Abstract
Agrobacterium tumefaciens is capable of transferring and integrating an oncogenic T-DNA (transferred DNA) from its tumor-inducing (Ti) plasmid into dicotyledonous plants. This transfer requires that the virulence genes (vir regulon) be induced by plant signals such as acetosyringone in an acidic environment. Salicylic acid (SA) is a key signal molecule in regulating plant defense against pathogens. However, how SA influences Agrobacterium and its interactions with plants is poorly understood. Here we show that SA can directly shut down the expression of the vir regulon. SA specifically inhibited the expression of the Agrobacterium virA/G two-component regulatory system that tightly controls the expression of the vir regulon including the repABC operon on the Ti plasmid. We provide evidence suggesting that SA attenuates the function of the VirA kinase domain. Independent of its effect on the vir regulon, SA up-regulated the attKLM operon, which functions in degrading the bacterial quormone N-acylhomoserine lactone. Plants defective in SA accumulation were more susceptible to Agrobacterium infection, whereas plants overproducing SA were relatively recalcitrant to tumor formation. Our results illustrate that SA, besides its well known function in regulating plant defense, can also interfere directly with several aspects of the Agrobacterium infection process.
Keywords: two-component system, tumorigenesis, defense response, rhizosphere, plant–microbe interaction
Agrobacterium tumefaciens, a member of the α-Proteobacteria, can transfer and integrate an oncogenic T-DNA (transferred DNA) from its tumor-inducing (Ti) plasmid into dicotyledonous plants, leading to the formation of crown gall tumors (1). This unique ability to transfer DNA forms the basis of plant molecular genetics. T-DNA transfer requires activation of the vir regulon on the Ti plasmid (1). vir genes are activated at an acidic pH, pH 5.5–6.0, which approximates the pH of the rhizosphere (2). Under acidic conditions, plant phenolic compounds together with many monosaccharide components of the plant cell wall are recognized by the sensor protein of a two-component regulatory system, VirA. After autophosphorylation, the VirA protein transfers the phosphate to the transcriptional regulator, VirG, which then binds to a specific 12-bp sequence (vir box) upstream of each of the Ti plasmid-encoded vir operons and promotes their transcription (1, 3). The plant signals also activate the transcription of the virA/G regulatory system. The 30 known vir genes include a gene, virD2, encoding an endonuclease that recognizes and cleaves the border sequences of the T-DNA, thereby releasing a single-stranded DNA molecule. This DNA (the T-strand) is exported through a type IV secretion system encoded by the virB operon (1). In addition to the vir genes, the transcription of the repABC operon that controls the copy number of the Ti plasmid is also activated by the VirA/G system (4).
The VirA/G system has been studied extensively (1). VirA exists as a dimer whose formation is independent of plant signals. The VirA protein is composed of four domains: the periplasmic domain, which binds the sugar-binding protein ChvE with its associated sugars and also detects acidic pH; the linker domain, which most likely interacts directly with the phenolic signal, such as acetosyringone; the kinase domain, involved in the autophosphorylation of the conserved histidine moiety; and the C-terminal receiver domain, whose function is unclear (5, 6).
In addition to the Ti plasmid-encoded vir genes, a number of chromosomally encoded genes, termed chv, are also important for virulence (1, 3). These genes include chvA and chvB, which are required for the attachment of Agrobacterium to plant cells. One of the most intensively studied is chvE (1). The ChvE protein binds a number of different sugars in the rhizosphere and then interacts with the periplasmic region of the VirA protein. The ChvG/I two-component system is required for virG gene expression and for growth under acidic conditions (1). None of the chromosomally encoded chv genes has a vir box in its upstream sequences, nor are they regulated by the VirA/G system.
Salicylic acid (SA), a plant phenolic metabolite, is a key signal molecule in regulating plant defense in response to a wide variety of pathogens (7–9). Upon infection, SA triggers either a localized or systemic acquired resistance response in which the plant gains long-lived resistance to pathogens (10). Studies on SA function have been focused primarily on defense mechanisms within the plant (10). However, some evidence suggests that SA may also directly affect bacteria. SA was shown to down-regulate fitness and virulence factor production in Pseudomonas aeruginosa PA14 (7). At concentrations that did not inhibit growth, SA also affected bacterial attachment and biofilm formation in this organism (7).
In this work, we demonstrate that SA directly affects the Agrobacterium infection process by inhibiting the induction of vir genes at concentrations that have little effect on growth. This phenolic compound also induces the expression of a lactonase, which degrades the quormone N-acylhomoserine lactone. Mutants and transgenic Arabidopsis plants whose metabolism of SA is modified show the predicted alterations in susceptibility to infection by Agrobacterium.
Results
SA Inhibits vir Gene Expression.
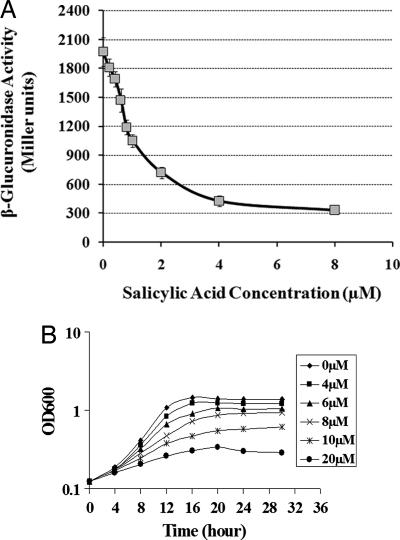
Numerous studies have shown that small molecules of plant origin play important roles in the interaction of Agrobacterium with plants. Phenolics, such as the many vir gene-inducing compounds, seem to be especially important in this regard (1). Indoleacetic acid (IAA), a plant hormone produced by T-DNA-encoded enzymes in transformed plants, can shut down vir gene induction (11). In the course of screening other phenolic compounds for their effect on vir gene induction, we tested SA, a phenolic compound important in plant defense. The expression of vir genes was compared in wild-type (WT) strain C58 grown in vir gene-inducing medium (pH 5.5) supplemented with varying concentrations of SA. The data in Fig. 1A show that SA significantly inhibited the expression of vir genes as monitored by assaying the promoter activity of a virB1::gusA transcriptional gene fusion. Two μM SA inhibited virB1 expression by >50%, and 8 μM SA exerted ≈90% inhibition. Moreover, at higher concentrations (>10 μM), SA also inhibited Agrobacterium growth significantly (Fig. 1B). The inhibition of vir gene expression and growth occurred at an acidic pH (pH 5.5), but at pH 7, growth was not inhibited (data not shown). Because the rhizosphere is typically acidic at the site of Agrobacterium–plant interactions, these data suggest that SA, under biologically relevant conditions, can directly affect the interaction of Agrobacterium with the plant either by repressing vir gene expression or, at slightly higher concentrations, by inhibiting bacterial growth. The observation that the plotted data (Fig. 1A) fit nicely to a sigmoidal logistic model suggests that the interaction of SA with its target is a simple, single-site interaction with no cooperativity and is specific and not the result of nonspecific mechanisms.
Fig. 1.
SA inhibits vir gene induction and, at higher concentrations, bacterial growth. (A) Effect of SA on Agrobacterium vir gene expression. Expression of a plasmid-borne virB1::gusA transcriptional gene fusion in Agrobacterium C58 grown in induction medium with acetosyringone (100 μM), carbenicillin (100 μg/ml), and various concentrations of SA (0–8 μM) is shown. After 16 h, β-glucuronidase activity was measured as described in Materials and Methods. (B) Effect of SA on Agrobacterium growth. Agrobacterium growth under various concentrations of SA (0–20 μM) is shown. Cells were grown in acidified AB minimal medium (pH 5.5) supplied with various concentrations of SA. Readings at A600 (OD600) are the mean of three independent experiments.
The two-component regulatory system, VirA/G, controls the expression of the vir regulon including virA/virG itself (1). To explore whether other members of the vir regulon were also inhibited by SA, we examined its effects on the expression of virA, virG, virD, virE, virH, as well as tzs (another VirA/G-regulated gene) (3). We also examined the expression of chvA, chvD, chvE, chvG, and chvI genes with or without SA treatment. We found that SA inhibited the expression of all of these virA/G-regulated vir genes but not chv genes as monitored by transcriptional gene fusions (Table 1). It seems highly unlikely that the inhibition of vir gene expression was the result of internal acidification of the bacteria because SA has a pKa of 2.97, whereas the induction medium was pH 5.5. Although the previous experiments were performed on the C58 strain, which induces the synthesis of the opine nopaline in tumors, the same results were also observed in strain A6, which induces octopine synthesis (data not shown). These results suggest that: (i) SA can shut down the transfer of T-DNA and virulence proteins by inhibiting the expression of the vir regulon but not chv genes, and (ii) this inhibition is mediated by repressing the expression of virA/G itself.
Table 1.
Effects of SA on expression of Agrobacterium vir and chv genes
| Gene fusion | AS, 0 μM | AS, 100 μM | AS, 100 μM SA, 8 μM |
|---|---|---|---|
| virD1::gusA | 193 | 4,215 | 439 |
| virE0::gusA | 261 | 4,927 | 523 |
| virH1::gusA | 349 | 2,782 | 531 |
| tzs::gusA | 247 | 1,658 | 455 |
| virA::gusA | 430 | 1,894 | 784 |
| virG::lacZ | 134 | 470 | 210 |
| chvA::gusA | 1,704 | 1,688 | 1,722 |
| chvD::gusA | 1,770 | 1,902 | 1,686 |
| chvE::gusA | 1,732 | 1,626 | 1,695 |
| chvG::gusA | 660 | 601 | 692 |
| chvI::gusA | 1,754 | 2,028 | 1,790 |
Agrobacterium cells harboring the indicated fusion plasmids were grown for 16 h in induction medium (pH 5.5) with or without 8 μM SA. The β-galactosidase or β-glucuronidase activity was determined as described in Materials and Methods. AS, acetosyringone.
Several SA derivatives were also tested for their effects on the expression of vir genes. At concentrations of 10 μM, both acetyl-SA (aspirin) and methyl-SA inhibited the expression of the virB1 gene, whereas salicylamide and benzoic acid had no effect (data not shown). We also tested catechol because SA is converted to catechol by salicylate hydroxylase (encoded by Atu1574) (12). Even at concentrations of 200 μM, catechol had no effect on the expression of vir genes, nor did it affect bacterial growth either at pH 5.5 or 7 (data not shown). Jasmonic acid, which also plays an important role in modulating plant defenses (10), had no effect on vir gene expresssion (data not shown).
Microarray Analysis of Cells Grown with SA.
To gain a global view of the effect of SA on Agrobacterium gene expression, we compared the transcriptome of C58 cells cultured at pH 5.5 (+100 μM acetosyringone) in the presence or absence of 6 μM SA for 6 h. This concentration of SA had no significant effect on cell growth under the conditions in which the cells were grown for RNA isolation (see Materials and Methods). Microarray analysis revealed that the transcription of 49 genes was inhibited by SA at least 2-fold. These genes included the 30 vir regulon members previously identified as being under the control of the VirA/G system (Table 2). Interestingly, none of the chv genes such as chvA, B, D, and E, which are not regulated by the virA/G system nor induced by phenolic compounds, was affected by SA, nor was the expression of another two-component system important in virulence, chvG/I. These data confirmed the gene expression data obtained by using transcriptional gene fusions (Table 1) and established that SA inhibited the expression of the vir regulon, but not other processes required for crown gall formation in which chv genes play essential roles (1, 3).
Table 2.
Selected list of microarray data for the vir regulon, rep genes, chv genes, and attKLM operon exhibiting altered gene expression in cells grown with salicylic acid (6 μM) for 7 h
| Gene | Ratio, log2 | Gene | Ratio, log2 | Gene | Ratio, log2 |
|---|---|---|---|---|---|
| tzs | −2.63 | virC2 | −1.39 | repA | −1.25 |
| virA | −1.25 | virD1 | −2.46 | repB | −1.18 |
| virB1 | −3.48 | virD2 | −1.66 | ||
| virB2 | −3.27 | virD3 | −2.16 | chvA | 0.21 |
| virB3 | −3.19 | virD4 | −1.35 | chvB | 0.21 |
| virB4 | −3.23 | virE0 | −2.66 | chvD | 0.07 |
| virB5 | −3.36 | virE1 | −3.08 | chvE | −0.14 |
| virB6 | −2.06 | virE2 | −2.93 | chvH | 0.61 |
| virB7 | −1.82 | virE3 | −1.97 | chvG | 0.32 |
| virB8 | −1.68 | virF | −1.94 | chvI | −0.2 |
| virB9 | −2.21 | virG | −0.98 | ||
| virB10 | −2.89 | virH1 | −2.21 | attK | 1.68 |
| virB11 | −2.17 | virH2 | −2.28 | attL | 1.85 |
| virC1 | −1.19 | virK | −2.54 | attM | 1.65 |
The ratio, log2, shows genes inhibited (negative) or induced (positive) by 6 μM SA.
The microarray analysis also revealed that SA repressed the expression of the repABC operon, which encodes the Ti plasmid replication and partitioning machinery (Table 2). Previous studies showed that the expression of this operon is under dual regulation. It is a member of the vir regulon induced by acetosyringone (4) and is also induced by the quorum-sensing signal 3OC8-HSL (13). The induction of the repABC operon elevates the Ti plasmid copy number, thereby increasing the number of copies of T-DNA and genes encoding the T-DNA export machinery located on the Ti plasmid. This action could enhance T-DNA transfer as well as conjugative transfer of the Ti plasmid (4, 13). The observation that SA repressed the expression of this repABC operon suggests that this plant defense molecule can reduce the increase in copy number of the Ti plasmid associated with vir gene induction.
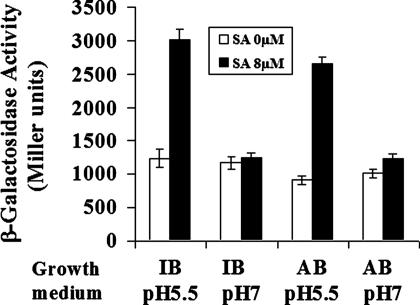
Microarray analysis further revealed that 36 genes were induced by SA at least 2-fold. Perhaps of greatest interest is the observation that SA stimulated the expression of the attKLM operon, which encodes a quormone degradation system (14). In Agrobacterium, the lactonase encoded by attM can hydrolyze the quorum-sensing signal 3OC8-HSL (14). Acylated homoserine lactones are a prevalent class of extracellular bacterial signals implicated in several important cell processes, including bacterial virulence (15). Recent studies have shown that γ-aminobutyric acid (GABA) and succinic semialdehyde activate the expression of the attKLM operon, resulting in quorum-sensing signal decay and reduced Agrobacterium virulence (16, 17). The microarray data were confirmed by demonstrating that 8 μM SA induced the expression of an attKLM::lacZ gene fusion (Fig. 2). Moreover, the SA-induced attKLM expression only occurred under acidic conditions and was not observed when cells were grown at pH 7 (Fig. 2). Acetosyringone did not affect the induction of the attKLM operon by SA (data not shown), which suggests that induction of this quormone-quenching system and inhibition of the vir regulon by SA are independent. The SA-activated expression of the attKLM operon is currently under investigation.
Fig. 2.
Effect of SA on the expression of the attKLM operon. Cells harboring the attKLM::lacZ gene fusion (16) were grown for 20 h with 0 μM or 8 μM SA at the indicated pH. IB, inducing medium containing 100 μM acetosyringone; AB, inducing medium without acetosyringone.
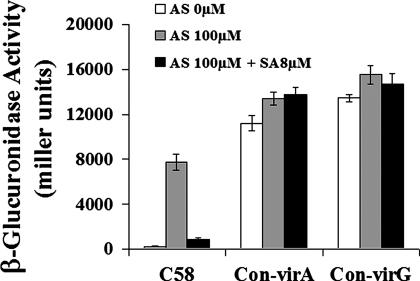
SA-Inhibited vir Gene Expression Can Be Rescued by Either virA or virG Mutants That Activate vir Gene Induction Independent of Acetosyringone.
An Agrobacterium virA mutant (G665D) has been isolated that does not require acetosyringone and monosaccharides to activate the vir regulon (18). A point mutation in virG (N54D) also renders vir regulon transcription independent of a phenolic signal and monosaccharides as well as VirA (19, 20). Although both the virA (G665D) and virG (N54D) mutants activate vir gene induction constitutively, acidic conditions are still required for the induction. To gain insight into the site of inhibition of the vir regulon by SA, we assessed whether vir gene expression in cells containing either the virA (G665D) or virG (N54D) was still inhibited by SA. The data in Fig. 3 show that the inhibitory effect of SA on vir gene expression was completely abolished in cells containing either a virA (G665D) or virG (N54D) plasmid. These observations further support the previous conclusion that SA inhibits vir regulon expression through the virA/G regulatory system. In addition, these results implicate both acetosyringone and VirA in the SA-inhibited expression of the vir regulon because neither acetosyringone-independent (either virA G665D or virG N54D mutants) nor virA-independent (virG N54D mutant) vir gene expression was inhibited by SA (18–20).
Fig. 3.
Constitutively active virA or virG rescues vir gene expression inhibited by SA. Cells containing a WT virA/virG allele and a virB1::gusA fusion plasmid are indicated as C58. The constitutive virA plasmid (indicated as Con-virA) or constitutive virG plasmid (indicated as Con-virG) were introduced into the C58 strain harboring the virB1::gusA fusion plasmid (pFUS1). Cells were grown for 24 h with the indicated concentrations of SA or acetosyringone (AS), and the β-glucuronidase activity was measured as described in Materials and Methods.
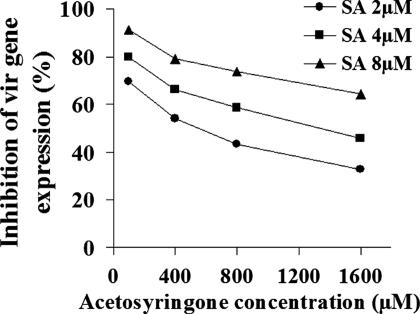
SA-Inhibited vir Gene Expression Can Be Partially Overcome by Increased Levels of Acetosyringone.
During Agrobacterium–plant host interactions, plant-released phenolic compounds such as acetosyringone interact with the sensory histidine kinase VirA to initiate the microbe–plant signaling process (1). To decipher the relationship between the plant-synthesized inhibitory signal molecule SA and the plant-released vir regulon-inducer acetosyringone, we assayed the degree of vir gene inhibition by SA when the levels of acetosyringone were modulated. Increasing the acetosyringone level significantly attenuated the inhibitory effects of SA on vir gene expression. With 4 μM SA, vir gene expression was inhibited >80% at 0.1 mM acetosyringone; in contrast, expression was only diminished 45% at 1.6 mM acetosyringone (Fig. 4). These data, together with the above observation that both acetosyringone-independent and virA-independent vir gene expression were not inhibited by SA, suggest that SA interferes with the activation of the VirA protein by acetosyringone.
Fig. 4.
Increasing acetosyringone attenuates the inhibition of vir gene expression by SA. Acetosyringone dose–response curves were obtained for cells grown in the absence or presence of the indicated concentrations of SA for 24 h, and the percentage inhibition in virB1::gusA expression was calculated. The data represent the averages of three independent determinations.
The VirA Kinase Domain Is the Site of SA Inhibition.
The results described in the preceding two sections indicate that SA inhibits vir gene expression through an effect on the VirA protein, possibly by competing with acetosyringone for a binding site on VirA. To explore this hypothesis, we compared the effect of 10 μM SA on vir gene expression that depended on VirA derived from pTiA6 and certain sections of that protein (Table 3). The expression and stability of the VirA protein fragments were confirmed previously (5, 6). The signal-transducing activity of WT (full-length VirA), PLK (periplasmic, linker, and kinase domains), LK (linker and kinase domains), and K (kinase domain) were all inhibited by 10 μM SA, regardless of the presence of acetosyringone. In agreement with the findings of others (5, 6), our assays indicate that the linker domain is required for a significant response to acetosyringone; while showing significant activity in the absence of acetosyringone, the PLK and LK versions of VirA are strongly stimulated by acetosyringone. Importantly, 10 μM SA inhibited the vir gene expression that depended on the acetosyringone-independent kinase (K) domain to roughly the same extent as that driven by the acetosyringone-responsive LK fragment, and the percentage of inhibition was not significantly reduced by added acetosyringone. Thus, SA apparently hinders the function of the VirA kinase domain rather than the ability of the protein to detect the phenolic inducer through the linker domain.
Table 3.
SA inhibits the function of the kinase domain of VirA
| VirA construct |
0 μM AS |
100 μM AS |
||||
|---|---|---|---|---|---|---|
| 0 μM SA | 10 μM SA | Average inhibition, % | 0 μM SA | 10 μM SA | Average inhibition, % | |
| WT | 97 (7) | 70 (4) | 28 | 3,310 (290) | 2,037 (231) | 38 |
| PLK | 2,746 (148) | 788 (145) | 71 | 6,045 (359) | 2,437 (326) | 60 |
| LK | 1,080 (59) | 495 (27) | 54 | 8,682 (324) | 5,324 (346) | 39 |
| K | 554 (36) | 263 (25) | 53 | 476 (36) | 282 (32) | 41 |
A136 cells expressed virG and all virA constructs from the constitutive PN25 promoter (21). VirA domains are indicated as WT, full-length virA; PLK, amino acids 1–711; LK, amino acids 285–711; and K, amino acids 426–711. Plasmid constructs also carry a virB::lacZ fusion and have been described previously (6). Assays of vir gene expression were performed after growth in acidified induction medium containing glucose and 0 or 100 μM acetosyringone with the indicated amounts of SA. The numbers in parentheses are the SD values of three independent samples.
Plants Overproducing SA Are More Resistant to Agrobacterium Infection, and Plants Defective in SA Accumulation Are More Susceptible.
To assess the role of SA in conferring resistance to Agrobacterium infection in planta, we assayed tumor formation on mutant or transgenic Arabidopsis plants that have altered levels of SA and SA-mediated signaling. Mutant cpr5-2 (22) and transgenic 35S-LOX2 (23) plants overaccumulate SA and express pathogenesis-related (PR) genes constitutively or in response to infection, respectively. Transgenic nahG plants, in contrast, convert endogenous SA to catechol, thereby reducing SA accumulation (24). SA-induced PR gene expression and systemic acquired resistance occur primarily through a signaling pathway involving the transcriptional activator NPR1. Mutant npr1-1 plants are defective in this signaling and exhibit decreased PR gene expression (25), although they accumulate at least WT SA levels in response to infection by various pathogens (26). We tested whether SA levels and NPR1-dependent activation of systemic acquired resistance affected the ability of WT Agrobacterium (strains A208 and A348) to form tumors on root segments. At a bacterial cell concentration of 106 cells per ml, cpr5-2 plants developed ≈8-fold fewer tumors than WT Col-0 plants, whereas NahG plants developed significantly more tumors than WT. Both LOX2 and npr1-1 plants showed a substantial decrease in tumor formation, although they were more susceptible than cpr5-2 plants [Table 4 and supporting information (SI) Fig. 5].
Table 4.
Tumor formation by A. tumefaciens on A. thaliana lines with altered SA
| Arabidopsis line | Bacterial cell density in cocultivation, cells per ml |
|||
|---|---|---|---|---|
| 108 | 107 | 106 | 105 | |
| Col-0 (WT) | 33% (n = 69) | 33% (n = 51) | 24% (n = 37) | 3% (n = 37) |
| NahG | 47% (n = 30) | 48% (n = 54) | 38% (n = 29) | 5% (n = 38) |
| npr1-1 | 17% (n = 35) | 2% (n = 42) | 9% (n = 23) | 0% (n = 23) |
| cpr5-2 | 22% (n = 36) | 18% (n = 76) | 3% (n = 40) | 0% (n = 33) |
| Col-0 (WT) | 34% (n = 61) | 19% (n = 63) | ||
| LOX2 | 9% (n = 171) | 9% (n = 155) | ||
Root segments were cocultivated with WT strain A208. Values represent the percentage of root segments with tumors. n, no. of root segments assayed. All tumors were approximately the same size, and each root segment had at most one tumor, regardless of the plant line used.
To probe whether altered SA production affected the plant's susceptibility to tumorigenesis because of the inhibition of vir gene expression or whether it acted at the level of responses within the plant (or both), we tested the ability of cells containing the constitutively active VirA to restore tumor formation on the most resistant Arabidopsis line, the cpr5-2 mutant (SI Fig. 5 and data not shown). Bacteria expressing the constitutive virA allele were highly virulent on the cpr5-2 roots, whereas WT C58 cells were unable to incite a single tumor on this plant line (Table 5). Together, these results indicate that SA inhibits tumor formation by Agrobacterium through an NPR1-independent mechanism and that the inhibition may be attributable primarily to the ability of SA to prevent vir gene induction in the bacterium.
Table 5.
Constitutive VirA restores tumor formation on A. thaliana lines with elevated salicylic acid content
| Arabidopsis line | Bacterium | Bacterial cell density, cells per ml |
|
|---|---|---|---|
| 108 | 106 | ||
| Col-0 (WT) | C58 | 8% (n = 50) | 9% (n = 32) |
| Con-VirA | 20% (n = 40) | 16% (n = 50) | |
| cpr5-2 | C58 | 0% (n = 28) | 0% (n = 31) |
| Con-VirA | 19% (n = 32) | 10% (n = 10) | |
Values represent percentage of root segments with tumors. C58 is the WT strain; Con-VirA indicates C58 with a plasmid carrying the constitutive allele of virA. n, no. of root segments assayed.
Discussion
It is well established that acetosyringone induces the expression of all vir genes and the Ti plasmid repABC operon (1, 4). Our observations that (i) SA significantly inhibited the expression of all vir genes and the Ti plasmid repABC operon; (ii) high levels of acetosyringone attenuated the SA-induced reduction in vir gene expression; and (iii) constitutively active virA or virG overcame the SA inhibition, led to the proposition that SA and acetosyringone play opposing roles during Agrobacterium–plant host interactions. SA interferes with the signaling process stimulated by acetosyringone, thereby blocking expression of the genes encoding the type IV secretion system and other virulence proteins required for tumorigenesis (1, 3). Previous studies have reported that at plant infection sites, SA accumulates to concentrations of 3–4 μM (27), approximately the level that we find affects vir regulon induction. Therefore, the SA concentration and the acidic conditions used in our study mimic the environment and are biologically relevant to the interaction of Agrobacterium with its plant hosts.
Although SA counteracts the virulence-promoting activity of acetosyringone, it appears to exert its effect at the kinase domain of VirA rather than directly blocking signal perception (Table 3). Two VirA functions that depend on the kinase region are autophosphorylation at the conserved histidine at codon 474 and transfer of the phosphate from that histidine to VirG. In vivo studies have shown that although the histidine on an LKR (linker–kinase–receiver) fragment appears to be constitutively phosphorylated, movement of the phosphate from VirA to VirG requires the addition of acetosyringone (21, 28). Thus, SA probably does not compete with detection of the phenolic inducer but may inhibit the phosphate transfer reaction that depends on its presence. The capacity of acetosyringone to overcome SA-mediated inhibition of vir gene expression (Fig. 4) may be the result of acetosyringone-mediated stimulation of phosphotransfer activities of VirA and simply reflect the combined effects of SA and acetosyringone.
In addition to inhibiting the vir regulon, SA induced the expression of the quormone degradation system encoded by the attKLM (Fig. 2). Recent studies have shown that GABA and succinic semialdehyde activate the expression of attKLM, resulting in quorum-sensing signal decay and reduced Agrobacterium virulence (16, 17). Thus, in addition to its role in orchestrating defense responses within the plant, SA provides the plant with additional defense strategies against Agrobacterium and perhaps other quorum-sensing bacteria in the rhizosphere.
Recent studies have shown that SA-treated P. aeruginosa accumulates less endogenous quormone than nontreated cells (29), although the molecular mechanisms by which SA affects quormone levels have yet to be defined. SA also affects the ability of Rhizobium to nodulate host plants (8, 9); whether this effect is mediated through quormone degradation remains obscure, although evidence exists that Rhizobium quorum sensing plays an essential role during symbiosis (30, 31).
In addition to SA, plants secrete other small molecules that inhibit VirA/G function. Among grasses, defensive benzoxazinones, such as the exuded product 2-hydroxy-4,7-dimethoxybenzoxazin-3-one (HDMBOA), also inhibit the VirA/G system (32, 33). Recent work suggests that the Agrobacterium vir genes are subject to feedback control (11). Upon expression of the integrated T-DNA genes, IAA inhibits the transcription of the vir regulon, presumably to save energy and metabolic resources. However, both microarray and gene fusion studies revealed that IAA does not activate expression of the attKLM operon (Z.-C.Y. and E.W.N., unpublished results). SA may serve to reinforce the feedback activity of IAA on vir gene expression. Thus, in an intriguing twist of evolution, SA produced by plants may have been hijacked by Agrobacterium to serve the pathogen by shutting off vir gene expression once these genes have served their purpose. Microarray analysis carried out at pH 5.5 demonstrated that unlike SA and IAA, GABA did not affect expression of the vir regulon (Z.-C.Y. and E.W.N., unpublished results).
Unexpectedly, the npr1-1 mutant was more resistant to Agrobacterium infection, even though npr1 mutants exhibit increased susceptibility to a wide range of pathogens, and conversely, overexpression of NPR1 in Arabidopsis improves resistance to several pathogens including Pseudomonas syringae (10). NPR1 activates the WRKY transcription factors involved in SA-dependent up-regulation of the defense-related PR genes (10). Our results indicate that NPR1 is not essential for Arabidopsis to mount a successful defense against Agrobacterium infection, suggesting that SA functions primarily by inhibiting induction of the vir regulon. This conclusion is further supported by our discovery that a constitutively active VirA conferred on C58 cells the ability to infect the highly resistant mutant plant, cpr5-2, which overaccumulates SA (Table 5). The enhanced resistance of the npr1-1 line is consistent with the observation that npr1 mutants overexpress the ICS1 gene required to synthesize and accumulate elevated levels of SA after infection relative to infected WT plants (10). Another potential explanation is that SA signaling also contributes in planta to resistance to Agrobacterium infection, but through an NPR1-independent pathway (10). It has been reported that Agrobacterium rapidly reduces the host systemic acquired resistance by down-regulating PR gene expression and decreasing the free SA level by 40% within 1 h after infection (34). It is conceivable that this effect is mediated through NPR1; an inactivating mutation in NPR1 would abrogate the pathogen's ability to disable the plant defenses, resulting in enhanced host resistance.
Taken together, our data suggest a model in which SA defends against Agrobacterium infection through direct effects on the transcription of the vir regulon and potentially also through an NPR1-independent (or partially NPR1-independent) signaling response that induces defense-related genes in the host. Such a combinatorial line of defense has also been implicated in Arabidopsis resistance to Staphylococcus aureus infection (26). The dramatic restoration of Agrobacterium virulence by a constitutively active VirA makes it highly unlikely that the level of SA produced by even the most resistant mutant is blocking tumorigenesis merely by affecting bacterial viability. However, at higher SA concentrations, inhibiting bacterial cell growth could be another strategy in the plant's defensive arsenal. Agrobacterium in turn appears to have evolved a mechanism to stymie the host defenses (34) and may even capitalize on the inhibitory effects of SA on vir gene expression to conserve its metabolic resources. This complex interplay between host and pathogen likely reflects an exquisite evolutionary balance, in which bacterial subversion of the host's intrinsic protection against disease counters the plant's multifaceted defense strategy mediated through SA.
Materials and Methods
Bacterial Strains, Growth Conditions, and Generating Transcriptional Gene Fusions.
A. tumefaciens strain C58, whose genome has been sequenced (12), was used to assay the expression of vir genes and the attKLM operon. The cells were grown in either MG/L or AB minimal medium (35) with arabinose as a carbon source at 28°C. A. tumefaciens strains A208, A348, and C58 were used to assay virulence on Arabidopsis thaliana roots. A208 is an Agrobacterium isolate that is more virulent on roots of A. thaliana than C58. A348 contains the same genome components as C58 except for the Ti plasmid, which originated from strain A6. The virB1 promoter was amplified and cloned into the pFUS1 vector to generate a virB1::gusA transcriptional gene fusion (36). Plasmid pWT160 (37) was used to measure virG expression. For generating gusA gene fusions to other vir and chv genes, a modified pJP2 gusA reporter vector was used (38). The primers used for amplifying all promoters are listed in SI Table 6. The reporter plasmids were introduced into Agrobacterium by electroporation.
Cell Culture and RNA Isolation.
Agrobacterium C58 cells were grown overnight in MG/L and then washed in induction medium (35) three times and subcultured in 25 ml of induction medium (with 100 μM acetosyringone) at an initial A600 of 0.15, with or without 6 μM SA. After 7 h, the A600 for the SA-treated culture was ≈0.65, whereas the A600 for the nontreated culture was ≈0.7. Four milliliters of the cultures were mixed with 8 ml of RNA Protect bacteria reagent (Qiagen, Valencia, CA) and processed as recommended by the manufacturer. Total bacterial RNA was isolated by using the Qiagen mini-RNA isolation kit according to the manufacturer's protocol. For the IAA and GABA microarray analyses, IAA was added at 40 μM; GABA was added at 1 mM.
Assaying Gene Fusions.
Cells harboring the gene fusion plasmids were grown overnight in MG/L supplemented with carbenicillin (100 μg/ml). Cells were washed with acidified AB minimal medium (pH 5.5) three times and inoculated into induction medium (pH 5.5) supplemented with 0.2% arabinose and 100 μM acetosyringone at an initial A600 of 0.1. Bacteria were incubated for 16–20 h and then assayed for β-galactosidase or β-glucuronidase activity as described previously (39). Unless otherwise indicated, the data presented represent the average of three independent determinations.
Microarray.
Unique 60-mer oligonucleotides representing each of the 5,419 predicted ORFs were selected by using the Featurama program designed at the Institute for Systems Biology in Seattle. Each oligonucleotide was synthesized in situ on 2.5- × 7.5-cm glass slides by Agilent Technologies (Santa Clara, CA). Each microarray experiment represents four biological replicates. cDNA was generated from 30 μg of total RNA by using random hexamer primers and SuperScript II (Invitrogen, Carlsbad, CA). Aminoallyl-modified dUTP was incorporated into the cDNA at a ratio of 4:1 aa-dUTP:dTTP, and cDNA was labeled with Cy3 or Cy5 monoreactive dyes (Amersham, Piscataway, NJ). Arrays were hybridized and washed according to the manufacturer's instructions (Agilent publication G4140-90030). Data acquisition was performed by using an Agilent G2565AA microarray scanner and extracted by using Agilent feature extraction software.
Microarray Data Analysis.
Initial data handling and visualization were done with the Matlab software (MathWorks, Natick, MA). All remaining data analysis was done in the R statistical computing environment by using the samr package in Bioconductor (40). Normalized data were analyzed to identify candidate differentially expressed genes. A t test statistic and a reference distribution were carried out.
Root Infection Assays.
The Arabidopsis WT Col-0 line and its derivatives NahG (24), 35S-LOX2 (23), npr1-1 (25), and cpr5-2 (22) were used in the root infection assays, which were performed as described previously (41).
Supplementary Material
Acknowledgments
We thank Drs. Matt Parsek and Amy Schaefer for helpful discussions and Drs. David Lynn and Justin Maresh (Emory University, Atlanta, GA) for virA plasmids and for suggestions on plotting and interpreting Fig. 1A. Dr. Xinnian Dong (Duke University, Durham, NC) kindly provided the transgenic Arabidopsis line nahG. We thank the Arabidopsis Stock Center (Columbus, OH) for the other Arabidopsis lines. We also thank Cassie Sather for helping with microarrays. We are grateful to Drs. Stan Gelvin and Janis Bravo for advice on the root infection assays. This work was supported by National Institutes of Health (NIH) Grant GM032618 and a U.S.–Israel Binational Agricultural Research and Development Research (BARD) Grant IS-3622-04R (to E.W.N.), by National Science Foundation Grant MCB-0416471 (to L.M.B.), and by NIH Grant GM47369 and the Carolyn Hoff Lynch Term Professor Fund (to A.N.B.).
Abbreviations
- IAA
indoleacetic acid
- PR
pathogenesis-related
- SA
salicylic acid
- T-DNA
portion of the Ti (tumor-inducing) plasmid that is transferred to plant cells.
Footnotes
The authors declare no conflict of interest.
This article contains supporting information online at www.pnas.org/cgi/content/full/0704866104/DC1.
References
- 1.McCullen CA, Binns AN. Annu Rev Cell Dev Biol. 2006;22:101–127. doi: 10.1146/annurev.cellbio.22.011105.102022. [DOI] [PubMed] [Google Scholar]
- 2.Chang CH, Winans SC. J Bacteriol. 1996;178:4717–4720. doi: 10.1128/jb.178.15.4717-4720.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Gelvin SB. Annu Rev Plant Physiol Mol Biol. 2000;51:223–256. doi: 10.1146/annurev.arplant.51.1.223. [DOI] [PubMed] [Google Scholar]
- 4.Cho H, Winans SC. Proc Natl Acad Sci USA. 2005;102:14843–14848. doi: 10.1073/pnas.0503458102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chang CH, Winans SC. J Bacteriol. 1992;174:7033–7039. doi: 10.1128/jb.174.21.7033-7039.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Gao R, Lynn DG. J Bacteriol. 2005;187:2182–2189. doi: 10.1128/JB.187.6.2182-2189.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Prithiviraj B, Bais HP, Weir T, Suresh B, Najarro EH, Dayakar BV, Schweizer HP, Vivanco JM. Infect Immun. 2005;73:5319–5328. doi: 10.1128/IAI.73.9.5319-5328.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Stacey G, McAlvin CB, Kim SY, Olivares J, Soto MJ. Plant Physiol. 2006;141:1473–1481. doi: 10.1104/pp.106.080986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Martínez-Abarca F. Mol Plant Microbe Interact. 1998;11:153–155. doi: 10.1094/MPMI.1998.11.8.839. [DOI] [PubMed] [Google Scholar]
- 10.Durrant WE, Dong X. Annu Rev Phytopathol. 2004;42:185–209. doi: 10.1146/annurev.phyto.42.040803.140421. [DOI] [PubMed] [Google Scholar]
- 11.Liu P, Nester EW. Proc Natl Acad Sci USA. 2006;103:4658–4662. doi: 10.1073/pnas.0600366103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wood DW, Setubal JC, Kaul R, Monks DE, Kitajima JP, Okura VK, Zhou Y, Chen L, Wood GE, Almeida NF, Jr, et al. Science. 2001;294:2317–2323. doi: 10.1126/science.1066804. [DOI] [PubMed] [Google Scholar]
- 13.Li PL, Farrand SK. J Bacteriol. 2000;182:179–188. doi: 10.1128/jb.182.1.179-188.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zhang HB, Wang LH, Zhang LH. Proc Natl Acad Sci USA. 2002;99:4638–4643. doi: 10.1073/pnas.022056699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Fuqua C, Parsek MR, Greenberg EP. Annu Rev Genet. 2001;35:439–468. doi: 10.1146/annurev.genet.35.102401.090913. [DOI] [PubMed] [Google Scholar]
- 16.Chevrot R, Rosen R, Haudecoeur E, Cirou A, Shelp BJ, Ron E, Faure D. Proc Natl Acad Sci USA. 2006;103:7460–7464. doi: 10.1073/pnas.0600313103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wang C, Zhang HB, Wang LH, Zhang LH. Mol Microbiol. 2006;62:45–56. doi: 10.1111/j.1365-2958.2006.05351.x. [DOI] [PubMed] [Google Scholar]
- 18.McLean BG, Greene EA, Zambryski PC. J Biol Chem. 1994;269:2645–2651. [PubMed] [Google Scholar]
- 19.Jin S, Song Y, Pan SQ, Nester EW. Mol Microbiol. 1993;7:555–562. doi: 10.1111/j.1365-2958.1993.tb01146.x. [DOI] [PubMed] [Google Scholar]
- 20.Pazour GJ, Ta CN, Das A. J Bacteriol. 1992;174:4169–4174. doi: 10.1128/jb.174.12.4169-4174.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wang Y, Mukhopadhyay A, Howitz V, Binns AN, Lynn DG. Gene. 2000;242:105–114. doi: 10.1016/s0378-1119(99)00541-7. [DOI] [PubMed] [Google Scholar]
- 22.Bowling SA, Guo A, Cao H, Gordon AS, Klessig DF, Dong X. Plant Cell. 1994;6:1845–1857. doi: 10.1105/tpc.6.12.1845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bell E, Creelman RA, Mullet JE. Proc Natl Acad Sci USA. 1995;92:8675–8679. doi: 10.1073/pnas.92.19.8675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lawton K, Weymann K, Friedrich L, Vernooij B, Uknes S, Ryals J. Mol Plant Microbe Interact. 1995;8:863–870. doi: 10.1094/mpmi-8-0863. [DOI] [PubMed] [Google Scholar]
- 25.Cao H, Glazebrook J, Clarke J, Volko S, Dong X. Cell. 1997;88:57–63. doi: 10.1016/s0092-8674(00)81858-9. [DOI] [PubMed] [Google Scholar]
- 26.Prithiviraj HP, Bais AK, Jha AK, Vivanco JM. Plant J. 2005;42:417–432. doi: 10.1111/j.1365-313X.2005.02385.x. [DOI] [PubMed] [Google Scholar]
- 27.Huang WE, Huang L, Preston GM, Naylor M, Carr JP, Li Y, Singer AC, Whiteley AS, Wang H. Plant J. 2006;46:1073–1083. doi: 10.1111/j.1365-313X.2006.02758.x. [DOI] [PubMed] [Google Scholar]
- 28.Mukhopadhyay A, Gao R, Lynn DG. ChemBioChem. 2004;5:1535–1542. doi: 10.1002/cbic.200300828. [DOI] [PubMed] [Google Scholar]
- 29.Bandara M, Zhu H, Sankaridurg PR, Willcox M. Invest Ophthalmol Visual Sci. 2006;47:4453–4460. doi: 10.1167/iovs.06-0288. [DOI] [PubMed] [Google Scholar]
- 30.Zheng H, Zhong Z, Lai X, Chen WX, Li S, Zhu J. J Bacteriol. 2006;188:1943–1949. doi: 10.1128/JB.188.5.1943-1949.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wisniewski-Dye F, Downie JA. Antonie Van Leeuwenhoek. 2002;81:397–407. doi: 10.1023/a:1020501104051. [DOI] [PubMed] [Google Scholar]
- 32.Maresh J, Zhang J, Lynn DG. ACS Chem Biol. 2006;25:165–175. doi: 10.1021/cb600051w. [DOI] [PubMed] [Google Scholar]
- 33.Zhang J, Boone L, Kocz R, Zhang C, Binns AN, Lynn DG. Chem Biol. 2000;7:611–621. doi: 10.1016/s1074-5521(00)00007-7. [DOI] [PubMed] [Google Scholar]
- 34.Gaspar YM, Nam J, Schultz CJ, Lee L, Gilson PR, Gelvin SB, Bacic A. Plant Physiol. 2004;135:2162–2171. doi: 10.1104/pp.104.045542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Cangelosi GA, Best EA, Martinetti G, Nester EW. Methods Enzymol. 1991;204:384–397. doi: 10.1016/0076-6879(91)04020-o. [DOI] [PubMed] [Google Scholar]
- 36.Reeve WG, Tiwari RP, Worsley PS, Dilworth MJ, Glenn AR, Howieson JG. Microbiology. 1999;145:1307–1316. doi: 10.1099/13500872-145-6-1307. [DOI] [PubMed] [Google Scholar]
- 37.Peng WT, Lee YW, Nester EW. J Bacteriol. 1998;180:5632–5638. doi: 10.1128/jb.180.21.5632-5638.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Karunakaran R, Mauchline TH, Hosie AHF, Poole PS. Microbiology. 2005;151:3249–3256. doi: 10.1099/mic.0.28311-0. [DOI] [PubMed] [Google Scholar]
- 39.Yuan ZC, Zaheer R, Finan TM. J Bacteriol. 2006;188:1089–1102. doi: 10.1128/JB.188.3.1089-1102.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Gentleman RC, Carey VJ, Bates DM, Bolstad B, Dettling M, Dudoit S, Ellis B, Gautier L, Ge Y, Gentry J. Genome Biol. 2004;5:R80.1–R80.16. doi: 10.1186/gb-2004-5-10-r80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Gelvin SB. In: Agrobacterium Protocols. 2nd Ed. Wang K, editor. Totowa, NJ: Humana; 2006. pp. 105–114. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.