Abstract
Asn-linked glycans (N-glycans) play important roles in the quality control (QC) of glycoprotein folding in the endoplasmic reticulum (ER) lumen and in ER-associated degradation (ERAD) of proteins by cytosolic proteasomes. A UDP-Glc:glycoprotein glucosyltransferase glucosylates N-glycans of misfolded proteins, which are then bound and refolded by calreticulin and/or calnexin in association with a protein disulfide isomerase. Alternatively, an α-1,2-mannosidase (Mns1) and mannosidase-like proteins (ER degradation-enhancing α-mannosidase-like proteins 1, 2, and 3) are part of a process that results in the dislocation of misfolded glycoproteins into the cytosol, where proteins are degraded in the proteasome. Recently we found that numerous protists and fungi contain 0–11 sugars in their N-glycan precursors versus 14 sugars in those of animals, plants, fungi, and Dictyostelium. Our goal here was to determine what effect N-glycan precursor diversity has on N-glycan-dependent QC systems of glycoprotein folding and ERAD. N-glycan-dependent QC of folding (UDP-Glc:glycoprotein glucosyltransferase, calreticulin, and/or calnexin) was present and active in some but not all protists containing at least five mannose residues in their N-glycans and was absent in protists lacking Man. In contrast, N-glycan-dependent ERAD appeared to be absent from the majority of protists. However, Trypanosoma and Trichomonas genomes predicted ER degradation-enhancing α-mannosidase-like protein and Mns1 orthologs, respectively, each of which had α-mannosidase activity in vitro. Phylogenetic analyses suggested that the diversity of N-glycan-dependent QC of glycoprotein folding (and possibly that of ERAD) was best explained by secondary loss. We conclude that N-glycan precursor length has profound effects on N-glycan-dependent QC of glycoprotein folding and ERAD.
Keywords: protein folding, protists, Trichomonas, Entamoeba
Animals, plants, most fungi, and social amoebae make a dolichol-PP-linked Asn-linked glycan (N-glycan) precursor composed of Glc3Man9GlcNAc2, which is transferred to Asn residues on the nascent polypeptide (1–3). Recently we took advantage of whole-genome sequencing of numerous protists and fungi to show that there is extensive secondary loss of sets of Asn-linked glycan (Alg) genes encoding enzymes that make dolichol-PP-linked precursors (Table 1) (4). For example, some protists and fungi make no N-glycans (Theileria and Encephalitozoon) or truncated N-glycans that are missing some mannose (Man) residues (Tetrahymena, Toxoplasma, and Cryptosporidium), lack glucose (Glc) (Entamoeba, Trichomonas, Leishmania, Cryptococcus, and Trypanosoma), or lack Man and Glc (Giardia and Plasmodium) (4).
Table 1.
N-glycan-dependent QC of glycoprotein folding and ERAD in representative eukaryotes
| Organisms | N-glycan | Folding |
Degradation |
|||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Gls2 | UGGT | CRT* | CNX* | ERGIC | Mns1† | EDEM† | Yos9 | PNGase | ||
| Sc/Sp | Glc3Man9GlcNAc2 | Y | N‡/Y | N | Y | Y | Y | Y | Y | Y |
| Hs/At/Dd | Glc3Man9GlcNAc2 | Y | Y | Y | Y | Y | Y | Y | Y | Y |
| Cn | Man9GlcNAc2 | Y | Y§ | N | Y | Y | Y | Y | Y | Y |
| Lm/Tb/Tc | Man7–9GlcNAc2 | Y | Y | Y | N | Y | N | N/Y/Y¶ | Y | N |
| Eh/Tv | Man5GlcNAc2 | Y‖ | Y§ | Y | N/Y | Y | N/Y** | N | N/Y | N/Y†† |
| Tt/Tg/Cp | Glc1–3Man5GlcNAc2 | Y | N | N | N/Y‡‡/N | N | N | N | N | N |
| Pf/Gl | GlcNAc2 | N | N | N | N | N | N | N | N | N |
| Ec/Ta | None | N | N | N | N | Y/N | N | N | N | N |
Organisms are grouped by confirmed or predicted N-glycan precursors (4). Sc, S. cerevisiae; Sp, S. pombe; Hs, Homo sapiens; At, Arabidopsis thaliana; Dd, D. discoideum; Cn, C. neoformans; Lm, L. major; Tb, T. brucei; Tc, T. cruzi; Eh, E. histolytica; Tv, T. vaginalis; Tt, T. thermophila; Tg, T. gondii; Cp, C. parvum; Pf, P. falciparum; Gl, G. lamblia; Ec, E. cuniculi; Ta, T. annulata.
*A phylogenetic analysis of CRT and CNX is shown in Fig. 3A.
†A phylogenetic analysis of Mns1 and EDEM is shown in Fig. 3B.
‡Sc Kre5 is orthologous to UGGT but does not have the same function (7).
¶The mannosidase activity of the Trypanosoma EDEM ortholog is shown in Fig. 6.
‖The function of the Trichomonas glucosidase 2 is demonstrated indirectly with its inhibitor, castanospermine, in Fig. 4B.
**The mannosidase activity of a Trichomonas Mns1 is shown in Fig. 7 B–D.
††The N-glycanase activity of the Trichomonas cytosolic PNGase is shown in Fig. 7A.
‡‡The Toxoplasma CNX is missing most of the arm that binds PDI [supporting information (SI) Fig. 8].
N-glycans play an important role in the quality control (QC) of glycoprotein folding in the endoplasmic reticulum (ER) lumen (1–3). Calreticulin (CRT) and calnexin (CNX) are homologous lectins, which bind glucosylated N-glycans in the lumen of the ER (Fig. 1A and Table 1) (5, 6). Both CRT and CNX have a Pro-rich arm that binds a protein disulfide isomerase, which participates in the refolding of glucosylated protein. N-glycans may derive their terminal Glc from a glucosylated dolichol-PP-linked precursor (primary glucosylation) or from the action of a UDP-Glc:glycoprotein glucosyltransferase (UGGT) that is specific for misfolded proteins (secondary glucosylation) (Fig. 1A and Table 1) (1–3, 7, 8). UGGT is functional in Trypanosoma, Schizosaccharomyces pombe, and humans but is not functional in Saccharomyces cerevisiae. After the terminal Glc is removed by glucosidases, Man residues on N-glycans of some well folded proteins are bound by homologous lectins (ERGIC-53, VIP36, and/or VIPL), and these glycoproteins are transported to the Golgi (1–3, 9, 10).
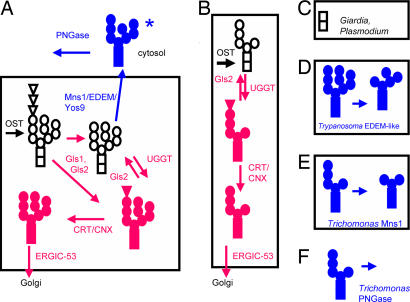
Fig. 1.
Whole gene sequences of numerous eukaryotes reveal great variations in the predicted N-glycan-dependent QC systems of glycoprotein folding and ERAD (see Table 1). (A) Animals, plants, most fungi, and Dictyostelium, which have an N-glycan precursor composed of Glc3Man9GlcNAc2 precursor, contain sets of proteins involved in N-glycan-dependent QC of glycoprotein folding (red) and ERAD (blue). Glycans are indicated for each glycoprotein, where squares are GlcNAc, circles are Man, and triangles are Glc. The asterisk on Man8BGlcNAc2 indicates that other mannosidase products may be present on misfolded glycoproteins dislocated into the cytosol. (B) Entamoeba and Trichomonas, which have a Man5GlcNAc2 precursor, contain set of proteins involved in N-glycan-dependent QC of glycoprotein folding (tested in Figs. 4 and 5). (C) Giardia and Plasmodium, which have a GlcNAc2 precursor, are missing all proteins involved in N-glycan-dependent QC control of glycoprotein folding and degradation. (D) Predicted mannosidase activity of the Trypanosoma EDEM-like protein (tested in Fig. 6). (E) Predicted mannosidase activity of Trichomonas Mns1 (tested in Fig. 7). (F) Predicted N-glycanase activity of Trichomonas cytosolic PNGase (tested in Fig. 7).
N-glycans may also play an important role in ER-associated degradation (ERAD) of proteins (Fig. 1A and Table 1) (1–3, 11). Experiments in yeast and mammalian cells suggest that an α-mannosidase (Mns1) and a second set of proteins called MnlI (mannosidase-like), Htm1 (homologous to mannosidase), or ER degradation-enhancing α-mannosidase-like protein (EDEM) are involved in selection of misfolded glycoproteins for dislocation into the cytosol for degradation by proteasomes (1–3, 12–16). One model is that Mns1 removes a single Man residue from the middle arm of Man9GlcNAc2 to make Man8BGlcNAc2, which in turn interacts with EDEM that has chaperone and/or lectin activity. This model is complicated by evidence that Mns1 has weak mannosidase activity in S. pombe (17); more than one Man residue may be removed from N-glycans of misfolded proteins in mammalian cells (13); N-glycan-dependent ERAD occurs in mutant cell lines that are missing the middle and upper Man arms on their N-glycans (18); and there are multiple EDEM paralogs, at least two of which (EDEM1 and EDEM3) have mannosidase activity in vivo (19–23). Yos9, which resembles the Man-6-P receptor, has also been implicated in N-glycan-dependent ERAD, likely as a lectin (24, 25). In some organisms, a cytosolic peptide-N-glycanase (PNGase) removes N-glycans from misfolded proteins, which are ubiquitinated and degraded in the proteasome (26).
Here we used bioinformatic and experimental approaches to determine what effects N-glycan precursor diversity has on N-glycan-dependent QC of glycoprotein folding and ERAD in selected protists and fungi (Fig. 1 and Table 1) (1–4). These results tested three predictions (Fig. 2). First, N-glycan-dependent QC factors for glycoprotein folding and degradation are likely absent from organisms that lack Man in their N-glycan precursors because there are no N-glycan substrates upon which the QC proteins might act (Figs. 1C and 2). Second, N-glycan-dependent folding is likely present and functional in organisms with at least Man5GlcNAc2 in their N-glycan precursors because the substrate for the UGGT is present (Figs. 1B and 2). Third, both N-glycan-dependent folding and degradation are likely present and functional in organisms with at least Man9GlcNAc2 in their precursors because substrates for both UGGT and Mns1 are present (Figs. 1A and 2).
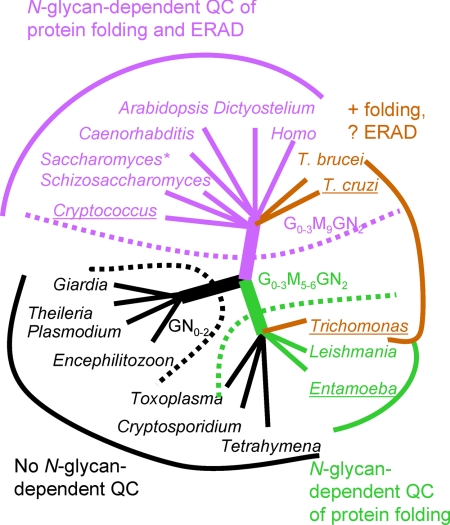
Fig. 2.
N-glycan precursors do not accurately predict the presence or absence of N-glycan-dependent QC systems for glycoprotein folding and ERAD. In this tree, organisms are grouped according to their N-glycan precursors (Table 1) (4). Encephalitozoon, Theileria, Plasmodium, and Giardia, which have N-glycan precursors composed of GN0–2, are predicted to have no N-glycan-dependent QC of glycoprotein folding and degradation (dotted black line). L. major, Trichomonas, Entamoeba, Tetrahymena, Cryptosporidium, and Toxoplasma, which have N-glycan precursors composed of Glc0–3Man5–6GlcNAc2, are predicted to have N-glycan-dependent QC of glycoprotein folding only (dotted green line). Saccharomyces, Schizosaccharomyces, Cryptococcus, Homo, Arabidopsis, Dictyostelium, T. brucei, and T. cruzi, which have N-glycan precursors composed of Glc0–3Man9GlcNAc2, are predicted to have N-glycan-dependent QC of glycoprotein folding and ERAD (dotted purple line). Results from protein predictions (Table 1), phylogenetic trees (Fig. 3), and experiments (Figs. 4–7) are shown with solid colored lines and names for each organism, where black again indicates no N-glycan-dependent QC, green indicates N-glycan-dependent QC of folding, and purple indicates N-glycan-dependent QC of folding and ERAD. Brown indicates organisms where the bioinformatic and experimental data demonstrate N-glycan-dependent QC of glycoprotein folding and suggest the possibility of N-glycan-dependent ERAD. Underlines beneath names of organisms indicate those that were included in in vitro or in vivo experiments.
Results and Discussion
For clarity, the bioinformatic results will be presented first, the experimental results second, and the phylogenetic inferences third.
N-Glycan-Dependent QC of Folding Is Present in Some Protists Containing Man5GlcNAc2 in Their N-Glycan Precursor.
The set of proteins involved in N-glycan-dependent QC of glycoprotein folding in animals, fungi, and plants includes the UGGT, which glucosylates misfolded protein, CRT and/or CNX, which bind and refold glucosylated proteins, glucosidase 2, which removes Glc, and ERGIC-53, which moves well folded glycoproteins to the Golgi (Figs. 1A, 2, and 3A and Table 1) (1–3, 5–10). As predicted, this set of proteins is present in Entamoeba, Dictyostelium, Trypanosoma, Leishmania, and Trichomonas, all of which have at least five Man in their N-glycan precursors and so synthesize a substrate for the UGGT (Figs. 1B, 2, and 3A and Table 1). Also as predicted, this set of proteins is absent from Giardia and Plasmodium, which lack Man residues in their N-glycans, and from Theileria and Encephalitozoon, which lack N-glycans altogether (Figs. 1C, 2, and 3A and Table 1) (4).
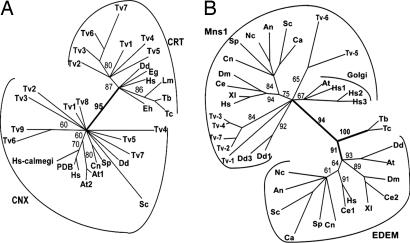
Fig. 3.
Phylogenetic methods distinguish CRT and CNX (A) and Mns1 and EDEM (B). (A) Phylogenetic reconstruction using the maximum likelihood method of representative CRT and CNX from organisms labeled as in Table 1 with the addition of Euglena gracilis (Eg). PDB refers to the CNX of Canis familiaris that has been crystallized, and calmegi refers to a second Homo CNX. (B) Phylogenetic reconstruction using the maximum likelihood method of representative Mns1, EDEM, and Golgi mannosidases. Organisms are labeled as in Table 1 with the addition of Candida albicans (Ca), Aspergillus nidulans (An), Neurospora crassa (Nc), Xenopus laevis (Xl), Drosophila melanogaster (Dm), and Caenorhabditis elegans (Ce). The mannosidase activities of recombinant of Trypanosoma EDEM-like and Trichomonas Mns1 are shown in Figs. 6 and 7.
The absence of N-glycan-dependent QC of glycoprotein folding in Toxoplasma, Cryptosporidium, and Tetrahymena was not expected, because these organisms have N-glycan precursors with Glc1–3Man5GlcNAc2 (Figs. 1–3 and Table 1) (4).
Finally, all eukaryotes studied appear to have N-glycan-independent QC of glycoprotein folding, as suggested by the presence of chaperones (Hsp70 and/or HSP90), protein disulfide isomerases, and peptidyl-prolyl cis-trans isomerases (SI Table 2) (1–3). Testing the function of N-glycan-independent QC of glycoprotein folding and ERAD (below) is beyond the scope of the present study.
N-Glycan-Dependent ERAD Appears to Be Absent from the Majority of Protists.
Mns1, EDEMs, Yos9, and PNGase, which are associated with N-glycan-dependent ERAD, are present in Dictyostelium, animals, plants, and most fungi, which have a Glc3Man9GlcNAc2 N-glycan precursor (Figs. 1A, 2, and 3B and Table 1) (1–3, 11–26). As predicted, these three proteins were absent from Entamoeba, Giardia, Tetrahymena, Plasmodium, Cryptosporidium, Toxoplasma, Encephalitozoon, and Theileria, which have truncated or no N-glycan precursors (Figs. 1 B and C, 2, and 3B and Table 1) (4).
Trypanosoma brucei and Trypanosoma cruzi, which make a Man9GlcNAc2 N-glycan precursor and so are predicted to have N-glycan-dependent ERAD, each contained a single EDEM-like protein and a Yos9 homolog (Figs. 1D, 2, and 3B and Table 1). To our surprise, Trichomonas, which makes a Man5GlcNAc2 N-glycan precursor and so was not expected to have N-glycan-dependent ERAD, contains putative Mns1-like mannosidases, Yos9, and cytosolic PNGases (Figs. 1 E and F and 3B and Table 1) (1–4, 11–26).
Finally, all eukaryotes examined appeared to have N-glycan-independent ERAD, as suggested by the presence of homologs of Der1, Cdc48, Np14, and Ufd1 (SI Table 2) (1–3, 11).
N-Glycan-Dependent QC of Glycoprotein Folding Is Functional in Trichomonas, Entamoeba, and Cryptococcus.
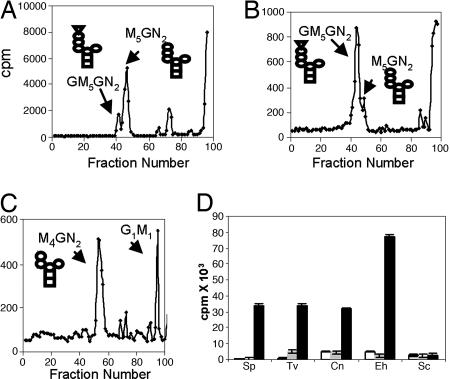
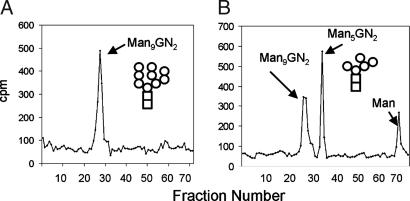
Because Trichomonas has a predicted N-glycan precursor (Man5GlcNAc2) that lacks Glc (Table 1 and ref. 4), the UGGT activity of Trichomonas was demonstrated in vivo by identification of a small amount of glucosylated N-glycan (GlcMan5GlcNAc2) (Fig. 4A) (2, 7). The glucosylated N-glycan of Trichomonas was dramatically increased when the protists were treated in vivo with castanospermine, which blocks the glucosidase II activity (Fig. 4B). The putative peaks containing GlcMan5GlcNAc2, the product of UGGT, was characterized by its digestion to GlcNAc2Man4 and ManGlc disaccharide using the Golgi endomannosidase and its failure to digest with Jackbean mannosidase (Fig. 4C) (27). Identical results were obtained with Entamoeba, and similar results were obtained with Cryptococcus (our unpublished data).
Fig. 4.
Trichomonas, Entamoeba, and Cryptococcus have functional UGGTs. N-glycans of Trichomonas were radiolabeled with Man in vivo for 10 min, released with PNGase, and separated on Biogel P4. (A) The major product of untreated Trichomonas was Man5GlcNAc2. (B) The major product of Trichomonas treated with castanospermine, which inhibits glucosidases, was GlcMan5GlcNAc2 (42). (C) Treatment of GlcMan5GlcNAc2 peak in B with a Golgi endomannosidase produced Man4GlcNAc2 and GlcMan (27). (D) In vitro glucosylation of thyroglobulin by membranes of Schizosaccharomyces (Sp), Trichomonas (Tv), Cryptococcus (Cn), Entamoeba (Eh), and Saccharomyces (Sc). Open bars are controls without addition of thyroglobulin; gray bars are after addition of native thyroglobulin, and black bars are with denatured thyroglobulin. Data show average ± standard deviation.
Membranes of Trichomonas, Entamoeba, and Cryptococcus glucosylated denatured thyroglobulin, an in vitro assay used previously to demonstrate UGGT function of Trypanosoma and Schizosaccharomyces (Fig. 4D) (7). In contrast, native thyroglobulin was not glucosylated by the UGGT of Trichomonas, Entamoeba, and Cryptococcus. As a control, Saccharomyces did not glucosylate denatured thyroglobulin (7).
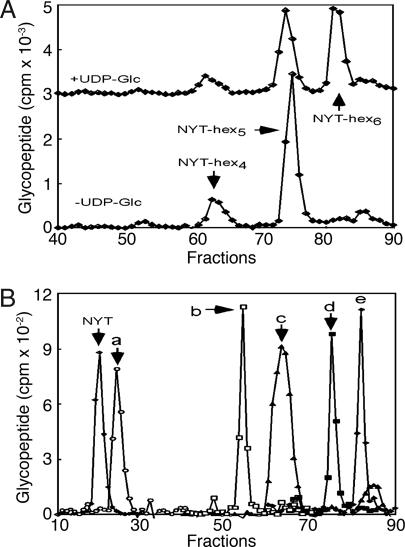
When Entamoeba membranes were incubated with a radiolabeled acceptor peptide in the presence of glucosidase and mannosidase inhibitors, the glycopeptide synthesized using the endogenous donor substrate was Man5GlcNAc2–NYT (Fig. 5A). A Hex6–NYT product was formed when the reaction was supplemented with UDP-Glc as a donor sugar (Fig. 5A). Glycosidase digestions showed that this product was GlcMan5GlcNAc2–NYT, and so was the product of the Entamoeba UGGT (Fig. 5B). In contrast, membranes from Alg6 mutants of CHO cells (1), which are lacking Glc on their N-glycan precursor, produced only a trace of GlcMan5GlcNAc2–NYT in parallel assays (data not shown). These results show that the UGGTs of mammals and Entamoeba have different properties with regard to some artificial targets of UGGT, even though the UGGTs of mammals and Entamoeba behaved the same way with thyroglobulin. A recombinant rat liver UGGT selectively glucosylates N-glycans on longer, more hydrophobic peptides (28).
Fig. 5.
Entamoeba membranes glucosylate Man5GlcNAc2 attached to an NYT peptide in a UDP-Glc-dependent manner. Glycopeptides produced by incubating Entamoeba membranes with a radiolabeled tripeptide acceptor (NYT, Nα-Ac-N-[125I]Y-T-NH2) were captured on ConA and resolved by HPLC. (A) In the absence of UDP-Glc the predominant product was NYT-hex5, whereas in the presence of UDP-Glc the predominant products were NYT-hex5 and NYT-hex6. (B) HPLC analysis of enzymatic digestion of the latter products showed NYT-hex5 was Man5GlcNAc2 and NYT-hex6 was GlcMan5GlcNAc2. NYT-hex5 (Man5GlcNAc2) was digested by α-1,2 mannosidase to NYT-hex3 (Man3GlcNAc2) (b). NYT-hex6 (GlcMan5GlcNAc2) was digested by N-glycanase to DYT (a), by the Golgi endomannosidase to NYT-hex4 (Man4GlcNAc2) (c), and by jack bean mannosidase to NYT-hex5 (GlcMan4GlcNAc2) (d). As expected, NYT-hex6 (GlcMan5GlcNAc2) was resistant to digestion by α-1,2 mannosidase (e).
The Trypanosoma EDEM Ortholog and the Trichomonas Mns1 Ortholog Each Have Mannosidase Activity.
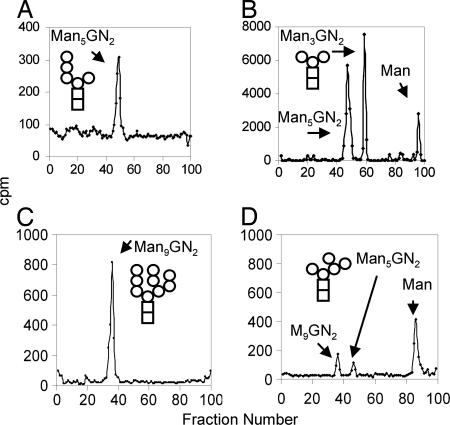
A T. cruzi EDEM-like protein, expressed in Pichia pastoris (29), cleaved four α-1,2-linked Man residues from Man9GlcNAc2 to make processed Man5GlcNAc2 (Figs. 1D and 6). The mannosidase activity of the Trypanosoma EDEM-like protein, which resembles that of mammalian Golgi mannosidase 1 (30), is consistent with the recent finding that human EDEM3 has mannosidase activity (20) and the previous demonstration that Trypanosoma membranes have mannosidase activity (31).
Fig. 6.
A recombinant EDEM-like enzyme of T. cruzi (see Fig. 3B) has α-1,2-mannosidase activity. Man9GlcNAc2 from Saccharomyces Δalg5 (A) was processed to Man5GlcNAc2 and Man (B) by the EDEM-like protein of T. cruzi, which was expressed as a recombinant secreted protein in Pichia. Note that the fraction sizes in Fig. 6 are different from those in Figs. 4 and 7, so that Man5GlcNAc2 elutes in a different fraction.
The Trichomonas Mns1, expressed in Saccharomyces, digested biosynthetic Man5GlcNAc2 to biantennary Man3GlcNAc2 with the release of free Man (Fig. 7B). The α-1,2 mannosidase activity of Trichomonas Mns1 was also shown by its ability to trim Man9GlcNAc2 to processed Man5GlcNAc2 and Man (Fig. 7 C and D). These results are consistent with our observation that some trichomonad N-glycans are trimmed in vivo to biantennary Man3GlcNAc2 (data not shown).
Fig. 7.
Trichomonas cytosolic PNGase has N-glycanase activity, whereas an Mns1-like enzyme of Trichomonas (see Fig. 3B) has α-1,2-mannosidase activity. (A) Recombinant Trichomonas PNGase, expressed as a GST fusion enzyme in E. coli, released Man5GlcNAc2 from Man-labeled glycopeptides of Trichomonas. (B) Recombinant Trichomonas Mns1, expressed in Saccharomyces, digested Man5GlcNAc2 to Man3GlcNAc2 and Man. The same Trichomonas Mns1 digested Man9GlcNAc2 from Saccharomyces (C) to processed Man5GlcNAc2 and Man (D).
The Trichomonas PNGase, which was expressed as a GST fusion enzyme in Escherichia coli (32), removed N-glycans from Trichomonas peptides labeled in vivo with Man and then trypsinized (Figs. 1F and 7A).
These in vitro results, as well as the presence of Yos9 in their genomes, suggest the possibility that Trypanosoma and/or Trichomonas have N-glycan-dependent ERAD in vivo (Fig. 2 and Table 1).
Argument for the Secondary Loss of N-Glycan-Dependent QC of Glycoprotein Folding.
N-glycan-dependent QC of glycoprotein folding was likely present in the common ancestor of extant eukaryotes and secondarily lost from selected eukaryotes (e.g., Giardia, Encephalitozoon, Tetrahymena, and Plasmodium) for the following reasons. First, in phylogenetic analyses, CRT and CNX form two distinct clades, which were supported by high bootstrap values (Fig. 3A). This means that CRT and CNX are paralogs, i.e., the product of gene duplication in a common eukaryotic ancestor. Second, the deep branching eukaryote Trichomonas contains both CNX and CRT (Fig. 3A and Table 1) (33). Plants, animals, and Dictyostelium also contain both CNX and CRT (1–3). Third, Entamoeba has CRT but has lost CNX, whereas fungi have CNX but have lost CRT (Fig. 3A and Table 1). Fourth, Toxoplasma has a truncated CNX that contains an intact Glc-binding domain but is missing most of the conserved arm that binds protein disulfide isomerase (SI Fig. 8). Fifth and finally, the Saccharomyces UGGT ortholog (Kre5) is deeply divergent (SI Fig. 9), so that it no longer glucosylates misfolded glycoproteins but is instead thought to be involved in β-1,6-glucan synthesis (7). Using similar arguments, we recently concluded that the present diversity of N-glycan precursors among extant eukaryotes also most likely resulted from secondary loss rather than primary absence (4).
Secondary loss may also explain the absence of N-glycan-dependent ERAD in numerous protists. First, in phylogenetic analyses, Mns1 and EDEM are present in two clades, suggesting that they were paralogs in a common eukaryotic ancestor (Fig. 2B). Second, Mns1 and EDEM are each present in a deep-branching eukaryote (Trichomonas and Trypanosoma, respectively) (33). On the other hand, primary absence is suggested by the absence of Mns1, EDEM, Yos9, and PNGase from so many of the protists examined here and by the inability of phylogenetic methods to accurately predict function (30). For example, are Mns1 of Trichomonas and EDEM of Trypanosoma involved in ERAD? Or are these mannosidases involved in making a biantennary Man3GlcNAc2 core for synthesis of complex N-glycans (34)?
Significance.
These results strongly support our hypothesis that the diversity of N-glycan precursors among various protists and fungi has profound effects on N-glycan-dependent QC of glycoprotein folding and degradation in the ER (Figs. 1 and 2). As predicted, Giardia, Plasmodium, Theileria, and Encephalitozoon, which have truncated or no N-glycans, are missing N-glycan-dependent QC proteins. In contrast, the absence of N-glycan-dependent QC of glycoprotein folding in Toxoplasma, Cryptosporodium, and Tetrahymena, which make Glc1–3Man5GlcNAc2, and the possible presence of N-glycan-dependent ERAD in Trichomonas, which makes Man5GlcNAc2, were not anticipated. It appears then that N-glycan precursor length does not always accurately predict the presence or absence of N-glycan-dependent QC systems for glycoprotein folding and ERAD (Figs. 1 and 2).
The experimental results demonstrate that the predicted set of proteins involved in N-glycan-dependent QC of glycoprotein folding in Entamoeba, Trichomonas, and Cryptococcus are indeed active (Figs. 1, 2, 4, and 5). The importance of N-glycan-dependent QC of glycoprotein folding is suggested by positive selection of sites for N-linked glycosylation (sequons) in secreted proteins versus cytosolic controls in these protists, as well as in metazoa, fungi, and plants (our unpublished data). In contrast, in protists lacking N-glycan-dependent QC of glycoprotein folding, there is no positive selection for sequons in secreted proteins.
Materials and Methods
Bioinformatic and Phylogenetic Methods.
Predicted proteins of protists (Giardia lamblia, Entamoeba histolytica, Trichomonas vaginalis, T. cruzi, T. brucei, Leishmania major, Tetrahymena thermophila, Plasmodium falciparum, Cryptosporidium parvum, Toxoplasma gondii, Theileria annulata, and Dictyostelium discoideum) and fungi (S. pombe, Encephalitozoon cuniculi, and Cryptococcus neoformans) were searched with S. cerevisiae proteins involved in N-glycan-dependent QC or glycoprotein folding or ERAD [glucosidase 1 and II, UGGT (Kre5p), CNX, ERGIC-53, α-mannosidase (Mns1p), EDEM (Htm1p), Yos9, and PNGase] (Table 1) (1–3, 35, 36). Alternatively, these same eukaryotes were searched with Saccharomyces proteins involved in N-glycan-independent QC of glycoprotein folding and degradation (Der1, CDC48, Npl4, Ufd1, Hrd1, Hrd3, Doa10, and Ire1) (SI Table 2) (11). N-terminal signal peptides and transmembrane helices were predicted by using software at www.cbs.dtu.dk/services/SignalP and www.cbs.dtu.dk/services/TMHMM, respectively (37, 38).
UGGT were identified by a length of >1,000 aa, a C-terminal glucosyltransferase domain, and a C-terminal ER retention signal. CRT and CNX were distinguished by the presence of a C-terminal transmembrane helix in the latter and by phylogenetic methods (Fig. 3A). ERGIC-53s were identified by a legume lectin domain and a C-terminal transmembrane helix, whereas glucosidase IIs were identified by their glycohydrolase domain. Mns1, Golgi mannosidases, and EDEM were distinguished by phylogenetic methods (Fig. 3B) (12–23, 30). Alignments of protein sequences were made by using ClustalW, and manual adjustments and trimming of the alignments were performed with Jalview (39). Phylogenetic trees were constructed from the positional variation with maximum likelihood using quartet puzzling (40, 41).
Tests of Glucosyltransferase and Glycosidase.
UGGTs that glucosylate N-glycans of misfolded proteins were demonstrated in three ways (7, 8). First, genome project strains of Entamoeba and Trichomonas were grown and labeled with [2]3H-Man in the presence or absence of 200 μg/ml of the glucosidase II inhibitor castanospermine (42). N-glycans were released with bacterial PNGase and characterized on a Biogel P-4 superfine column as described (4). A putative GlcMan5GlcNAc2 peak was digested with a Golgi endomannosidase (27).
Second, thyroglobulin, which contains N-glycans composed primarily of Man9GlcNAc2, is an excellent target for testing the UGGT activity of membranes (2, 7). Membranes of Entamoeba and Trichomonas were incubated with tritiated UDP-Glc and 20 μg of thyroglobulin, as described (7).
Third, the ability of Entamoeba UGGT to glucosylate Man5GlcNAc2 attached to a tripeptide was tested by using a modified in vitro oligosaccharyltransferase assay (4, 43). Entamoeba membranes were incubated for 2–30 min at 37°C with the membrane-permeable tripeptide acceptor Nα-Ac-Asn-[125I]-Tyr-Thr-NH2 (NYT) at 5 μM in the presence of deoxynojiromycin and swainsonine to ensure that the glycopeptide products were not degraded by glucosidases or mannosidases, respectively (42). UGGT activity was detected by analyzing glycopeptides synthesized in the presence or absence of 1 mM UDP-Glc. Glycopeptide products were collected by binding to immobilized Con A and separated on HPLC using standards from Saccharomyces (44). As a control, membranes from an Alg6 CHO cell mutant (1), which synthesizes Man9GlcNAc2, were tested for their UGGT activity.
Tests of Mannosidases.
The T. cruzi EDEM-like protein gene was cloned in the pPICZα vector and expressed by using an EasySelect Pichia expression kit (Invitrogen, Carlsbad, CA) (29). The Trichomonas Mns1 gene was cloned in pYES2.1/V5-His-TOPO vector (Invitrogen) and expressed in the yeast strain BY4741 under a Gal1 promoter. In each case, the recombinant protein, which contained a polyhistidine tag, was purified from yeast cell extract using Probond Purification System (Invitrogen). Recombinant Trypanosoma EDEM-like protein and Trichomonas Mns1 were tested by using Man9GlcNAc2, which was obtained from Man-labeled S. cerevisiae ΔAlg5 (1).
Test of Trichomonas PNGase.
A representative Trichomonas PNGase gene was cloned into the pGEX-6T vector to make a recombinant GST-TvPNGase fusion enzyme in E. coli (32). Target N-glycans, which were generated by labeling Trichomonas with 3H-Man and trypsinizing delipidated glycoproteins, digested with 5 μg of recombinant GST-TvPNGase at 37°C for 72 h in 50 mM Tris·HCl (pH 6.8) containing 2 mM DTT. A negative control was GST alone. Released sugars were purified by passage through a Glycoclean R cartridge (Prozyme, San Leandro, CA) before loading on a Biogel P-4 column.
Supplementary Material
Acknowledgments
This work was supported in part by National Institutes of Health Grants AI44070 (to J.S.), AI48082 (to J.S.), GM31318 (to P.W.R.), and GM43768 (to R.G.).
Abbreviations
- ER
endoplasmic reticulum
- CNX
calnexin
- CRT
calreticulin
- EDEM
ER degradation-enhancing α-mannosidase-like protein
- ERAD
ER-associated degradation
- QC
quality control
- UGGT
UDP-Glc:glycoprotein glucosyltransferase
- PNGase
peptide-N-glycanase
- Glc
glucose
- Man
mannose.
Footnotes
The authors declare no conflict of interest.
This article contains supporting information online at www.pnas.org/cgi/content/full/0704862104/DC1.
References
- 1.Helenius A, Aebi M. Annu Rev Biochem. 2004;73:1019–1049. doi: 10.1146/annurev.biochem.73.011303.073752. [DOI] [PubMed] [Google Scholar]
- 2.Trombetta ES, Parodi AJ. Annu Rev Cell Dev Biol. 2003;19:649–676. doi: 10.1146/annurev.cellbio.19.110701.153949. [DOI] [PubMed] [Google Scholar]
- 3.Moreman KW, Molinari M. Curr Opin Struct Biol. 2006;16:592–599. doi: 10.1016/j.sbi.2006.08.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Samuelson J, Banerjee S, Magnelli P, Cui J, Kelleher DJ, Gilmore R, Robbins PW. Proc Natl Acad Sci USA. 2005;102:1548–1553. doi: 10.1073/pnas.0409460102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Schrag JD, Bergeron JJ, Li Y, Borisova S, Hahn M, Thomas DY, Cygler M. Mol Cell. 2001;8:633–644. doi: 10.1016/s1097-2765(01)00318-5. [DOI] [PubMed] [Google Scholar]
- 6.Leach MR, Cohen-Doyle MF, Thomas DY, Williams DB. J Biol Chem. 2002;277:29686–29697. doi: 10.1074/jbc.M202405200. [DOI] [PubMed] [Google Scholar]
- 7.Fernandez FS, Trombetta SE, Hellman U, Parodi AJ. J Biol Chem. 1994;269:30701–30706. [PubMed] [Google Scholar]
- 8.Conte I, Labriola C, Cazzulo JJ, Docampo R, Parodi AJ. Mol Biol Cell. 2003;14:3529–3540. doi: 10.1091/mbc.E03-04-0228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Appenzeller-Herzog C, Roche AC, Nufer O, Hauri HP. J Biol Chem. 2004;279:12943–12950. doi: 10.1074/jbc.M313245200. [DOI] [PubMed] [Google Scholar]
- 10.Veloso LM, Svensson K, Schneider G, Pettersson RF, Lindqvist Y. J Biol Chem. 2002;277:15979–15984. doi: 10.1074/jbc.M112098200. [DOI] [PubMed] [Google Scholar]
- 11.Kostova Z, Wolf DH. EMBO J. 2003;22:2309–2317. doi: 10.1093/emboj/cdg227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Jakob CA, Burda P, Roth J, Aebi MJ. Cell Biol. 1998;142:1223–1233. doi: 10.1083/jcb.142.5.1223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hosokawa N, Tremblay LO, You Z, Herscovics A, Wada I, Nagata K. J Biol Chem. 2003;278:26287–26294. doi: 10.1074/jbc.M303395200. [DOI] [PubMed] [Google Scholar]
- 14.Nakatsukasa K, Nishikawa S, Hosokawa N, Nagata K, Endo T. J Biol Chem. 2001;276:8635–8638. doi: 10.1074/jbc.C100023200. [DOI] [PubMed] [Google Scholar]
- 15.Jakob CA, Bodmer D, Spirig U, Battig P, Marcil A, Dignard D, Bergeron JJ, Thomas DY, Aebi M. EMBO Rep. 2001;2:423–430. doi: 10.1093/embo-reports/kve089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hosokawa N, Wada I, Hasegawa K, Yorihuzi T, Tremblay LO, Herscovics A, Nagata K. EMBO Rep. 2001;2:415–422. doi: 10.1093/embo-reports/kve084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Movsichoff F, Castro OA, Parodi AJ. Mol Biol Cell. 2005;16:4714–4724. doi: 10.1091/mbc.E05-03-0246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ermonval M, Kitzmuller C, Mir AM, Cacan R, Ivessa NE. Glycobiology. 2001;11:565–576. doi: 10.1093/glycob/11.7.565. [DOI] [PubMed] [Google Scholar]
- 19.Mast SW, Diekman K, Karaveg K, Davis A, Sifers RN, Moremen KW. Glycobiology. 2005;15:421–436. doi: 10.1093/glycob/cwi014. [DOI] [PubMed] [Google Scholar]
- 20.Hirao K, Natsuka Y, Tamura T, Wada I, Morito D, Natsuka S, Romero P, Sleno B, Tremblay LO, Herscovics A, et al. J Biol Chem. 2006;281:9650–9658. doi: 10.1074/jbc.M512191200. [DOI] [PubMed] [Google Scholar]
- 21.Olivari S, Galli C, Alanen H, Ruddock L, Molinari M. J Biol Chem. 2005;280:2424–2428. doi: 10.1074/jbc.C400534200. [DOI] [PubMed] [Google Scholar]
- 22.Lederkremer GZ, Glickman MH. Trends Biochem Sci. 2005;30:297–303. doi: 10.1016/j.tibs.2005.04.010. [DOI] [PubMed] [Google Scholar]
- 23.Olivari S, Molinari M. FEBS Lett. 2007 doi: 10.1016/j.febslet.2007.04.070. [DOI] [PubMed] [Google Scholar]
- 24.Buschhorn BA, Kostova Z, Medicherla B, Wolf DH. FEBS Lett. 2004;577:422–426. doi: 10.1016/j.febslet.2004.10.039. [DOI] [PubMed] [Google Scholar]
- 25.Szathmary R, Bielmann R, Nita-Lazar M, Burda P, Jakob CA. Mol Cell. 2005;19:765–775. doi: 10.1016/j.molcel.2005.08.015. [DOI] [PubMed] [Google Scholar]
- 26.Suzuki T, Park H, Hollingsworth NM, Sternglanz R, Lennarz WJ. J Cell Biol. 2000;149:1039–1052. doi: 10.1083/jcb.149.5.1039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Spiro MJ, Bhoyroo VD, Spiro RG. J Biol Chem. 1997;272:29356–29363. doi: 10.1074/jbc.272.46.29356. [DOI] [PubMed] [Google Scholar]
- 28.Taylor SC, Thibault P, Tessier DC, Bergeron JJ, Thomas DY. EMBO Rep. 2003;4:405–411. doi: 10.1038/sj.embor.embor797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hamilton SR, Bobrowicz P, Bobrowicz B, Davidson RC, Li H, Mitchell T, Nett JH, Rausch S, Stadheim TA, Wischnewski H, et al. Science. 2003;301:1244–1246. doi: 10.1126/science.1088166. [DOI] [PubMed] [Google Scholar]
- 30.Herscovics A. Biochim Biophys Acta. 1999;1473:96–107. doi: 10.1016/s0304-4165(99)00171-3. [DOI] [PubMed] [Google Scholar]
- 31.Parodi AJ, Lederkremer GZ, Mendelzon DH. J Biol Chem. 1983;258:5589–5595. [PubMed] [Google Scholar]
- 32.Smith DB, Johnson KS. Gene. 1998;67:31–40. doi: 10.1016/0378-1119(88)90005-4. [DOI] [PubMed] [Google Scholar]
- 33.Bapteste E, Brinkmann H, Lee JA, Moore DV, Sensen CW, Gordon P, Durufle L, Gaasterland T, Lopez P, Muller M, Philippe H. Proc Natl Acad Sci USA. 2002;99:1414–1419. doi: 10.1073/pnas.032662799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Acosta-Serrano A, O'Rear J, Quellhorst G, Lee SH, Hwa KY, Krag SS, Englund PT. Eukaryot Cell. 2004;3:255–263. doi: 10.1128/EC.3.2.255-263.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Altschul SF, Madden TL, Schaffer AA, Zhang J, Zhang Z, Miller W, Lipman DJ. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Mewes HW, Albermann K, Bahr M, Frishman D, Gleissner A, Hani J, Heumann K, Kleine K, Maierl A, Oliver SG, et al. Nature. 1997;387:7–65. doi: 10.1038/42755. [DOI] [PubMed] [Google Scholar]
- 37.Nielsen H, Brunak S, von Heijne G. Protein Eng. 1999;12:3–9. doi: 10.1093/protein/12.1.3. [DOI] [PubMed] [Google Scholar]
- 38.Krogh A, Larsson B, von Heijne G, Sonnhammer EL. J Mol Biol. 2001;305:567–580. doi: 10.1006/jmbi.2000.4315. [DOI] [PubMed] [Google Scholar]
- 39.Thompson JD, Higgins DG, Gibson TJ. Nucleic Acids Res. 1994;22:4673–4680. doi: 10.1093/nar/22.22.4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Jones DT, Taylor WR, Thornton JM. Comput Appl Biosci. 1992;8:275–282. doi: 10.1093/bioinformatics/8.3.275. [DOI] [PubMed] [Google Scholar]
- 41.Strimmer K, Von Haeseler A. Proc Natl Acad Sci USA. 1997;94:6815–6819. doi: 10.1073/pnas.94.13.6815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Elbein AD. FASEB J. 1991;5:3055–3063. doi: 10.1096/fasebj.5.15.1743438. [DOI] [PubMed] [Google Scholar]
- 43.Kelleher DJ, Kreibich G, Gilmore R. Cell. 1992;69:55–65. doi: 10.1016/0092-8674(92)90118-v. [DOI] [PubMed] [Google Scholar]
- 44.Kelleher DJ, Karaoglu D, Gilmore R. Glycobiology. 2001;11:321–333. doi: 10.1093/glycob/11.4.321. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.