growth. eIF4GI-silenced cells fail to respond to nutritional signals, indicating that eIF4GI links nutrient sensing to cell proliferation, probably as an mTOR effector.
K. Nielsen (Aarhus, Denmark) presented the crystal structure of the exon-junction complex, which contains the eIF4A-like eIF4AIII bound to RNA and with ATP poised in the prehydrolysis state. This structure provides information on the mechanism of action of eIF4A. The α/β domains of eIF4AIII are in a closed conformation and both contribute to interactions with RNA, accounting for the higher RNA affinity of the ATP-bound conformation. The sharp kink in the RNA induced by DEAD-box motif 1b is incompatible with binding of a double-stranded RNA, suggesting a function for this motif and the post-motif II region as wedges that separate the strands of RNA duplexes. Structural comparison of eIF4AIII in the prehydrolysis state and VASA—a Drosophila DEAD-box RNA helicase—in its active state suggests that minor conformational changes in motif 1 of eIF4A would allow ATP hydrolysis to occur.
In addition to cap-dependent initiation, alternative modes of initiation have evolved that allow translation to occur during conditions in which cap-dependent initiation is compromised. Therefore, IRESs are mRNA elements that promote ribosomal binding to internal areas in 5′ untranslated regions (UTRs) independently of eIF4E and the 5′-end of mRNA (Hellen & Sarnow, 2001). Important progress has been made in elucidating the functional mechanism of the IRES of the hepatitis C virus (HCV). P. Lukavsky (Cambridge, UK) reported that IRES domain II is required to promote eIF5-induced hydrolysis of eIF2-bound GTP and eIF2·GDP release, which are both prerequisites for ribosomal subunit joining. Domain II induces conformational changes in the 40S subunit that might be responsible for its influence on this step in initiation. Bioinformatic analyses (C. Hellen, New York, NY, USA) identified HCV-like IRESs in picornaviruses from several genera, suggesting that IRESs have exchanged between unrelated flaviviruses and picornaviruses by recombination. Initiation on the foot-and-mouth disease virus IRES depends on binding of eIF4G/eIF4A·ATP, and dimethyl sulphate footprinting reported by E. Martínez-Salas (Madrid, Spain) suggests that this interaction occurs in vivo.
The HCV IRES is a promising target for antiviral drugs because its initiation mechanism differs from that used by cellular mRNAs. To facilitate the development of small-molecule screening strategies, T. Masek (Prague, Czech Republic) reported a new bicistronic reporter system that functions in living yeast. Instead of directly assaying translation of the second cistron, Masek's system enhances the signal by using the GAL4 transcription factor as the second cistron and a host yeast strain that expresses a reporter gene controlled by a GAL4-inducible promoter. The HCV IRES was active in yeast cells and can therefore be subjected to screening by this new system.
Groups of viral IRESs have similar structures despite their low sequence identity; however, no common discernable characteristics that could identify IRESs in cellular mRNAs have been identified. M. Mokrejs (Prague, Czech Republic) presented a bioinformatics approach that allows predictions of IRESs in cellular mRNAs. This study did not identify any common properties of cellular IRESs, including the previously suggested ‘Y' structure. Similar observations were presented by M. Holcik (Ottawa, Ontario, Canada) who also reported on a genome-wide RNA structure search that identified new IRESs that have limited structural similarity with the X-linked inhibitor of apoptosis IRES. These data suggest that, unlike viral IRESs, cellular IRESs are not defined by overall structure, but instead depend on short sequence motifs and trans-acting factors for their function.
Several initiation factors are universally conserved. A. La Teana (Ancona, Italy) presented a detailed characterization of the ribosomal interaction of Escherichia coli IF2, an eIF5B homologue, which mediates ribosomal subunit joining and binding of initiator transfer RNA (tRNA) to the 30S subunit (Laursen et al, 2005). The nonconserved N2 domain of IF2 is required for a high-affinity but functionally dispensable interaction with 30S subunits that is independent of GTP and IF1. Conversely, the weaker but functional interaction of IF2 with the 30S subunit involves the β-barrel module G3 located within the G domain of IF2 and strongly depends on both GTP and IF1. However, N-terminally truncated IF2 could bind 30S subunits even in the absence of GTP or IF1 when initiator tRNA was present, suggesting that after GTP hydrolysis and release of IF1, tRNA could still stabilize IF2/70S binding. La Teana suggested that hydrolysis of IF2-bound GTP is required for the conformational transition that allows the complete dissociation of IF2 from the 3′-acceptor end of initiator tRNA followed by the release of IF2 from the 70S ribosome.
Binding of eIF6 to eukaryotic 60S subunits prevents their association with 40S subunits. eIF6 phosphorylation at distinct sites regulates its ribosomal association and subcellular localization. P. Londei (Bari, Italy) reported that Sulfolobus solfataricus aIF6 is tightly but substoichiometrically bound to 50S ribosomal subunits during normal growth and, similar to eIF6, prevents subunit association and represses in vitro translation. Its expression is upregulated in response to thermal stress, indicating that it might have a regulatory function. Phosphorylation of aIF6 in vivo does not induce its release from 50S subunits, suggesting an alternative mechanism of reactivation of sequestered 50S subunits that can be triggered in vitro by high concentrations of Na+.
Termination of eukaryotic translation is triggered by peptide release factors eRF1 and eRF3. eRF1 recognizes all three termination codons and induces hydrolysis of peptidyl tRNA, but the function of eRF3 remained unclear. T. Pestova (New York, NY, USA) reported that GTP hydrolysis by eRF3 couples stop codon recognition and peptidyl-tRNA hydrolysis mediated by eRF1. Therefore, binding of eRF1, eRF3 and GTP to pre-termination complexes yields a complex that requires further rearrangement, induced by GTP hydrolysis, for correct positioning of the GGQ loop of eRF1 in the peptidyl transferase centre to induce peptide release. The mechanism of recycling of eukaryotic post-termination ribosomal complexes that contain mRNA and P-site deacylated tRNA is completely unknown. Pestova also reported that eIF3, eIF1 and eIF1A are able to promote recycling of these complexes, and that a complete set of eIFs can promote post-termination ribosomal recycling and subsequent initiation on the released mRNA.
Translation regulation
Protein synthesis accounts for a large proportion of the energy consumed by cells and is therefore tightly regulated to cope with the reduced energy availability that occurs during stress (Fig 2; Holcik & Sonenberg, 2005). The important recovery phase of the cellular stress response also often involves reprogramming of the translation machinery. P. Sunnerhagen (Göteborg, Sweden) presented microarray analysis of polysomes of yeast cells undergoing hyperosmotic stress, and showed that certain classes of mRNAs are upregulated primarily at the post-transcriptional level and at discrete times after induction of stress. Mutations in members of stress-activated MAP kinase (SAPK) pathways, in particular radiation sensitivity complementation kinase (Rck) 2, impaired the ability of the cells to control the translational response to hyperosmotic stress.
Figure 2.
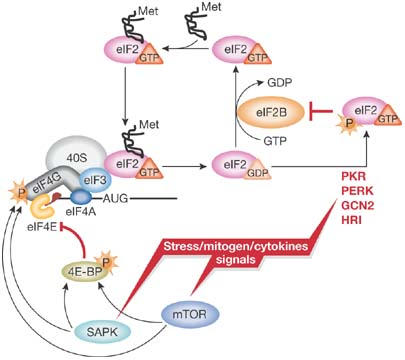
A simplified model of translation regulation by stress. Various stress, mitogen and cytokine signals evoke activation of distinct signalling pathways resulting in the reduction of protein synthesis. Activation of eIF2α kinases (PKR-like ER kinase (PERK); haem-regulated inhibitor kinase (HRI); double-stranded RNA-activated protein kinase (PKR); general control non-derepressible-2 (GCN2)) results in the reduction of available ternary complex, whereas activation of the mammalian target of rapamycin (mTOR) and/or stress-activated MAP kinase pathways (SAPK) impinges on the activation of messenger RNA and its recruitment into the 48S initiation complex. For simplicity, only selected factors are shown. Met, methionine; P, phosphate.
The GCN2 kinase, which is activated by nutrient deprivation, phosphorylates the eIF2α subunit, thereby reducing ternary complex levels and downregulating translation of most mRNAs. GCN2 regulates feeding behaviour, learning and memory in mice; however, GCN2 activation in the brain is inhibited by a highly expressed protein known as imprinted and ancient (IMPACT). B. Castilho (Sao Paulo, Brazil) reported on detailed mapping of the expression of IMPACT in the mouse brain and found that it is expressed in distinct areas, predominantly in the hypothalamus. Interestingly, IMPACT is not found in hippocampal dentate gyrus granule cells and CA3 pyramidal neurons, suggesting that GCN2 is required for hippocampal-dependent memory.
Global regulation of translation involves modulation of the activity of general initiation factors, whereas mRNA-specific control is mediated by regulatory proteins or noncoding RNAs that bind to the 3′ and/or 5′ UTRs of target mRNAs. Male-specific lethal 2 (MSL-2) is a part of the dosage compensation complex of Drosophila melanogaster, which ensures increased transcription of the single male X chromosome. Repression of translation of msl-2 mRNA in females constitutes a paradigm for specific control of translation by RNA-binding proteins (Hentze et al, 2007). In the first part of his keynote lecture, M. Hentze (Heidelberg, Germany) reported that robust msl-2 repression in females requires binding of the female-specific sex-lethal (SXL) protein to both 5′ and 3′UTRs of msl-2 mRNA. SXL mediates repression through a ‘failsafe' mechanism by targeting two initiation steps: SXL bound to the msl-2 3′UTR inhibits recruitment of 43S complexes to the 5′UTR in a cap-independent manner, whereas SXL bound to the 5′UTR inhibits their scanning to the initiation codon. Repression by SXL binding to the 3′UTR requires additional proteins including upstream of N-ras (UNR), which is recruited by SXL. The possibility that poly(A)-binding protein (PABP) and UNR might interact indicates that UNR might inhibit one or more activities of PABP in initiation.
MicroRNAs (miRNAs), which are approximately 21–23 nucleotide-long regulators of gene expression, base-pair to target mRNAs and, depending on complementarity, either cause mRNA cleavage or repress translation (Pillai et al, 2007). The mechanism by which miRNAs repress translation has not been resolved: different studies implicate a post-initiation step, inhibition of eIF4E/cap and poly(A) function, and accumulation of repressed mRNAs in processing bodies. In the second part of his talk, Hentze reported that to address the biochemical basis for repression, his laboratory developed an in vitro system based on Drosophila cell-free translation extracts that recapitulated repression without affecting mRNA stability. The system contains the abundant endogenous miRNA miR-2 and a capped, polyadenylated luciferase reporter mRNA, which contains in its 3′UTR several miR-2 binding sites from Reaper mRNA, a validated miR-2 target. Repression of translation of this mRNA was specific and was alleviated by antisense oligonucleotides targeted against miR-2. Sucrose density gradient centrifugation analysis revealed that miR-2 inhibits initiation at the stage of 43S complex recruitment in a cap-dependent manner. Intriguingly, repressed mRNA that failed to enter 48S or 80S initiation complexes did not appear in the messenger ribonucleoprotein fractions commonly found at the top of the gradients, but was instead engaged in heavy complexes named ‘pseudopolysomes', with a sedimentation coefficient of more than 80S (Thermann & Hentze, 2007). A. Fatica (Rome, Italy) showed that miR-223 represses translation of NF1-A, a transcription factor involved in haematopoiesis. Interestingly, the repressed NF1-A mRNA was found in heavy complexes, perhaps also engaged in pseudopolysomes.
B. Vecerek (Vienna, Austria) and D. Lin (New York, NY, USA) presented other examples of regulation by noncoding RNAs. RyhB noncoding RNA, the transcription of which is induced in iron-depleted E. coli, binds to mRNAs that encode iron-binding proteins to induce their degradation. Vecerek reported that RyhB also represses synthesis of the ferric uptake regulator Fur by base-pairing to the initiation codon region of a short open reading frame (uof) upstream to the fur cistron; the coupled translation of uof and fur is consequently coordinately repressed. Brain cytoplasmic RNA1 (BC1) noncoding RNA is a translational repressor that is specifically delivered to postsynaptic dendritic microdomains in the central nervous system; it presumably regulates protein synthesis at the synapse and contributes to long-term synaptic plasticity. Lin reported that BC1 RNA interacts with eIF4A and PABP, and data indicating that it inhibits the helicase activity of eIF4A but stimulates its ATPase activity suggest that the BC1-eIF4A interaction represses initiation by uncoupling ATP hydrolysis from RNA unwinding.
Concluding remarks
Although translation has been studied for more than 50 years, new concepts continue to emerge. This meeting provided excellent opportunities to discuss new data and hypotheses leading towards a more complete understanding of molecular mechanisms of protein synthesis and the role of its regulation in the context of the living organism.
Acknowledgments
We thank M. Pospisek, L. Valasek and their colleagues for organizing the conference, and the Federation of European Microbiological Societies (FEMS), The RNA Society, Roche, Eppendorf, Medesa, Beckman Coulter, Bio-Rad, BioTech and Promega for financial support. We apologize to those whose data were not discussed owing to space limitations.
References
- Hellen CU, Sarnow P (2001) Internal ribosome entry sites in eukaryotic mRNA molecules. Genes Dev 15: 1593–1612 [DOI] [PubMed] [Google Scholar]
- Hentze MW, Gebauer F, Preiss T (2007) cis-regulatory sequences and trans-acting factors in translational control. In Translational Control in Biology and Medicine, MB Mathews, N Sonenberg, JWB Hershey (eds), pp 269–298. New York, USA: Cold Spring Harbor Laboratory Press [Google Scholar]
- Hinnebusch AG, Dever TE, Asano K (2007) Mechanism of translation in the yeast Saccharomyces cerevisiae. In Translational Control in Biology and Medicine, MB Mathews, N Sonenberg, JWB Hershey (eds), pp 225–268. New York, USA: Cold Spring Harbor Laboratory Press [Google Scholar]
- Holcik M, Sonenberg N (2005) Translational control in stress and apoptosis. Nat Rev Mol Cell Biol 6: 318–327 [DOI] [PubMed] [Google Scholar]
- Laursen BS, Sorensen HP, Mortensen KK, Sperling-Petersen HU (2005) Initiation of protein synthesis in bacteria. Microbiol Mol Biol Rev 69: 101–123 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pestova TV, Lorsch JR, Hellen CU (2007) The mechanism of translation initiation in eukaryotes. In Translational Control in Biology and Medicine, MB Mathews, N Sonenberg, JWB Hershey (eds), pp 87–128. New York, USA: Cold Spring Harbor Laboratory Press [Google Scholar]
- Pillai RS, Bhattacharyya SN, Filipwicz W (2007) Repression of protein synthesis by miRNAs: how many mechanisms? Trends Cell Biol 17: 118–126 [DOI] [PubMed] [Google Scholar]
- Thermann R, Hentze MW (2007) Drosophila miR2 induces pseudo-polysomes and inhibits translation initiation. Nature [doi: 10.038/nature05878] [DOI] [PubMed] [Google Scholar]


