Abstract
Mitochondrial heat-shock protein 70 (mtHsp70) and its partner proteins drive protein import into the matrix. Tim15/Zim17/Hep1 is a mtHsp70 partner protein on the matrix side of the inner mitochondrial membrane. We determined the nuclear magnetic resonance (NMR) structure of the core domain of Tim15. On the basis of the NMR structure, we created Tim15 mutants and tested their ability to complement the functional defects of Tim15 depletion and to suppress self-aggregation of mtHsp70 in vivo. A pair of basic residues, Arg 106 and His 107, conserved Asp 111 and flexible loop 133–137, and were important (Arg 106–His 107 pair and Asp 111) or partly important (the loop 133–137) for yeast cell growth, mitochondrial protein import and the suppression of mtHsp70 aggregation. Therefore, the function of Tim15 in yeast cell growth is well correlated with its ability to suppress mtHsp70 aggregation, although it is still unknown whether inhibition of mtHsp70 aggregation is the primary function of Tim15.
Keywords: chaperone, Hsp70, mitochondria, protein import, Tim15
Introduction
Mitochondria are essential double-membrane organelles in eukaryotic cells and contain approximately 1,000 different proteins. Mitochondrial functions rely on the correct transport of resident proteins synthesized in the cytosol to mitochondria. Protein import into mitochondria is mediated by membrane protein complexes—protein translocators—in the outer and inner mitochondrial membranes, in cooperation with their assistant proteins in the cytosol, intermembrane space and matrix (Endo et al, 2003; Wiedemann et al, 2004; Neupert & Herrmann, 2007). Proteins destined for the mitochondrial matrix cross the outer membrane with the aid of the outer membrane translocator—the TOM40 complex—and then the inner membrane with the aid of the inner membrane translocator—the TIM23 complex—and finally mitochondrial motor and chaperone (MMC) proteins including mitochondrial heat-shock protein 70 (mtHsp70), and translocase in the inner mitochondrial membrane (Tim)15, which is also known as zinc finger motif (Zim)17 or mtHsp 70 escourt protein (Hep)1 in the matrix (Burri et al, 2004; Sichting et al, 2005; Szklarz et al, 2005; Yamamoto et al, 2005).
Hsp70s are a family of ubiquitous molecular chaperones of approximately 70 kDa with ATPase activities (Bukau & Horwich, 1998). They bind to and release unfolded protein substrates quickly in their ATP-bound states, but in their ADP-bound states substrate binding and release are slow. Hsp70s generally function in cooperation with their partner proteins or co-chaperones. In the yeast mitochondrial protein import systems (van der Laan et al, 2006; Neupert & Herrmann, 2007), mtHsp70 functions in cooperation with MMC proteins: Yge1p/Mge1p functions as a nucleotide-exchange factor for mtHsp70, stress seventy subfamily C (Ssc1p); Tim14/presequence translocase-associated protein import motor (Pam18), a J protein, stimulates the ATPase activity of mtHsp70; and Tim16/Pam16 facilitates docking of mtHsp70 to another MMC protein, Tim44 of the TIM23 complex. Tim44, a subunit of the TIM23 complex, allows two mtHsp70 molecules to bind to newly translocating precursor proteins at the exit of the protein-conducting channel of the TIM23 complex, presumably in a hand-over-hand manner. Tim15, a peripheral inner membrane protein facing the matrix, assists mtHsp70 in protein import into mitochondria both in vitro and in vivo (Burri et al, 2004; Yamamoto et al, 2005), although it is not associated with the TIM23 complex (Szklarz et al, 2005). Tim15 has two zinc-finger motifs that are typical of a class of J proteins and essential for Tim15 functions (Yamamoto et al, 2005), whereas Tim14 lacks a zinc-finger motif; therefore Tim15 might function in trans for a J protein Tim14 to facilitate, for example, the substrate binding for mtHsp70. Alternatively, because depletion of Tim15 leads to the aggregation of mtHsp70, it has been proposed that the primary function of Tim15 could be as a mtHsp70-specific chaperone to prevent its self-aggregation (Sichting et al, 2005; Szklarz et al, 2005).
To gain further insight into the functions of Tim15, we determined the nuclear magnetic resonance (NMR) structure of the core domain of Tim15. On the basis of the structure determined, we created several Tim15 mutants to identify the region that is important for its function, including the ability to prevent self-aggregation of mtHsp70.
Results And Discussion
The core domain of Tim15/Zim17
To gain structural information on Tim15/Zim17, we purified the 15N-labelled mature part of Tim15 without a presequence (residues 43–174) from Escherichia coli cells for NMR spectral analyses. Tim15 contains two zinc-finger motifs (C75XXC78 and C100XXC103; Fig 1A); therefore, we recorded a [1H,15N] heteronuclear single quantum correlation (HSQC) NMR spectrum of Tim15 after extensive dialysis against EDTA-containing buffer to remove Zn2+ (Fig 1A). The HSQC spectrum of Tim15 without Zn2+ showed NH resonances with little chemical-shift dispersion. However, on the addition of Zn(OAc)2, the HSQC spectrum of Tim15 became well resolved and showed NH peaks with large chemical-shift dispersion (Fig 1B). Therefore, Tim15 only has a stable, well-ordered tertiary structure in the presence of Zn2+, suggesting a structural role for the two zinc-finger motifs chelating Zn2+ in the conformation of Tim15.
Figure 1.
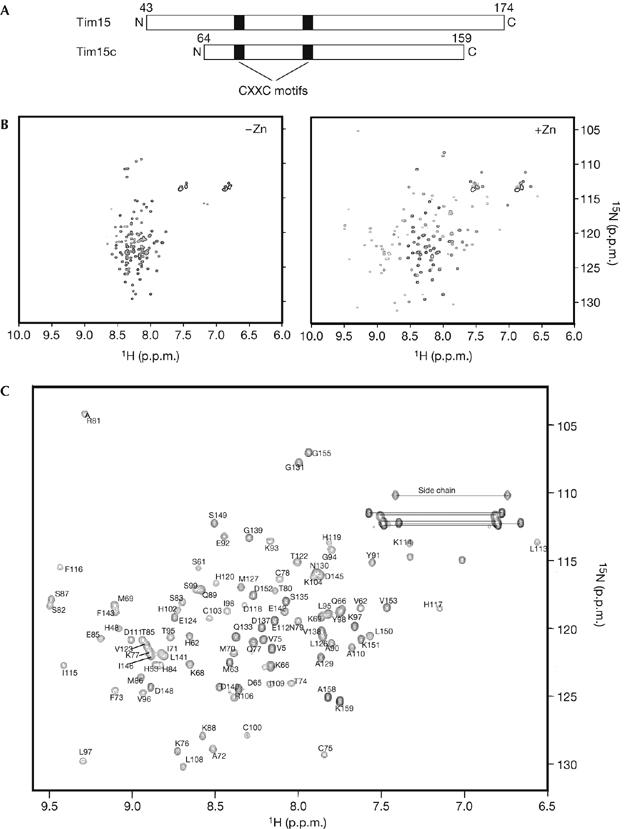
[1H,15N]HSQC spectra of Tim15/Zim17 and Tim15c. (A) Domain organization of Tim15 and Tim15c. Black boxes indicate zinc-finger motifs. (B) Purified Tim15 in 20 mM Tris–HCl (pH 7.4), 50 mM NaCl, 1 mM DTT and 30 μM Zn(OAc)2 was treated with 2 mM EDTA overnight at 4°C, dialysed twice against 20 mM sodium acetate (pH 4.5), 50 mM NaCl and 1 mM DTT at 4°C to remove EDTA, and concentrated to 0.2 mM. [1H,15N]HSQC spectra were recorded before (left) and after (right) the addition of 1 mM Zn(OAc)2. (C) [1H,15N]HSQC spectrum of 0.7 mM Tim15c with assignments for backbone amide crosspeaks. Side chain amide groups are indicated with horizontal lines. The spectra were recorded in 20 mM sodium acetate (pH 4.5), 50 mM NaCl, 1 mM DTT, 30 μM Zn(OAc)2 and 95% H2O/5% D2O at 299 K. DTT, dithiothreitol; HSQC, heteronuclear single quantum correlation.
Limited digestion with trypsin followed by mass spectrometry measurements of the tryptic peptides showed that Tim15 contains a trypsin-resistant core domain (Tim15c), consisting of residues 64–159 (supplementary Fig S1 online). A comparison of [1H,15N]HSQC NMR spectra and circular dichroism (CD) spectra of Tim15c and full-length Tim15 showed that residues 64–159 have similar conformations in both proteins (supplementary Fig S2 online), and that Tim15c could functionally replace Tim15 in yeast cells (supplementary Fig S3 online). Thus we decided to determine the NMR structure of the 96-residue Tim15c instead of the full-length Tim15 (Fig 1B).
Structure of Tim15c
NMR spectra of purified Tim15c in aqueous solution with 30 μM Zn(OAc)2 were recorded at 299 K. The backbone and side-chain resonances of Tim15c were assigned from standard heteronuclear three-dimensional NMR experiments (Fig 1C). The three-dimensional structure of Tim15c was determined using the programme CYANA based on 1,656 nuclear Overhauser enhancement-derived distance and 100 dihedral angle restraints (Table 1). A total of 100 NMR structures were calculated and the 20 structures with the lowest energy were selected. The final 20 structures were superimposed onto the mean coordinate position for the backbone atoms (N, Ca, C′) of residues 68–130 and 140–155 (Fig 2A). The overall structure of Tim15c is well defined by NMR data, except for the amino-terminal residues 64–67, the carboxy-terminal residues 156–159 and the loop segment spanning residues 131–139, which are not structured. The r.m.s. displacement to the mean coordinate position is 0.36 Å for the backbone atoms of residues 68–130 and 140–155 (Table 1).
Table 1.
Structural statistics for 20 final nuclear magnetic resonance structures
| Distance restraints | |
|---|---|
| All | 1,656 |
| Short range (∣i−j∣⩽1) | 911 |
| Medium range (1<∣i−j∣<5) | 259 |
| Long range (∣i−j∣⩾5) | 486 |
| Hydrogen bonds* | 36 |
| Zinc distance restraints | 12 |
| Dihedral angle constraints† | |
| φ | 50 |
| ψ | 50 |
| Mean CYANA target function | 1.27 |
| C. Ramachandran plot statistics % | |
| Most favoured regions | 78.4 |
| Additionally allowed regions | 21.0 |
| Generously allowed regions | 0.4 |
| Disallowed regions | 0.1 |
| R.m.s.d.s to mean structure (residues) (Å) | |
| Backbone atoms (N, Cα, C′, O) | 0.36 |
| All heavy atoms | 0.81 |
*Hydrogen-bond constraints were derived from HD exchange experiments and implemented as two upper (HN–O 2.3 Å, N–O 3.3 Å) and two lower (HN–O 1.8 Å, N–O 2.8 Å) distance constraints for each hydrogen bond. †Backbone φ and ψ dihedral constraints determined with TALOS.
Figure 2.
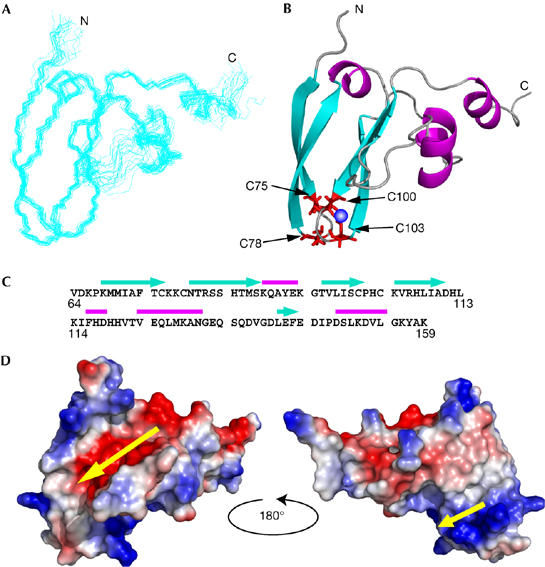
Nuclear magnetic resonance solution structure of Tim15c. (A) Superposition of the 20 lowest energy structures of Tim15c. (B) A ribbon representation of the lowest energy structure of Tim15c, showing the distribution of regular secondary structure elements and location of the two zinc-finger motifs. Side chains of the zinc ligands are shown in red and the zinc atom as a blue sphere. (C) The amino-acid sequence of Tim15c showing the locations of secondary structure elements: α-helices are indicated by magenta bars and β-strands by cyan arrows. (D) Electrostatic potential mapped onto the surface of Tim15c (left) and after rotation by 180° as shown (right). Red and blue regions represent localization of negative and positive charge, respectively. Yellow arrows indicate an acidic groove (left) and a basic groove (right).
The determined NMR structure (Fig 2A,B) shows that Tim15c is an L-shaped molecule. The two zing-finger motifs are located at the end of the L, and are sandwiched by two-stranded antiparallel β-sheets consisting of segments 68–75 and 79–87, and segments 95–100 and 104–110 (Fig 2B,C). Two short α-helices (residues 123–130 and 147–153) constitute another leg of the L (Fig 2B,C). The overall folding of Tim15c does not resemble that of the cysteine-rich domain of DnaJ (Martinez-Yamout et al, 2000). Fig 2D shows the electrostatic potential of Tim15c mapped onto the molecular surface and viewed from opposite angles. The outer (convex) face of the L (Fig 2D, left) has a large acidic groove, which is lined with five acidic residues, Asp 140, Glu 142, Glu 144, Asp 145 and Asp 148, whereas the inner (concave) face of the L (Fig 2D, right) has two positively charged residues, Arg 106 and His 107, next to the zinc-finger motifs.
Mutagenesis of Tim15/Zim17
On the basis of the NMR structure of Tim15c, we created several mutants of Tim15/Zim17 to delineate functionally important residues or regions of Tim15. Although tim15Δ cells with chromosomal disruption of the TIM15 gene could not grow on specific carbohydrate diet (SCD; Fig 3B), a plasmid carrying the TIM15 gene could complement the TIM15 disruption, allowing the cells to grow at 30°C and 37°C (Fig 3B). tim15Δ cells transformed with a plasmid carrying a gene for Tim155DE, which lacks negative charges in the acidic groove of the convex face of the L-shaped molecule by five combined mutations of D140A, E142A, E144A, D145A and D148A (Fig 3A, top left), could also grow on SCD at 30°C and 37°C (Fig 3B). Therefore, negative charges in the acidic groove are not essential for the function of Tim15. Conversely, charge neutralization of the opposite concave face of the L-shaped molecule by the R106A and H107A mutations (2RH; Fig 3A, top right) did not allow Tim15 to complement the growth defects of tim15Δ cells at either 30°C or 37°C (Fig 3B). Replacement of Asp 111, which is located near the concave face (Fig 3A, top right) and is conserved among Tim15 homologues in various organisms, with Ala (Tim15D111A) also rendered Tim15 defective in complementing the TIM15 deletion in cell growth (Fig 3B). Tim15 contains a flexible loop connecting the zinc-finger motifs (residues 100–103) and α-helix (residues 123–128); therefore, we also tested the role of this loop in Tim15 functions. Interestingly, a plasmid with a gene for Tim15Δ133−137, which lacks the loop (Fig 3A, bottom), could only partly complement the TIM15 gene disruption, showing growth defects at the elevated temperature (37°C) on SCD, and these growth defects became prominent on nonfermentable SCG (see Methods; Fig 3B). Using CD measurements we confirmed that those mutations did not significantly alter the overall folding of Tim15 (supplementary Fig S4 online). Therefore, a pair of basic residues, Arg 106 and His 107, conserved Asp 111, and the loop spanning residues 133–137 (only in part) are important for yeast cell viability.
Figure 3.
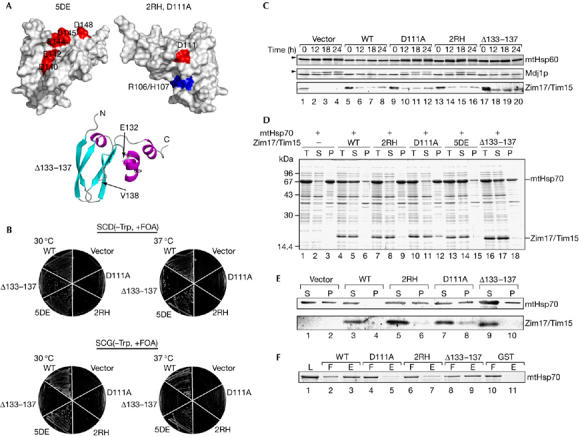
Functions of Tim15/Zim17 mutants. (A) The positions of mutations are indicated on the surface of Tim15c for 5DE (top left, D140A/E142A/E144A/D146A/D148A), 2RH (top right, R106A/H107A) and D111A (top right), and on the ribbon model of Tim15c for Δ133–137 (bottom; deletion of residues 133–137). (B) Plasmids (pRS314) carrying the genes for wild-type Tim15 (WT), Tim15D111A (D111A), Tim152RH (2RH), Tim155DE (5DE) or Tim15Δ133−137 (Δ133–137) or without an inserted gene (vector) were introduced into the tim15Δ strain supplied with the TIM15 gene from a single-copy plasmid of the URA3 selection marker (Δtim15/pRS316-Tim15). The resulting transformants were grown on SCD(−Trp) and SCG(−Trp) plates containing 0.1% 5-FOA at 30°C or 37°C for 2 days (SCD; see Methods) or 3 days (SCG; see Methods). (C) The GAL-TIM15 strain (Yamamoto et al, 2005) transformed with the plasmids (pRS314) carrying the genes for Tim15 or Tim15 mutants as in (B) was first grown in SCGal(−Trp) at 30°C, then transferred to SCD(−Trp) and grown at 30°C for the indicated time points, and cell extracts were prepared. Proteins were analysed by SDS–PAGE and immunoblotted with antibodies against the indicated proteins. Arrowheads indicate the precursor forms of mtHsp60 and Mdj1p. (D) The Escherichia coli BL21(DE3) cells containing a plasmid expressing mtHsp70 alone, or mtHsp70 with Tim15 derivatives as in (B) were grown in 50 ml of TB medium supplemented with 50 μg/ml ampicillin at 37°C to an OD600≈0.6–0.8. Protein expression was induced with 1 mM isopropyl-β-D-thiogalactopyranoside for 2 h at 37°C. Cells were collected, resuspended in 20 mM Tris–HCl (pH 7.4) and 0.3 M NaCl and broken open by sonication. The soluble and insoluble fractions were separated by centrifugation at 18,000g for 10 min. Proteins were analysed by SDS–PAGE and CBB R250 staining. P, pellet; S, supernatant; T, total lysate. (E) Mitochondria were isolated from the strains as in (C) after cultivation in SCD(−Trp) for 12 h at 30°C. Mitochondria were incubated at 37°C for 5 min and lysed with 20 mM Tris–HCl (pH 7.4), 300 mM NaCl and 1% Triton X-100. The soluble and insoluble fractions were separated by centrifugation at 18,000g for 20 min. Proteins were analysed by SDS–PAGE and immunodecoration with antibodies against mtHsp70 and Tim15. P, pellet; S, supernatant. (F) A 1 μM portion of GST, GST–Tim15 (WT), GST–Tim152RH (2RH), GST–Tim15D111A (D111A) or GST–Tim15Δ133−137 was incubated with mtHsp70 (1 μM), which had been extensively dialysed against nucleotide-free buffer, in binding buffer (20 mM Tris–HCl, pH 7.4, 100 mM KCl, 1 mM DTT, 100 μM ZnSO4) containing 100 μl of glutathione-Sepharose (Amersham) for 30 min at 25°C. The resin was then washed with binding buffer and the bound proteins were eluted with 20 mM glutathione. Proteins in the loaded fraction (L, 5% of the total), flow-through fraction (F, 5% of the total) and eluted fraction (E, 20% of the total) were analysed by SDS–PAGE and Coomassie brilliant blue R-250 staining. GST, glutathione-S-transferase; DTT, dithiothreitol; mtHsp60, mitochondrial Hsp60; OD, optical density; SDS–PAGE, SDS–polyacrylamide gel electrophoresis.
Previously, we showed that depletion of Tim15 led to the accumulation of uncleaved precursor forms of mtHsp60 and Mdj1p, suggesting that Tim15 has a role in mitochondrial protein import (Yamamoto et al, 2005). The genes for Tim15, Tim15D111A, Tim152RH or Tim15Δ133−137 were thus introduced into the GAL-TIM15 strain, in which the promoter of Tim15 was replaced by the inducible GAL7 promoter. After a shift from galactose-containing medium to galactose-free medium, cell extracts were prepared and analysed for the accumulation of precursor forms of Hsp60 and Mdj1p. Cells with Tim15D111A, Tim152RH or Tim15Δ133−137 (only slightly), but not wild-type Tim15, showed accumulation of the precursor forms 12 h after the shift. Therefore, the ability of Tim15 derivatives, especially wild-type Tim15, Tim15D111A and Tim152RH, to restore the growth of tim15Δ cells were well correlated with their ability to facilitate mitochondrial protein import in vivo (Fig 3C).
We then tested the ability of mutant Tim15 to prevent self-aggregation of mtHsp70 by expression experiments using E. coli cells (Sichting et al, 2005). We transformed E. coli cells with a plasmid for mtHsp70 or both mtHsp70 and a Tim15 mutant and analysed the protein distribution between soluble and insoluble fractions by centrifugation of the total cell lysates. When mtHsp70 was expressed alone in E. coli cells at 37°C, it was recovered in the insoluble fraction, indicating self-aggregation of mtHsp70 (Fig 3D, lanes 1–3). However, on coexpression of wild-type Tim15, both Tim15 and mtHsp70 were recovered in the soluble fraction (Fig 3D, lanes 4–6). Tim155DE produced similar effects on the solubility of mtHsp70 in E. coli cells (Fig 3D, lanes 13–15), whereas expression of Tim152RH or Tim15D111A with mtHsp70 failed to keep mtHsp70 soluble and Tim152RH and Tim15D111A remained soluble (Fig 3D, lanes 7–12). By contrast, expression of Tim15Δ133−137 with mtHsp70 in E. coli cells only partly maintained the solubility of mtHsp70, whereas Tim15Δ133−137 itself remained soluble (Fig 3D, lanes 16–18). We also tested the ability of the Tim15 mutants to affect self-aggregation of mtHsp70 in yeast mitochondria. Mitochondria with Tim15 or its mutants were isolated and solubilized with Triton X-100 after 5 min at 37°C. Then, insoluble materials were separated from the soluble material by centrifugation. In the presence of wild-type Tim15, mtHsp70 was recovered in the soluble fraction, whereas 40% of mtHsp70 became insoluble in the absence of Tim15 (Fig 3E, lanes 1–4). Similarly, 40%, 50% and 30% of mtHsp70 became insoluble in mitochondria in the presence of Tim15D111A, Tim152RH or Tim15Δ133−137, respectively, instead of Tim15 (Fig 3E, lanes 5–10).
Next, we tested directly the ability of Tim15 mutants to interact with mtHsp70. To test the interactions, we created fusion proteins between glutathione S-transferase (GST) and the Tim15 derivatives and subjected them to pull-down assays by using glutathione-Sepharose beads. When GST or GST–Tim15 fusion proteins were eluted from the beads using 20 mM glutathione, mtHsp70 was eluted with GST–Tim15 or partly with GST–Tim15Δ133−137 (Fig 3F, lanes 2, 3, 8 and 9), but not with GST, GST–Tim15D111A or GST–Tim152RH (Fig 3F, lanes 4–7, 10 and 11). These results suggest that the basic concave face—the region around Asp 111—might form a binding site on Tim15 for mtHsp70 and that the association with mtHsp70 seems to maintain solubility of mtHsp70, which is well correlated with the ability to complement the TIM15 gene deletion in cell growth.
Is solubilization of Hsp70 specific to Tim15/Zim17?
How does Tim15/Zim17 maintain the solubility of mtHsp70? The nucleotide-free state of mtHsp70 has a high tendency to self-aggregate (Sichting et al, 2005); therefore, Tim15 could interact with the aggregation-prone nucleotide-free state of mtHsp70 to prevent its aggregation. To test this hypothesis, we examined the nucleotide-binding state of mtHsp70 that is favourable for Tim15 binding. The results show that Tim15 in the form of the GST fusion protein binds to the nucleotide-free state as well as the ADP state or ATP-bound state (in the presence of nonhydrolisable ATP analogues, ATP-γS and AMP-PNP) of mtHsp70 (Fig 4A).
Figure 4.
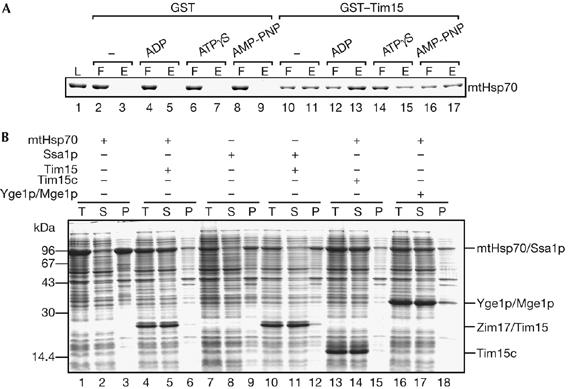
The ability to maintain solubility of mtHsp70 is not limited to Tim15/Zim17. (A) A 1 μM portion of GST and GST–Tim15 (Yamamoto et al, 2005) was incubated with nucleotide-free mtHsp70 (1 μM) and glutathione-Sepharose with or without 10 mM MgCl2 and 5 mM ADP, adenosine 5′-O-(thiotriphosphate) (ATP-γS) or adenosine 5′-(β, γimino)-triphosphate (AMP-PNP) for 30 min at 25°C. The resin was then washed once and the bound proteins were eluted using 20 mM glutathione. Proteins in the loaded fraction (L, 5% of the total), flow-through fraction (F, 5% of the total) and eluted fraction (E, 20% of the total) were analysed by SDS–PAGE and Coomassie brilliant blue R-250 staining. (B) mtHsp70 or Ssa1p was expressed with Tim15, Tim15c or Yge1p/Mge1p in E. coli cells at 37°C and analysed as in Fig 3C. GST, glutathione-S-transferase; mtHsp70, mitochondrial Hsp70; P, pellet; S, supernatant; SDS–PAGE, SDS–polyacrylamide gel electrophoresis; T, total lysate.
Yge1p/Mge1p, a nucleotide exchange factor, binds to the ADP-bound state of mtHsp70 to release ADP, and subsequent binding of ATP to mtHsp70 dissociates Yge1p from mtHsp70. This means that Yge1p remained bound to the nucleotide-free state of mtHsp70 after ADP release until ATP binding to mtHsp70. Therefore, we tested the ability of Yge1p to maintain solubility of mtHsp70 in E. coli cells. As a control, coexpression of Tim15 or Tim15c prevents mtHsp70 from self-aggregation in E. coli cells (Fig 4B, lanes 4–6 and 13–15). On coexpression of Yge1p, both Yge1p and mtHsp70 were recovered in the soluble fraction (Fig 4B, lanes 16–18). This raises the possibility that any protein that interacts with and stabilizes the nucleotide-free form of mtHsp70 can suppress self-aggregation of mtHsp70. As binding of Yge1p to mtHsp70 did not compete with that of GST–Tim15 (supplementary Fig S5 online), Tim15 or Yge1p does not simply mask the small aggregation-causing region on the mtHsp70 molecule.
Is self-aggregation at elevated temperature specific to mtHsp70? To investigate this question, we transformed E. coli cells with a plasmid that encoded yeast cytosolic Hsp70, Ssa1p. Interestingly, Ssa1p expressed in E. coli cells at 37°C was recovered in the insoluble fraction after centrifugation of the cell lysates, indicating the self-aggregation of Ssa1p irrespective of coexpression of Tim15 (Fig 4B, lanes 7–12). This in turn means that, in yeast cells, aggregation of Ssa1p must be suppressed by its partner proteins, including nucleotide exchange factors.
These results indicate that a class of Hsp70s is prone to self-aggregation at high temperatures. Self-aggregation of Hsp70s can be prevented by interactions with their partner proteins, but, for example, promotion of nucleotide exchange, and not the ability to inhibit aggregation, is the primary function of the partner protein, Yge1p, of mtHsp70s. Therefore, although Tim15 does not have a nucleotide-exchange activity for mtHsp70 (Sichting et al, 2005), it remains unknown as to whether Tim15 has a primary function other than maintaining the solubility of mtHsp70.
Methods
Yeast growth conditions. Yeast cell growth was compared at 30°C and 37°C on SCG (−Trp; 0.67% yeast nitrogen base without amino acids, 0.5% vitamin assay casamino acids and 2% glycerol without tryptophan supplementation) and SCD (−Trp; 0.67% yeast nitrogen base without amino acids, 0.5% vitamin assay casamino acids and 2% glucose without tryptophan supplementation).
NMR measurements and structural determination. NMR spectra were recorded on a Bruker AVANCE600 NMR spectrometer. NMR samples were either in 95% H2O/5% D2O or in 99.9% D2O and contained 20 mM sodium acetate (pH 4.5), 50 mM NaCl, 1 mM dithiothreitol and 30 μM ZnSO4 or Zn(OAc)2; the protein concentration was 0.5–2.0 mM. 1H, 13C and 15N resonances of the protein were assigned with standard triple resonance experiments. Details of NMR structural determination of Tim15c are described in the supplementary information online. The coordinates of ensembles of the 20 conformers have been deposited in the Protein Data Bank (www.pdb.org; PDB ID code 2E2Z).
Supplementary information is available at EMBO reports online (http://www.emboreports.org).
Supplementary Material
supplementary Figures and Information
Acknowledgments
We acknowledge the support of this work by grants for Scientific Research from the Japan Ministry of Education, Culture, Sports, Science and Technology and a grant from Japan Science and Technology Agency. We thank members of the Endo laboratory for discussions and comments.
References
- Bukau B, Horwich AL (1998) The chaperone machines. Cell 82: 351–359 [DOI] [PubMed] [Google Scholar]
- Burri L, Vascotto K, Fredersdorf S, Tiedt R, Hall MN, Lithgow T (2004) Zim17, a novel zinc finger protein essential for protein import into mitochondria. J Biol Chem 279: 50243–50249 [DOI] [PubMed] [Google Scholar]
- Endo T, Yamamoto H, Esaki M (2003) Functional cooperation and separation of translocators in protein import into mitochondria, the double-membrane bounded organelles. J Cell Sci 116: 3259–3267 [DOI] [PubMed] [Google Scholar]
- Martinez-Yamout M, Legge GB, Zhang O, Wright PE, Dyson HJ (2000) Solution structure of the cysteine-rich domain of the Escherichia coli chaperone protein DnaJ. J Mol Biol 300: 805–818 [DOI] [PubMed] [Google Scholar]
- Neupert W, Herrmann JM (2007) Translocation of proteins into mitochondria. Annu Rev Biochem, (doi:10.1146/annurev.biochem.76.052705.163409) [DOI] [PubMed] [Google Scholar]
- Sichting M, Mokranjac D, Azem A, Neupert W, Hell K (2005) Maintenance of structure and function of mitochondrial Hsp70 chaperones requires the chaperone Hep1. EMBO J 24: 1046–1056 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szklarz LKS et al. (2005) Inactivation of the mitochondrial heat shock protein Zim17 leads to aggregation of matrix hsp70s followed by pleiotropic effects on morphology and protein biogenesis. J Mol Biol 351: 206–218 [DOI] [PubMed] [Google Scholar]
- van der Laan M, Rissler M, Rehling P (2006) Mitochondrial preprotein translocases as dynamic molecular machines. FEMS Yeast Res 6: 849–861 [DOI] [PubMed] [Google Scholar]
- Wiedemann N, Frazier AE, Pfanner N (2004) The protein import machinery of mitochondria. J Biol Chem 279: 14473–14476 [DOI] [PubMed] [Google Scholar]
- Yamamoto H, Momose T, Yatsukawa Y, Ohshima C, Ishikawa D, Sato T, Tamura Y, Ohwa Y, Endo T (2005) Identification of a novel member of yeast mitochondrial Hsp70-associated motor and chaperone proteins that facilitates protein translocation across the inner membrane. FEBS Lett 579: 507–511 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
supplementary Figures and Information


