Abstract
To control the quality of RNA biogenesis in the nucleus, cells use sophisticated molecular machines. These machines recognize and degrade not only RNA trimmings—the leftovers of RNA processing—but also incorrectly processed RNAs that contain defects. By using this mechanism, cells ensure that only high-quality RNAs are engaged in protein synthesis and other cellular processes. The exosome—a complex of several exoribonucleolytic and RNA-binding proteins—is the central 3′-end RNA degradation and processing factor in this surveillance apparatus. The exosome operates with auxiliary factors that stimulate its activity and recruit its RNA substrates in the crowded cellular environment. In this review, we discuss recent structural and functional data related to the nuclear quality-control apparatus, including the long-awaited structure of the human exosome and its activity.
Keywords: RNA decay, exoribonuclease, exosome, structure, nuclear polyadenylation, TRAMP
Introduction
Cellular RNAs are diverse in length and shape, and their functions range from simple messengers to regulators and enzymes involved in gene expression. In eukaryotes, RNAs are produced by one of three RNA polymerases (Pol I–III), which are subsequently processed to their mature form and their trimmings recycled. This concerted RNA biogenesis, which is mediated by many RNA-binding proteins, ribonucleases and other enzymes, is overseen by RNA quality-control mechanisms that ensure only correctly processed RNAs are exported to the cytoplasm, where another set of cellular factors modulate the rate of RNA degradation (Garneau et al, 2007).
The exosome is the main RNA degrader in the nucleus. It was initially identified as a component in the processing and maturation of ribosomal RNA precursors but it is in fact involved in the processing and degradation of most nuclear RNAs (reviewed by Houseley et al, 2006). The exosome localizes to important sites of RNA biogenesis, such as actively transcribed genes (Andrulis et al, 2002; Hieronymus et al, 2004) or the nucleolus (Dez et al, 2006), and is engaged in RNA surveillance (Dez et al, 2006; Torchet et al, 2002) possibly through its interactions with other RNA-binding proteins. Although the role of the exosome was described more than 10 years ago, it is only recently that structural and functional studies have provided information on the biochemistry and the molecular organization of this large complex. Furthermore, an additional RNA quality-control pathway involving the exosome and a newly described complex, known as the Trf4 or Trf4–Air2–Mtr4 polyadenylation (TRAMP) complex (Kadaba et al, 2006; LaCava et al, 2005; Vanacova et al, 2005; Wyers et al, 2005), has been discovered. Here, we highlight the recent data on the structural and biochemical differences between the human and yeast exosomes, and summarize the current knowledge of the pathways that control the quality of RNA in the eukaryotic nucleus. Although the new studies have provided a tremendous amount of data in the field of nuclear RNA surveillance, there are still a few contradictory issues, which is typical of any young discipline.
Function of the exosome in the nucleus
The exosome is required for the processing and degradation of pre-ribosomal RNAs, pre-small nuclear/small nucleolar RNAs and pre-transfer RNAs (Allmang et al, 1999; Hilleren et al, 2001; Houseley et al, 2006; Kadaba et al, 2004; Kuai et al, 2004; Mitchell et al, 1997; Torchet et al, 2002; van Hoof et al, 2000) and is also involved in nuclear degradation of aberrant pre-messenger RNAs that result from mutations in various 3′-end processing, splicing and export factors (Bousquet-Antonelli et al, 2000; Das et al, 2003; Libri et al, 2002; Torchet et al, 2002). It also degrades messenger RNAs with mutations in their coding sequence in a process known as degradation of RNA in the nucleus (Das et al, 2006). Exosome-mediated RNA surveillance is linked directly to and might regulate other processes in the cell. Surveillance of different types of RNAs occurs at distinct nuclear, nucleolar or subnucleolar regions (reviewed by Fasken & Corbett, 2005; Houseley et al, 2006; Jensen et al, 2003). In yeast, the exosome components interact with transcription elongation factors and are recruited directly to transcriptionally induced genes (Hieronymus et al, 2004; Hilleren et al, 2001; Vasiljeva & Buratowski, 2006). In this way, the exosome co-transcriptionally monitors the messenger ribonucleoprotein state of nascent transcripts and retains messenger RNAs that are aberrant or otherwise export-incompetent (Fasken & Corbett, 2005; Hilleren et al, 2001; Thomsen et al, 2003). In yeast mutants that have defective exosome subunits, such RNAs are released from transcription sites and can either be exported to the cytoplasm or are sequestered to specific subnucleolar foci to be degraded (Carneiro et al, 2007; Dez et al, 2006; Hilleren et al, 2001).
The use of mutants defective in RNA degradation in combination with microarray analysis led to the identification of cryptic unstable transcripts with no obvious or, as yet, identified function (Davis & Ares, 2006; Wyers et al, 2005). Notably, the use of tiling microarrays has recently uncovered transcription from many previously unannotated regions of the yeast and the mammalian genomes (David et al, 2006; Davis & Ares, 2006; Wyers et al, 2005). All these apparently cryptic transcripts are transcribed by the Pol II machinery, although it is unknown why only some of them are highly prone to degradation. It was proposed that the efficient degradation of cryptic unstable transcripts limits the genomic noise resulting from the transcription of inappropriate promoter-like regions (Wyers et al, 2005). However, some of the RNAs might be functional or, alternatively, the regulation of their transcription and stability might affect the rate of transcription of the surrounding genes through a transcription-interference mechanism (Davis & Ares, 2006).
Molecular architecture of exosomes
The exosome is conserved from yeast to humans, and is also found in several archaea. In addition, the core of the exosome has a structural homologue in bacteria—the polynucleotide phosphorylase—which is part of the degradosome (Symmons et al, 2000). The overall molecular architecture of the exosome has evolved from an ancient ring-like fold of the bacterial phosphorolytic exoribonuclease RNase PH, which is a hexameric ring of three homodimers (Choi et al, 2004).
In archaea, the exosome core forms a hexameric ring structure with a central hole involving two types of RNase PH-like subunit, ribosomal RNA processing factor (Rrp)41 and Rrp42 (Buttner et al, 2005; Lorentzen et al, 2005). The ring is assembled such that the two subunits interact to form a trimer of Rrp41–Rrp42 heterodimers (Fig 1). The three Rrp41 exosome subunits have phosphorolytic activity, whereas the three Rrp42 subunits are inactive. The catalytic sites are located inside the chamber of the ring-like exosome core structure (Fig 1). The Rrp42 non-catalytic subunits have a structural role as they provide a platform for the ring formation and assist in the binding of RNA substrates (Lorentzen & Conti, 2005). In addition, they are involved in binding to Rrp4 or cep1 synthetic lethality (Csl)4, which are RNA-binding proteins with amino-terminal, K-homology and S1 domains (Rrp4) or zinc-ribbon domains (Csl4; Table 1; Buttner et al, 2005), and to other protein factors (Walter et al, 2006). Three copies of Rrp4 and/or Csl4 associate on top of the exosome core structure and form an extension of the exosomal entry tunnel (Fig 1). Recent structural studies rationalized that such an architectural arrangement allows the regulation of RNA entry into the processing chamber (Lorentzen et al, 2007). Conti and colleagues also revealed how the exosome core binds to RNA in the vicinity of the catalytic site (Lorentzen et al, 2007; Lorentzen & Conti, 2005). The RNA substrates are recognized by a network of base-non-specific and ribose-specific interactions between the sugar-phosphate backbone and the predominantly arginine side-chains of the Rrp41–Rrp42 interface, which forms an electropositive binding pocket (Fig 1).
Figure 1.
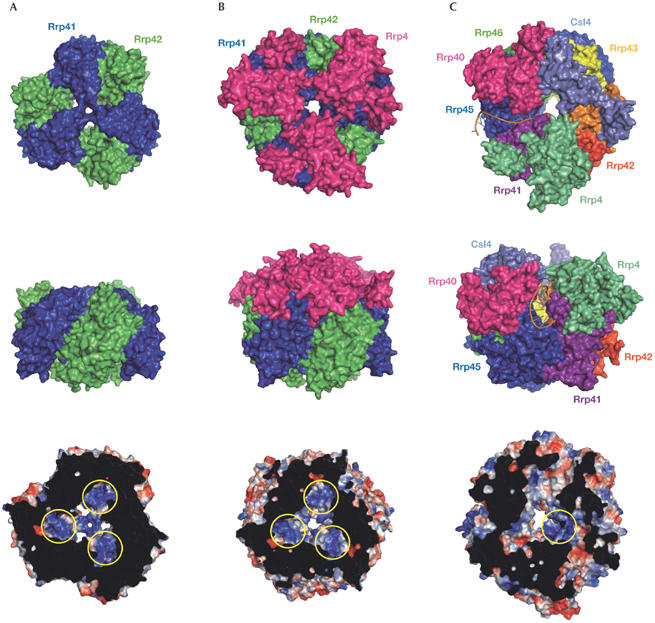
Molecular architecture of the exosomes. (A) Surface representation of the archaeal exosome (Buttner et al, 2005; Lorentzen et al, 2005); (B) the archaeal exosome with the ‘cap' formed by Rrp4 (Buttner et al, 2005; Lorentzen et al, 2005); and (C) the eukaryotic exosome (Dziembowski et al, 2007; Liu et al, 2006). Top views (top row), side views (middle row) and bottom views (bottom row). (C) A model of an RNA substrate highlights a putative path of RNA into the exosome through the cleft that is specific to the human exosome (Dziembowski et al, 2007; Liu et al, 2006). In the bottom row, a slice view on the electrostatic surface potentials (blue, positive; red, negative) shows the RNA-binding pockets in the vicinity of the catalytic sites as indicated by the yellow circles. Graphics were prepared with PyMOL (http://pymol.sourceforge.net). Csl, cep1 synthetic lethality; Rrp, ribosomal RNA processing factor.
Table 1.
Subunits of the eukaryotic and archaeal exosomes
| Human | Yeast | Archaea | Domains |
|---|---|---|---|
| Core | |||
| Rrp41* | Rrp41 | Rrp41* | RNase PH |
| Rrp45 | Rrp45 | — | RNase PH |
| Mtr3 | Mtr3 | — | RNase PH |
| Rrp42 | Rrp42 | Rrp42 | RNase PH |
| Rrp46 | Rrp46 | — | RNase PH |
| Rrp43 | Rrp43 | — | RNase PH |
| Csl4 | Csl4 | — | NT, S1, zinc-ribbon |
| Rrp4 | Rrp4 | — | NT, S1, KH |
| Rrp40 | Rrp40 | — | NT, S1, KH |
| Associated proteins | |||
| — | — | Csl4 | NT, S1, zinc-ribbon |
| — | — | Rrp4 | NT, S1, KH |
| Rrp6* | Rrp6* | — | RNase D |
| — | Rrp44* (Dis3) | — | RNase R |
| — | Rrp47 | — | |
*denotes active subunits. Note that for activity, the Rrp41 subunit requires Rrp42 and Rrp45 subunits in archaea and human, respectively. Csl, cep1 synthetic lethality; KH, K-homology; Mtr, mRNA transport; NT, N-terminal; PH, phosphate-dependent; Rrp, ribosomal RNA processing factor.
In contrast to archaea, the eukaroytic exosome uses six different subunits to form the ring structure. However, these subunits can be classified as Rrp41-like—Rrp41, Rrp46, and Mtr3—and Rrp42-like—Rrp45 (polymyositis/scleroderma (PM/Scl)-75 human), Rrp43 (opa-interacting protein (OIP)2 human), and Rrp42—based on their similarities to their archaeal counterparts. Interestingly, the eukaryotic six-member ring does not assemble into a stable structure in the absence of Rrp4, Rrp40 and Csl4 (Liu et al, 2006), indicating that the eukaryotic exosome core is composed of nine subunits. The human exosome is asymmetrical and shows a more complex architecture compared with its archaeal counterpart (Fig 1). Each Rrp41-like–Rrp42-like heterodimer binds only the correct heterodimer partner and does not form trimers of the same heterodimers. It is highly likely that the eukaryotic exosome has evolved to include different subunits that are necessary to bind various auxiliary factors involved in RNA degradation and processing. The human exosome core has only one catalytic site in the Rrp41–Rrp45 heterodimer (Liu et al, 2006), a characteristic that was previously suspected (Lorentzen & Conti, 2005). The human Rrp41–Rrp45 heterodimer in isolation also has RNA-degradation activity that results in a degradation pattern identical to the pattern of the complete exosome. This could be a result of the asymmetrical arrangement in which the RNA entry tunnel of the human exosome is significantly widened by a large cleft between Rrp4 and Rrp40 subunits. This cleft is located exactly over the Rrp41–Rrp45 heterodimer and could act as an entry site for the RNA (Fig 1). In this scenario, the Rrp41–Rrp45 heterodimer would use its surface within the cleft to bind to RNA. This would explain the virtually identical RNA degradation patterns of the Rrp41–Rrp45 heterodimer and the human exosome. Consistently, the RNase PH domain of human Rrp45 in isolation binds to U-rich and AU-rich sequences (Anderson et al, 2006). To understand how RNA is threaded into the human exosome, the structure of the exosome in complex with RNA will need to be solved.
Interestingly, the structural studies by Conti, Sattler and colleagues show that the N-terminal region of Saccharomyces cerevisiae Rrp40 is unstructured in its free form, whereas the S1 and K-homology domains are essentially identical to human RRP40 in the exosome core structure (Oddone et al, 2007). This indicates that the exosome core formation includes an induced-fit mechanism in which certain domains undergo a conformational change on finding the ‘correct' binding partner. These authors also show that the weak RNA-binding affinity of individual subunits becomes significantly attenuated in the context of the assembled exosome.
In eukaryotes, the nuclear exosome associates with the auxiliary factor Rrp6, which participates in both RNA processing and quality control. The structure of S. cerevisiae Rrp6 has a conserved RNase D core with a flanking helicase and RNase D carboxy-terminal domain, and an N-terminal domain that is proposed to mediate the interaction with the exosome core (Midtgaard et al, 2006).
Clearly, further structures of the eukaryotic exosome core that bind to additional protein factors—particularly the TRAMP complex—and to RNA substrates need to be solved to decipher how this quality-control machinery operates.
Degradation mechanism of exosomes
The exosome core associates with additional protein factors in a compartment- and organism-specific manner. In yeast, the exosome core has an additional stable subunit, the hydrolytic exonuclease Rrp44 (also known as Dis3; Mitchell et al, 1997), and in the nucleus it binds to two more subunits: the hydrolytic ribonuclease Rrp6, and Rrp47 (Mitchell et al, 2003). By contrast, in humans, Rrp44 was not detected in the affinity-purified exosomes and the Rrp6 homologue (PM/Scl-100) was also found in the cytoplasmic fractions (Chen et al, 2001).
The reasons for the differences in the composition of the human and yeast exosomes have become apparent from recent studies (Fig 2). It was shown that the human core exosome has one catalytically active site, within the Rrp41–Rrp45 dimer (Dziembowski et al, 2007; Liu et al, 2006), whereas the yeast nine-subunit core has no catalytically active site (Dziembowski et al, 2007; Liu et al, 2006). Instead, the hydrolytic nuclease Rrp44 is the crucial enzyme for RNA degradation in yeast (Dziembowski et al, 2007; Liu et al, 2006). Therefore, eukaryotic exosomes have human-like and yeast-like subtypes with phosphorolytic and hydrolytic activities, respectively. The former subtype degrades RNA inside the exosome chamber, whereas the latter subtype uses the hydrolytic enzyme that is most likely anchored on the surface of the exosome core assembly (Hernandez et al, 2006). It is not known why yeast switched to the hydrolytic mode of RNA degradation; however, it is still possible that under specific conditions and/or on specific RNA substrates the yeast Rrp41 also becomes active. This is a question that needs to be investigated in the future. Similarly, the contribution of the human homologues of the hydrolytic nucleases, RRP6 and RRP44, to the exosome activities in vivo must be assessed further.
Figure 2.
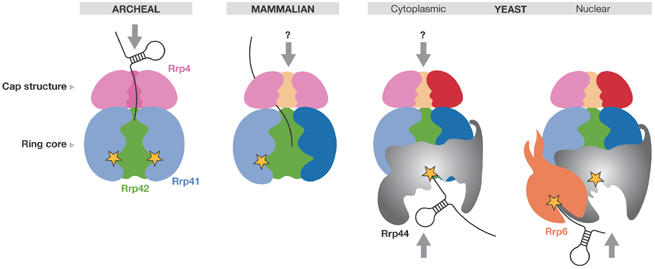
Schematic diagram of the exosome topologies and RNA threading. The arrows indicate the confirmed or hypothetical (indicated by a question mark) pathways of an RNA substrate in various forms of the exosomes (Dziembowski et al, 2007; Liu et al, 2006; Lorentzen et al, 2007). Yellow stars indicate catalytically active sites. The predicted positions of the yeast hydrolytic exonucleases Rrp44 and Rrp6 are indicated. Rrp, ribosomal RNA processing factor.
The recombinant yeast Rrp44 is highly processive, even on structured RNAs; however, its activity is significantly reduced on binding to the exosome core (Liu et al, 2006). This is perhaps to prevent high activity levels that could interfere with the stability of many functional RNAs in the cell. At present, it is not clear whether Rrp44 undergoes a conformational change or whether the active site is partly masked on binding to the exosome core. The observation that Rrp44 is responsible for yeast exosome activity suggests that the substrate RNA might not pass through the central channel of the ring-like structure. Further structural and functional analyses are required to determine the exact path of the RNA substrate.
Rrp6, the second nucleus-specific hydrolytic exonuclease in yeast, has distributive exonucleolytic activity on unstructured and poly(A)-extended RNAs, and displays only weak activity on structured substrates (Liu et al, 2006). Its activity is not affected on assembly with the exosome core and Rrp44. Although Rrp6 is a non-essential gene, its deletion results in many RNA-processing defects (Allmang et al, 1999; Davis & Ares, 2006; Egecioglu et al, 2006; Kadaba et al, 2004; Kuai et al, 2004; Liu et al, 2006; van Hoof et al, 2000; Wyers et al, 2005). This suggests that although partly overlapping in function, Rrp44 and Rrp6 have different substrate specificities in vivo. The function of human RRP6 has not been analysed in detail and the extent to which it is specific for nuclear RNA metabolism is unclear.
Other players in nuclear RNA surveillance
Although the reconstituted exosomes efficiently degrade RNAs in vitro (Dziembowski et al, 2007; Liu et al, 2006), in vivo RNAs often form complex secondary and tertiary conformations, acquire nucleotide modifications and/or assemble into specific ribonucleoprotein particles (RNPs) that need to be protected from nucleases. Accessory factors are then required to help the exosome recognize aberrant RNAs or RNPs that need to be discarded or further processed (reviewed by Olesen et al, 2005). Two accessory protein complexes were shown to stimulate the exosome in the yeast nucleus. First is the polyadenylation complex called TRAMP4 and/or TRAMP5 depending on whether it contains the poly(A) polymerase Trf4 or Trf5 (Houseley & Tollervey, 2006; Kadaba et al, 2004, 2006; LaCava et al, 2005; Vanacova et al, 2005; Wyers et al, 2005). TRAMP adds short poly(A) tails to aberrant or unstable transcripts, forming a favourable substrate for the exosome (Fig 3). The identity of the exonuclease within the yeast nuclear exosome that degrades these polyadenylated targets is still unknown. Rrp6 shows higher activity on unstructured poly(A)-extended molecules and Rrp44 can degrade more structured RNAs (Liu et al, 2006); therefore, it is possible that Rrp6 initiates digestion and, by inducing some conformational changes, it hands the RNA over to Rrp44. The helicase Mtr4 might help to unwind the structured portions of the RNAs when Rrp44 is in its less active conformation. Mtr4 might also act as a scaffolding protein because it interacts with both the TRAMP and the exosome complexes (de la Cruz et al, 1998; LaCava et al, 2005; Vanacova et al, 2005; Wyers et al, 2005). It is not known how TRAMP recognizes aberrant RNAs. One possibility is that it monitors the conformation status of RNAs. For example, it can specifically polyadenylate and target transfer RNAs that are incorrectly folded for degradation (Kadaba et al, 2004; Vanacova et al, 2005). Further structural and functional studies are required to understand fully the mechanism of substrate recognition.
Figure 3.
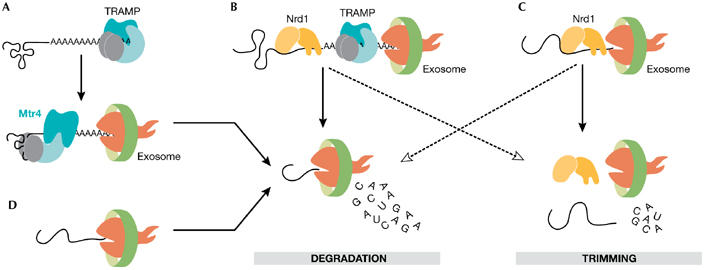
Exonucleolytic activity of the exosome is stimulated by accessory protein complexes in vitro and in vivo. (A) The Trf4–Air2–Mtr4 polyadenylation (TRAMP) complex tags aberrant RNAs with short stretches of oligo(A)s, which initiates RNA digestion by the exosome (LaCava et al, 2005; Vanacova et al, 2005). (B) Mtr4 helicase of the TRAMP complex unwinds the structured parts of RNAs. The TRAMP complex associates with the Nrd1 complex that binds to short sequence elements on a subset of nuclear RNAs (Vasiljeva & Buratowski, 2006). The interaction between the specific RNA recognition mediated by the Nrd1 complex and the polyadenylation activity mediated by the TRAMP complex acts as the initiation step for RNA degradation by the exosome. (C)The Nrd1 complex can stimulate exosome activity on RNAs with the Nrd1 complex-specific binding sites (Vasiljeva & Buratowski, 2006). This often leads to partial digestion of the RNA (trimming), but can also cause RNA degradation. (D) The exosome destroys the leftovers of RNA processing, such as the products of endonucleolytic cleavage, apparently by itself. Air, arginine methyltransferase-interacting RING finger protein; Mtr, mRNA transport; Nrd, nuclear pre-mRNA down-regulation; Trf, topoisomerase one-related function.
Both the exosome and the TRAMP4 complexes interact with an additional RNA-binding complex called the nuclear pre-mRNA down-regulation (Nrd)1 complex (Vasiljeva & Buratowski, 2006), which consists of the RNA helicase Sen1, and the proteins Nrd1 and nuclear polyadenylated RNA-binding (Nab)3 that recognize specific sequence motifs on RNAs (Carroll et al, 2004; Steinmetz & Brow, 1998). This complex is required for transcription termination of small nuclear RNA and small nucleolar RNA genes (Conrad et al, 2000; Steinmetz & Brow, 1996, 1998; Steinmetz et al, 2001). In vitro, it can directly stimulate exosome degradation of substrates with Nrd1- and Nab3-binding motifs (Vasiljeva & Buratowski, 2006). In vivo, it probably helps to bring the exosome to specific RNA substrates. However, it is not clear whether the Nrd1 complex acts by itself or whether it requires the association with TRAMP4 to stimulate the exosome (Vasiljeva & Buratowski, 2006).
Concluding remarks and open questions
The recent structural information on the human exosome core supports the existence of a common basis for RNA-degradation machineries in prokaryotes, eukaryotes and archaea. It is also striking how the RNA-degradation pathway mediated by the exosome resembles the features of protein degradation by proteasomes, a concept first envisioned by van Hoof & Parker (1999). Despite the increasing amount of data on exosomes, there are still many questions to be answered. How does the quality-control machinery distinguish its RNA substrates from other RNA molecules? What allows the exosome to switch from the degradation mode to the processing mode? How exactly is the exosome stimulated by the TRAMP and Nrd1 complexes, and what other protein factors are required for its proper function in vivo? Many homologues of Trf4-like proteins exist in higher eukaryotes; therefore, it will be interesting to see whether similar poly(A)-mediated degradation pathways operate in the nuclei of metazoa. To conclude, there is no doubt that in the near future we will witness considerable efforts to unravel the details of the RNA surveillance apparatus—a universal machinery with the fundamental quest to seek and destroy damaged molecules.

Stepanka Vanacova

Richard Stefl
Acknowledgments
We apologize to those authors whose work could not be cited owing to space constraints. We thank W. Keller for support, and C. Rammelt, M.-J. Schmidt, D. Ladle and L. Krejci for critical reading of the manuscript and for helpful comments. The authors are supported by the Ministry of Education of the Czech Republic (MSM0021622413), the University of Basel and the Swiss National Science Fund. R.S. is in receipt of a European Molecular Biology Organization/Howard Hughes Medical Institute Start-up Grant and a Human Frontier Science Program Career Development Award.
References
- Allmang C, Kufel J, Chanfreau G, Mitchell P, Petfalski E, Tollervey D (1999) Functions of the exosome in rRNA, snoRNA and snRNA synthesis. EMBO J 18: 5399–5410 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anderson JR, Mukherjee D, Muthukumaraswamy K, Moraes KC, Wilusz CJ, Wilusz J (2006) Sequence-specific RNA binding mediated by the RNase PH domain of components of the exosome. RNA 12: 1810–1816 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andrulis ED, Werner J, Nazarian A, Erdjument-Bromage H, Tempst P, Lis JT (2002) The RNA processing exosome is linked to elongating RNA polymerase II in Drosophila. Nature 420: 837–841 [DOI] [PubMed] [Google Scholar]
- Bousquet-Antonelli C, Presutti C, Tollervey D (2000) Identification of a regulated pathway for nuclear pre-mRNA turnover. Cell 102: 765–775 [DOI] [PubMed] [Google Scholar]
- Buttner K, Wenig K, Hopfner KP (2005) Structural framework for the mechanism of archaeal exosomes in RNA processing. Mol Cell 20: 461–471 [DOI] [PubMed] [Google Scholar]
- Carneiro T, Carvalho C, Braga J, Rino J, Milligan L, Tollervey D, Carmo-Fonseca M (2007) Depletion of the yeast nuclear exosome subunit Rrp6 results in accumulation of polyadenylated RNAs in a discrete domain within the nucleolus. Mol Cell Biol 27: 4157–4165 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carroll KL, Pradhan DA, Granek JA, Clarke ND, Corden JL (2004) Identification of cis elements directing termination of yeast nonpolyadenylated snoRNA transcripts. Mol Cell Biol 24: 6241–6252 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen CY, Gherzi R, Ong SE, Chan EL, Raijmakers R, Pruijn GJ, Stoecklin G, Moroni C, Mann M, Karin M (2001) AU binding proteins recruit the exosome to degrade ARE-containing mRNAs. Cell 107: 451–464 [DOI] [PubMed] [Google Scholar]
- Choi JM, Park EY, Kim JH, Chang SK, Cho Y (2004) Probing the functional importance of the hexameric ring structure of RNase PH. J Biol Chem 279: 755–764 [DOI] [PubMed] [Google Scholar]
- Conrad NK, Wilson SM, Steinmetz EJ, Patturajan M, Brow DA, Swanson MS, Corden JL (2000) A yeast heterogeneous nuclear ribonucleoprotein complex associated with RNA polymerase II. Genetics 154: 557–571 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Das B, Butler JS, Sherman F (2003) Degradation of normal mRNA in the nucleus of Saccharomyces cerevisiae. Mol Cell Biol 23: 5502–5515 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Das B, Das S, Sherman F (2006) Mutant LYS2 mRNAs retained and degraded in the nucleus of Saccharomyces cerevisiae. Proc Natl Acad Sci USA 103: 10871–10876 [DOI] [PMC free article] [PubMed] [Google Scholar]
- David L, Huber W, Granovskaia M, Toedling J, Palm CJ, Bofkin L, Jones T, Davis RW, Steinmetz LM (2006) A high-resolution map of transcription in the yeast genome. Proc Natl Acad Sci USA 103: 5320–5325 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davis CA, Ares M Jr (2006) Accumulation of unstable promoter-associated transcripts upon loss of the nuclear exosome subunit Rrp6p in Saccharomyces cerevisiae. Proc Natl Acad Sci USA 103: 3262–3267 [DOI] [PMC free article] [PubMed] [Google Scholar]
- de la Cruz J, Kressler D, Tollervey D, Linder P (1998) Dob1p (Mtr4p) is a putative ATP-dependent RNA helicase required for the 3′ end formation of 5.8S rRNA in Saccharomyces cerevisiae. EMBO J 17: 1128–1140 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dez C, Houseley J, Tollervey D (2006) Surveillance of nuclear-restricted pre-ribosomes within a subnucleolar region of Saccharomyces cerevisiae. EMBO J 25: 1534–1546 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dziembowski A, Lorentzen E, Conti E, Seraphin B (2007) A single subunit, Dis3, is essentially responsible for yeast exosome core activity. Nat Struct Mol Biol 14: 15–22 [DOI] [PubMed] [Google Scholar]
- Egecioglu DE, Henras AK, Chanfreau GF (2006) Contributions of Trf4p- and Trf5p-dependent polyadenylation to the processing and degradative functions of the yeast nuclear exosome. RNA 12: 26–32 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fasken MB, Corbett AH (2005) Process or perish: quality control in mRNA biogenesis. Nat Struct Mol Biol 12: 482–488 [DOI] [PubMed] [Google Scholar]
- Garneau NL, Wilusz J, Wilusz CJ (2007) The highways and byways of mRNA decay. Nat Rev Mol Cell Biol 8: 113–126 [DOI] [PubMed] [Google Scholar]
- Hernandez H, Dziembowski A, Taverner T, Seraphin B, Robinson CV (2006) Subunit architecture of multimeric complexes isolated directly from cells. EMBO Rep 7: 605–610 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hieronymus H, Yu MC, Silver PA (2004) Genome-wide mRNA surveillance is coupled to mRNA export. Genes Dev 18: 2652–2662 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hilleren P, McCarthy T, Rosbash M, Parker R, Jensen TH (2001) Quality control of mRNA 3′-end processing is linked to the nuclear exosome. Nature 413: 538–542 [DOI] [PubMed] [Google Scholar]
- Houseley J, Tollervey D (2006) Yeast Trf5p is a nuclear poly(A) polymerase. EMBO Rep 7: 205–211 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Houseley J, LaCava J, Tollervey D (2006) RNA-quality control by the exosome. Nat Rev Mol Cell Biol 7: 529–539 [DOI] [PubMed] [Google Scholar]
- Jensen TH, Dower K, Libri D, Rosbash M (2003) Early formation of mRNP: license for export or quality control? Mol Cell 11: 1129–1138 [DOI] [PubMed] [Google Scholar]
- Kadaba S, Krueger A, Trice T, Krecic AM, Hinnebusch AG, Anderson J (2004) Nuclear surveillance and degradation of hypomodified initiator tRNAMet in S. cerevisiae. Genes Dev 18: 1227–1240 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kadaba S, Wang X, Anderson JT (2006) Nuclear RNA surveillance in Saccharomyces cerevisiae: Trf4p-dependent polyadenylation of nascent hypomethylated tRNA and an aberrant form of 5S rRNA. RNA 12: 508–521 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuai L, Fang F, Butler JS, Sherman F (2004) Polyadenylation of rRNA in Saccharomyces cerevisiae. Proc Natl Acad Sci USA 101: 8581–8586 [DOI] [PMC free article] [PubMed] [Google Scholar]
- LaCava J, Houseley J, Saveanu C, Petfalski E, Thompson E, Jacquier A, Tollervey D (2005) RNA degradation by the exosome is promoted by a nuclear polyadenylation complex. Cell 121: 713–724 [DOI] [PubMed] [Google Scholar]
- Libri D, Dower K, Boulay J, Thomsen R, Rosbash M, Jensen TH (2002) Interactions between mRNA export commitment, 3′-end quality control, and nuclear degradation. Mol Cell Biol 22: 8254–8266 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Q, Greimann JC, Lima CD (2006) Reconstitution, activities, and structure of the eukaryotic RNA exosome. Cell 127: 1223–1237 [DOI] [PubMed] [Google Scholar]
- Lorentzen E, Conti E (2005) Structural basis of 3′ end RNA recognition and exoribonucleolytic cleavage by an exosome RNase PH core. Mol Cell 20: 473–481 [DOI] [PubMed] [Google Scholar]
- Lorentzen E, Walter P, Fribourg S, Evguenieva-Hackenberg E, Klug G, Conti E (2005) The archaeal exosome core is a hexameric ring structure with three catalytic subunits. Nat Struct Mol Biol 12: 575–581 [DOI] [PubMed] [Google Scholar]
- Lorentzen E, Dziembowski A, Lindner D, Seraphin B, Conti E (2007) RNA channelling by the archaeal exosome. EMBO Rep 8: 470–476 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Midtgaard SF, Assenholt J, Jonstrup AT, Van LB, Jensen TH, Brodersen DE (2006) Structure of the nuclear exosome component Rrp6p reveals an interplay between the active site and the HRDC domain. Proc Natl Acad Sci USA 103: 11898–11903 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitchell P, Petfalski E, Shevchenko A, Mann M, Tollervey D (1997) The exosome: a conserved eukaryotic RNA processing complex containing multiple 3′-->5′ exoribonucleases. Cell 91: 457–466 [DOI] [PubMed] [Google Scholar]
- Mitchell P, Petfalski E, Houalla R, Podtelejnikov A, Mann M, Tollervey D (2003) Rrp47p is an exosome-associated protein required for the 3′ processing of stable RNAs. Mol Cell Biol 23: 6982–6992 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oddone A, Lorentzen E, Basquin J, Gasch A, Rybin V, Conti E, Sattler M (2007) Structural and biochemical characterization of the yeast exosome component Rrp40. EMBO Rep 8: 63–69 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olesen JR, Libri D, Jensen TH (2005) A link between transcription and mRNP quality in Saccharomyces cerevisiae. RNA Biol 2: 45–48 [DOI] [PubMed] [Google Scholar]
- Steinmetz EJ, Brow DA (1996) Repression of gene expression by an exogenous sequence element acting in concert with a heterogeneous nuclear ribonucleoprotein-like protein, Nrd1, and the putative helicase Sen1. Mol Cell Biol 16: 6993–7003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steinmetz EJ, Brow DA (1998) Control of pre-mRNA accumulation by the essential yeast protein Nrd1 requires high-affinity transcript binding and a domain implicated in RNA polymerase II association. Proc Natl Acad Sci USA 95: 6699–6704 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steinmetz EJ, Conrad NK, Brow DA, Corden JL (2001) RNA-binding protein Nrd1 directs poly(A)-independent 3′-end formation of RNA polymerase II transcripts. Nature 413: 327–331 [DOI] [PubMed] [Google Scholar]
- Symmons MF, Jones GH, Luisi BF (2000) A duplicated fold is the structural basis for polynucleotide phosphorylase catalytic activity, processivity, and regulation. Structure 8: 1215–1226 [DOI] [PubMed] [Google Scholar]
- Thomsen R, Libri D, Boulay J, Rosbash M, Jensen TH (2003) Localization of nuclear retained mRNAs in Saccharomyces cerevisiae. RNA 9: 1049–1057 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Torchet C, Bousquet-Antonelli C, Milligan L, Thompson E, Kufel J, Tollervey D (2002) Processing of 3′-extended read-through transcripts by the exosome can generate functional mRNAs. Mol Cell 9: 1285–1296 [DOI] [PubMed] [Google Scholar]
- van Hoof A, Parker R (1999) The exosome: a proteasome for RNA? Cell 99: 347–350 [DOI] [PubMed] [Google Scholar]
- van Hoof A, Lennertz P, Parker R (2000) Yeast exosome mutants accumulate 3′-extended polyadenylated forms of U4 small nuclear RNA and small nucleolar RNAs. Mol Cell Biol 20: 441–452 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vanacova S, Wolf J, Martin G, Blank D, Dettwiler S, Friedlein A, Langen H, Keith G, Keller W (2005) A new yeast poly(A) polymerase complex involved in RNA quality control. PLoS Biol 3: e189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vasiljeva L, Buratowski S (2006) Nrd1 interacts with the nuclear exosome for 3′ processing of RNA polymerase II transcripts. Mol Cell 21: 239–248 [DOI] [PubMed] [Google Scholar]
- Walter P, Klein F, Lorentzen E, Ilchmann A, Klug G, Evguenieva-Hackenberg E (2006) Characterization of native and reconstituted exosome complexes from the hyperthermophilic archaeon Sulfolobus solfataricus. Mol Microbiol 62: 1076–1089 [DOI] [PubMed] [Google Scholar]
- Wyers F et al. (2005) Cryptic pol II transcripts are degraded by a nuclear quality control pathway involving a new poly(A) polymerase. Cell 121: 725–737 [DOI] [PubMed] [Google Scholar]


