Abstract
Trehalose is potentially a useful cryo- or anhydroprotectant molecule for cells and biomolecules such as proteins and nucleotides. A major obstacle to application is that cellular membranes are impermeable to trehalose. In this study, we isolated and characterized the functions of a facilitated trehalose transporter [trehalose transporter 1 (TRET1)] from an anhydrobiotic insect, Polypedilum vanderplanki. Tret1 cDNA encodes a 504-aa protein with 12 predicted transmembrane structures. Tret1 expression was induced by either desiccation or salinity stress. Expression was predominant in the fat body and occurred concomitantly with the accumulation of trehalose, indicating that TRET1 is involved in transporting trehalose synthesized in the fat body into the hemolymph. Functional expression of TRET1 in Xenopus oocytes showed that transport activity was stereochemically specific for trehalose and independent of extracellular pH (between 4.0 and 9.0) and electrochemical membrane potential. These results indicate that TRET1 is a trehalose-specific facilitated transporter and that the direction of transport is reversible depending on the concentration gradient of trehalose. The extraordinarily high values for apparent Km (≥100 mM) and Vmax (≥500 pmol/min per oocyte) for trehalose both indicate that TRET1 is a high-capacity transporter of trehalose. Furthermore, TRET1 was found to function in mammalian cells, suggesting that it confers trehalose permeability on cells, including those of vertebrates as well as insects. These characteristic features imply that TRET1 in combination with trehalose has high potential for basic and practical applications in vivo.
Keywords: anhydrobiosis, insect, trehalose transport, Polypedilum vanderplanki dessication-inducible gene
Sugars act primarily as energy and carbon sources for living organisms. Trehalose [α-d-glucopyranosyl-(1,1)-α-d-glucopyranoside, Glc(α1–1α)Glc], however, has multiple functions that distinguish it from other common disaccharides. These functions include the following: (i) protection as a “chemical chaperone” or an antioxidant against stresses such as desiccation, heat, low temperature, and high and low oxygen (1–5); (ii) prevention of osteoporosis by disturbing osteoclast differentiation (6); (iii) alleviation of polyglutamine and polyalanine diseases such as Huntington's disease (7) and oculopharyngeal muscular dystrophy (8), respectively, probably by preventing abnormal protein aggregation; (iv) induction of the mammalian target of rapamycin-independent autophagy, causing the clearance of mutant proteins associated with Huntington's disease and Parkinson's disease (9); and (v) control of growth and inflorescence blanching in plants by acting as a signal molecule (10–12).
Human platelets have been successfully freeze-dried after incorporation of trehalose by pinocytosis (13, 14), showing that trehalose plays an important role as a cryo- and anhydroprotectant. Pinocytosis is not applicable to red and white blood cells, however, because the cells are impermeable to trehalose. Spontaneous uptake by fluid-phase endocytosis is largely dependent on cell characteristics so that uptake of a large amount of trehalose is not expected.
Certain transporters promote permeation of trehalose across cellular membranes. As examples, MalEFGK2 (TC: 3.A.1.1) for archaea and bacteria (15) and MAL11/AGT1 (TC: 2.A.1.1.11) for yeast (16–18) are active sugar transporters requiring either ATP hydrolysis or a favorable membrane potential for transport (15, 16, 18). Substrate selectivity of these transporters is relatively broad and includes α-glucosides such as trehalose, sucrose [β-d-fructofuranosyl-α-d-glucopyranoside, Fruf(β2–1α)Glc], and maltose [4-O-α-d-glucopyranosyl-1-α-d-glucose, Glc(α1–4)Glc] (15, 17). To our knowledge, these transporters have not been used for practical applications because it is difficult to coordinately express the four different genes needed for MalEFGK2, and the optimum pH for MAL11/AGT1 is acidic rather than neutral. For both transporters, the direction of trehalose transport is only inward. However, we reasoned that facilitated sugar transporters might solve these problems. The facilitated glucose transporter (GLUT)/SLC2A family of facilitated glucose transporters allows glucose to cross the cell membrane (19, 20). This transport is bidirectional between the external environment and cytosol, is governed by a single gene product, and is independent of pH and membrane potential (19, 20). We are not aware of any previous reports of a facilitated trehalose-specific transporter.
Trehalose is the major hemolymph sugar in most insects. It is predominantly synthesized in the fat body and released into the hemolymph (21). Although 44 homologues containing a sugar transporter motif exist in Drosophila melanogaster, according to the protein family database (Pfam; www.sangar.ac.uk/Software/Pfam/), thus far no trehalose transporter has been identified because it is not possible to estimate the substrate specificity from the primary structure alone.
Larvae of the sleeping chironomid, Polypedilum vanderplanki, a temporary rock pool dweller, undergo complete dehydration during dry periods followed by rehydration and resumption of activity when moisture is available. This biological state of tolerance to extreme desiccation is referred to as “cryptobiosis” or “anhydrobiosis” (22). During desiccation or salt stress, larvae accumulate ≤20% of their dry mass as trehalose (23–25). These observations indicate strongly that trehalose transporter genes must be highly expressed in the fat body and that it should be feasible to isolate trehalose transporter genes from P. vanderplanki larvae.
In this article, we report the cloning of a gene (Tret1) from P. vanderplanki that encodes a high-capacity facilitated trehalose transporter [trehalose transporter 1 (TRET1)], and we describe its biochemical properties. Because TRET1 is highly specific for trehalose, it provides a simple and specific way to introduce trehalose into a variety of cells.
Results
Molecular Cloning of the Tret1 Gene from P. vanderplanki.
Candidates for trehalose transporter genes were identified in our original P. vanderplanki EST (Pv-EST) database by using total RNA from larvae desiccated for 0, 12, or 36 h (26). We then identified a subset of six EST clones that form a single cluster annotated as a sugar transporter [supporting information (SI) Fig. 8A]. Based on these data and followed by 5′- and 3′-RACE, we obtained full-length cDNA (≈2.3 kb) designated as Tret1 of which a single ORF encodes a 55-kDa protein of 504 aa (SI Fig. 8A). TRET1 has a domain for sugar (and other) transport (Pfam accession no. PF00083) located at amino acid residues 46 and 484 (SI Fig. 8B), for which the E value was 2.8e-97. The family of sugar (and other) transporters contains the GLUT/SLC2A family and belongs to the major facilitator superfamily (MFS). From the prediction of secondary structure of membrane proteins using SOSUI analysis, TRET1 is thought to form a 12-transmembrane structure (SI Fig. 8C), which is typical for the MFS. An additional site for N-linked glycosylation is located in the first loop at position 73 of TRET1 (SI Fig. 8C), suggesting that the loop is extracellular. TRET1, however, showed low homology with the GLUT/SLC2A family; the highest score was 35.6% identity with the GLUT8/SLC2A8 protein, indicating that TRET1 is a new member of MFS.
Expression of the Tret1 Gene in P. vanderplanki.
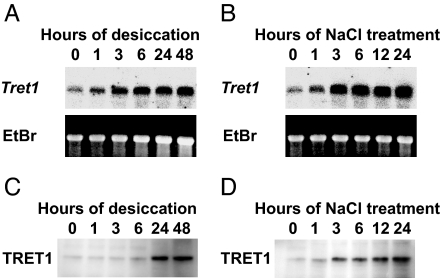
Northern and Western blot analyses showed that both accumulation of mRNA and protein for TRET1 were increased by desiccation and salinity stresses (Fig. 1). This expression pattern is in accordance with the known pattern of trehalose accumulation in these larvae (24, 25). Western blot analysis indicates that the molecular mass of TRET1 is ≈58 kDa (Fig. 1 C and D). The difference from the value deduced from cDNA (55 kDa) is probably due to N-glycosylation of TRET1 in vivo. We performed in situ hybridization to determine which tissues expressed Tret1 in larvae of P. vanderplanki en route to anhydrobiosis. As shown in Fig. 2, Tret1 was mainly expressed in the fat body and not in other tissues including the midgut, muscle, and integuments after 24 h of dehydration.
Fig. 1.
Expression of the Tret1 gene and protein are induced by both desiccation and salinity stress in larvae of P. vanderplanki. (A and B) Total RNA was isolated from the larvae at various times during either desiccation (A) or 1% (wt/vol) NaCl treatment (B). Northern blot analyses were performed by using the full-length TRET1 cDNA as a probe, which revealed a single 2.3-kb transcript. EtBr staining shows 28S rRNA after electrophoresis. (C and D) Protein extracts from the larvae treated alike with desiccation (C) or 1% (wt/vol) NaCl (D) were subjected to Western blot analysis using anti-PvTRET1–1 antibody, which revealed a 58-kDa protein.
Fig. 2.
The Tret1 gene is expressed mainly in the fat body of larvae of P. vanderplanki. In situ hybridization was carried out by using either antisense (A, C, E, and G) or sense (B, D, F, and H) riboprobes for Tret1 mRNA to hybridize to cross-sections of a dehydrating larvae in the thorax (A, B, E, and F) and posterior abdomen (C, D, G, and H). (Magnifications: A–D, ×100; E–H, ×400.) Rectangles in A–D indicate trimming areas for E–H. FB, fat body; Mg, midgut; Mu, muscle; SG, salivary gland.
TRET1 Is a Trehalose-Specific Transporter.
When expressed in Xenopus oocytes, the TRET1::AcGFP1 fusion protein was clearly localized to the membrane (Fig. 3A Center and SI Fig. 9), and the translation product of the Tret1 gene actually transported trehalose (Fig. 3A Bottom). Uptake of trehalose into oocytes expressing TRET1 increased linearly for at least 6 h when incubated in 105 mM trehalose (SI Fig. 10). The recognition of substrate was highly specific. Maltose, sucrose, and lactose [4-O-β-d-galactopyranosyl-d-glucose, Gal(β1–4)Glc] were not transported, and methyl-α-glucoside and 2-deoxyglucose were transported at a much lower rate (Fig. 3B). For comparison, the Glut1 (TC: 2.A.1.1.28) gene product transported only 2-deoxyglucose (Fig. 3C). Interestingly, TRET1 did not transport even anomers of trehalose such as neotrehalose [α-d-glucopyranosyl-(1,1)-β-d-glucopyranoside, Glc(α1–1β)Glc] and isotrehalose [β-d-glucopyranosyl-(1,1)-β-d-glucopyranoside, Glc(β1–1β)Glc] (Fig. 3D). TRET1 transported only the homodimer and monomer of α-glucopyranoside, but not β-glucopyranoside, fructofuranoside, and galactoside, indicating that TRET1 precisely recognizes the stereochemical structure of disaccharides like trehalose.
Fig. 3.
The Tret1 gene encodes a trehalose-specific transporter. (A) Cellular localization of TRET1 in Xenopus oocytes. Oocytes injected with either the TRET1::AcGFP1 fusion or AcGFP1 cRNA or water as sham were observed under the fluorescence microscope with normal images (Top), and images using haze reduction software to eliminate blurring (Middle). Using comparable conditions, injected oocytes were assayed for trehalose uptake in 105 mM trehalose after 3 h at 15°C (Bottom). (B and C) Substrate selectivities of TRET1 (B) and human GLUT1 (C) for disaccharides and glucose derivatives are also shown. Xenopus oocytes expressing either TRET1 or GLUT1 were incubated in 105 mM of various sugars for 3 h. Tre, trehalose; Mal, maltose; Suc, sucrose; Lac, lactose; MAG, methyl-α-glucopyranoside; 2-DOG, 2-deoxy-glucose. Each value is the mean ± SEM (n = 3). (D) To examine substrate selectivity, Xenopus oocytes expressing TRET1 were incubated in 25 mM trehalose anomers for 3 h. Each value is the mean ± SEM (n = 3).
TRET1 Is a Facilitated Transporter.
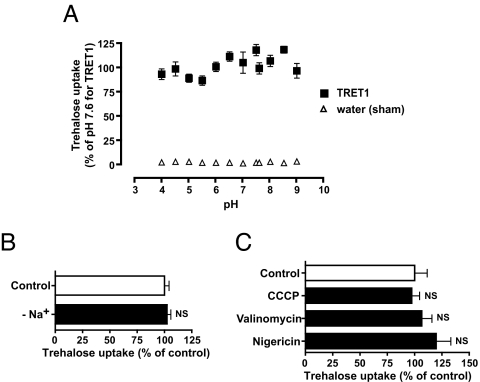
MFS contains two subclasses: facilitated transporters and secondary active transporters. Activities of secondary active transporters depend on the electrochemical membrane potential resulting from the distribution of ions such as H+ or Na+ for bacteria, yeast, and plants or animals. For example, proton-dependent MAL11/AGT1 can act only under acidic extracellular conditions (16). In contrast, facilitated transporters function independently of the electrochemical membrane potential. TRET1 acted over a wide extracellular pH range, between 4.0 and 9.0 (Fig. 4A) and in a Na+-free medium in which choline chloride replaced NaCl (Fig. 4B). Reduction of the electrochemical membrane potential and ATP synthesis caused by ionophores and an uncoupler did not significantly affect the transport activity of TRET1 (Fig. 4C). These properties indicate that TRET1 is a facilitated transporter.
Fig. 4.
TRET1 is a facilitated transporter. (A) The pH dependency of TRET1. Xenopus oocytes expressing TRET1 were incubated in 105 mM trehalose at various pH conditions for 3 h. Each value is the mean ± SEM (n = 3). (B) The Na+ dependency of TRET1. Xenopus oocytes expressing TRET1 were incubated in 105 mM trehalose in either normal buffer (control) or Na+-free buffer (−Na+) for 3 h. Each value is the mean ± SEM (n = 3). (C) The effects of ionophores and an uncoupler on TRET1 activity. Xenopus oocytes expressing TRET1 were preincubated in either control buffer (0.1% acetone) or buffer containing 10 μM nigericin (Na+, K+ ionophore), 10 μM valinomycin (K+ ionophore), or 10 μM carbonylcyanide m-chlorophenylhydrazone (CCCP) (H+ ionophore, uncoupler) for 15 min, and then incubated in 105 mM trehalose with the corresponding ionophores or an uncoupler for 3 h. Each value is the mean ± SEM (n = 3).
TRET1 Can Transport Trehalose Bidirectionally.
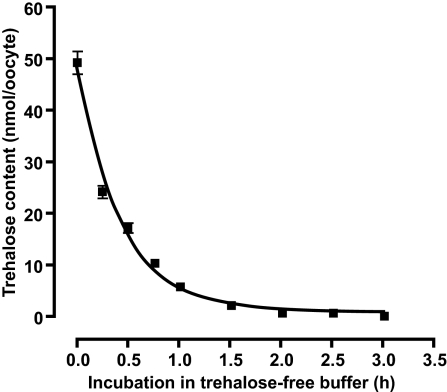
In general, facilitated transporters enable their substrates to flow across membranes down concentration gradients. By using the Xenopus oocyte expression system, we found that the transport of trehalose was shifted from an inward to outward direction when its concentration gradient was reversed between the cytosol and external media (Fig. 5). This characteristic feature enables cells to easily discharge excess trehalose by reducing its concentration in the extracellular environment.
Fig. 5.
Bidirectional trehalose transport activity of TRET1. Xenopus oocytes expressing TRET1 were first incubated in 105 mM trehalose for 3 h and then transferred into trehalose-free buffer. Trehalose content was examined over time. Each value is the mean ± SEM (n = 3).
TRET1 Is a High-Capacity Transporter.
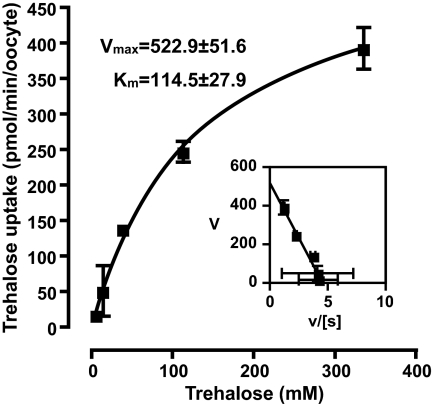
Kinetic analyses showed that apparent Km and Vmax values of TRET1 activity for trehalose were 114.5 ± 27.9 mM and 522.9 ± 51.6 pmol/min per oocyte, respectively (Fig. 6). This Km is exceptionally high and shows a low affinity for substrate compared with typical sugar transporters such as GLUT1 (≈3 mM glucose), GLUT2 (≈17 mM glucose), and GLUT4 (≈6.6 mM glucose) (27). The Vmax for TRET1 was considerably higher, indicating that TRET1 is a high-capacity trehalose transporter.
Fig. 6.
Kinetics analysis of zero-trans activity of TRET1 for trehalose. Xenopus oocytes expressing TRET1 were incubated in various concentrations of trehalose for 15 min. Uptake data were fitted to the Michaelis–Menten and Eadie–Hofstee (Inset) equations. Apparent Km and Vmax were calculated by nonlinear approximation. Each value is the mean ± SEM (n = 3).
TRET1 Acts Independently of Cell Type.
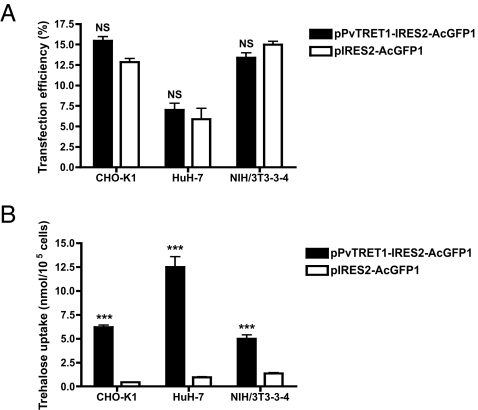
Next, we introduced the Tret1 gene into mouse fibroblasts (NIH/3T3), Chinese hamster ovary cells (CHO-K1), and human hepatoma cells (HuH-7). In all of these Tret1-transfected lines, trehalose uptake was significantly increased 4- to 14-fold higher than those transfected with vector alone (Fig. 7). Taken with the results on expression in Xenopus oocytes, these data show that TRET1 confers trehalose permeability in cells from other vertebrates, including mammals. Recently, we succeeded in generating a TRET1-stable expression cell line in which proliferation and cell shape did not change even after >20 passages of the cultured cells, indicating that expression of TRET1 does not impair cell viability.
Fig. 7.
TRET1 allows mammalian cells to increase trehalose uptake. Either the TRET1 expression vector (pPvTRET1-IRES2-AcGFP1) or vector only (pIRES2-AcGFP1) were transfected into CHO-K1, HuH-7, or NIH/3T3-3-4 cells. (A) Transfection efficiency was estimated from the ratio of AcGFP1-expressing cells analyzed by a flow cytometer. Each value is the mean ± SEM (n = 3). (B) The transfected cells were incubated in medium containing 100 mM trehalose for 3 h to determine trehalose uptake. Each value is the mean ± SEM (n = 3).
Discussion
Until now, there has been no evidence to our knowledge that the MFS from muticellular organisms can transport disaccharides. This study demonstrates a facilitated transporter specific to trehalose.
Thus far, several trials introducing trehalose into cells have been made and brought a certain degree of success. For example, introduction of bacterial trehalose biosynthetic enzyme genes (otsA and otsB) into human fibroblasts increases intracellular trehalose concentration and results in enhanced desiccation tolerance (28). In this system, however, it is hard to eliminate trehalose even when it is no longer necessary. This retention could cause ill effects because trehalose can prevent refolding of denatured proteins (29). In another approach, engineered switchable pores or extracellular nucleotide-gated channels (engineered α-hemolysin or P2X7 purinergic receptor pore) were created in cellular membranes to allow trehalose uptake (30, 31). That approach enabled trehalose to move from the extracellular fluid into cells; however, undesired influx and efflux of other molecules probably occur simultaneously.
TRET1 has several advantages for these applied studies. One significant advantage is that high intracellular trehalose concentrations can be maintained because of this high-capacity transporter. The intracellular trehalose concentration can also be easily controlled by changing the extracellular trehalose concentration without an undesirable influx and/or efflux of other molecules at neutral pH. In addition, because TRET1 is a single gene product, transgenesis into cells should not be difficult.
In larvae of P. vanderplanki, desiccation stress simultaneously induces trehalose synthesis (23, 24) and gene expression of Tret1 in the fat body. This expression profile suggests that the Tret1 gene is involved in transporting trehalose synthesized in the fat body into the hemolymph during desiccation. Expression of Tret1 also was induced by hypersalinity, supporting our hypothesis that salinity stress can mimic desiccation in its effects on larval gene transcription (26). The extraordinarily high values for both Km and Vmax for trehalose are physiologically reasonable because TRET1 can retain a high capacity for transport activity even when trehalose is highly concentrated in the dehydrating larval body during the final stage of entry into anhydrobiosis (23–25).
Based on the physiological role of TRET1, this research might eventually generate a method to store cells in the dry state by transporting trehalose into them. We should point out that certain rotifers (micrometazoans) have recently been shown to have excellent desiccation tolerance, but lack detectable amounts of trehalose or any other sugar (32). This interesting exception demonstrates that there is much to learn about the cellular and molecular basis of anhydrobiosis in organisms adapted to do so. However, this exception does not diminish the rationale of using trehalose to confer desiccation tolerance on nonadapted cells. Indeed, the introduction of trehalose into mammalian cells significantly enhanced their tolerance to dehydration (28, 30, 31, 33), in some cases coupled with the transgenic expression of p26, a small heat shock protein (33). Therefore, expression of TRET1 alone, or coupled with other molecules related to desiccation resistance, can render cells desiccation tolerant.
TRET1 might be useful in a variety of areas, including medical biology, cryobiology, anhydrobiology, and food preservation. For example, an increase in trehalose content in brain or muscle by exogenously expressing TRET1 is expected to alleviate Huntington's disease or oculopharyngeal muscular dystrophy, respectively. Likewise, cells and organs loaded with a high concentration of trehalose may be cryopreserved more safely and might even be lyophilized for preservation such as the case for human platelets (13, 14). In addition to these applications, there are benefits for glycotechnology because strict substrate specificity of TRET1 provides major advantages in the screening of newly synthesized trehalose analogs. By using TRET1, we can investigate biological functions of trehalose as a chemical chaperone, a radical scavenger, and a signal molecule in vivo.
Much evidence indicates that trehalose is an especially interesting sugar. We believe that TRET1 will open many possibilities in which trehalose can be used to achieve basic and applied goals.
Materials and Methods
Animal and Cell Culture.
P. vanderplanki was reared on a milk agar diet under controlled light conditions (13-h light/11-h dark) at 27°C (23, 24). To induce expression of anhydrobiosis-related genes, final instar larvae (each ≈1 mg of wet body mass) were desiccated by a procedure described in a previous report (25).
CHO-K1 (RCB0285), HuH-7 (RCB1366), or NIH/3T3-3-4 (RCB1862) cells were provided by the RIKEN Cell Bank (Tsukuba, Japan). CHO-K1 cells were cultured in a Ham's F-12 medium (Sigma–Aldrich, St. Louis, MO) containing 10% FBS (Tissue Culture Biologicals, Tulare, CA), 100 units/ml penicillin, and 100 μg/ml streptomycin (Sigma–Aldrich) at 37°C, 5% CO2, and 95% relative humidity. HuH-7 and NIH/3T3-3-4 cells were cultured in DMEM (Sigma–Aldrich) supplemented with 10% FBS, 100 units/ml penicillin, and 100 μg/ml streptomycin (Sigma–Aldrich) at 37°C, 5% CO2, and 95% relative humidity.
Tret1 cDNA Cloning from P. vanderplanki.
In the Pv-EST database (26), six EST clones annotated as sugar transporter-like genes were assembled into one cluster designated as Tret1 (SI Fig. 8). The full-length Tret1 cDNA was obtained by 5′- and 3′-RACE using a SMART RACE cDNA Amplification kit (Clontech, Mountain View, CA) with specific primers as follows: PvTRET1–5RACE-R1, 5′-TGCATCTGCCTACACCTCACCTGCA-3′; PvTRET1–5RACE-R2, 5′-GGCAGATG C A A ATCCAACTACCATTGAACCC-3′; PvTRET1–3RACE-F1, 5′-TGTACCAGAGACACAGGGCAAATCATTAG A A GAGA-3′; and PvTRET1–3RACE-F2, 5′-CGAGTAAGACGCATGTCATCAGTCGCGA-3′.
The full-length Tret1 cDNA was subcloned into pCR4Blunt-TOPO (Invitrogen, Carlsbad, CA) to produce pCR-TRET1. DNA sequences were analyzed with GENETYX-MAC (Genetyx, Tokyo, Japan). Motif analysis was performed with Pfam (www.sanger.ac.uk/Software/Pfam/), and secondary structures of membrane proteins were predicted by using the SOSUI system (bp.nuap.nagoya-u.ac.jp/sosui/).
Northern Blot Analysis.
Total RNA from the larvae either undergoing dehydration or submerged in 1% (wt/vol) NaCl was isolated with Trizol (Invitrogen). Fifteen micrograms of total RNA was run on a 1% agarose/20 mM guanidine isothiocyanate gel and transferred onto Hybond N-Plus membrane (GE Healthcare Bio-Sciences, Piscataway, NJ). Hybridization was carried out at 42°C in 5× standard saline phosphate/EDTA [0.18 M NaCl, 10 mM phosphate (pH 7.4), and 1 mM EDTA] containing 0.5% SDS and 50% formamide. For hybridization probes, full-length Tret1 cDNA fragments were synthesized from pCR-TRET1 by PCR and labeled with [α-32P]dATP by using the Strip-Ez labeling kit (Ambion, Austin, TX). The membranes were analyzed by BAS 2500 (Fuji Film, Tokyo, Japan).
Western Blot Analysis.
Proteins were extracted from larvae as for Northern blot analysis with a 20-fold volume of tissue-protein extraction reagent (T-PER; Pierce, Rockford, IL) containing a protease inhibitor mixture (Complete; Roche Diagnostics, Basel, Switzerland). A 7.5-μg protein sample was subjected to SDS/PAGE by using 5–20% gradient gels and subsequently transferred onto Hybond-P membranes (GE Healthcare Bio-Sciences). Membranes were treated with anti-PvTRET1–1 polyclonal antibody and then goat anti-rabbit IgG (H+L) conjugated with horseradish peroxidase (American Qualex, La Mirada, CA). Anti-PvTRET1–1 antibody was raised in a rabbit against the synthesized peptide corresponding to the TRET1 sequence at positions 242–254 (LRGKKADVEPELK) and purified by affinity chromatography using the peptide (Sigma–Aldrich). Immunoreacted proteins were detected with Immobilon Western Chemiluminescent HRP Substrate (Millipore, Billerica, MA) and analyzed by LAS-3000 (Fuji Film).
In Situ Hybridization.
In situ hybridization was performed under contract with Genostaff (Tokyo, Japan). A larva of P. vanderplanki dehydrated for 24 h was fixed with tissue fixative (Genostaff), embedded in paraffin by their proprietary procedures, and sectioned at 6 μm. The tissue sections were dewaxed with xylene and rehydrated through an ethanol series and PBS, fixed in 4% paraformaldehyde in PBS for 15 min, and then treated with 15 μg/ml proteinase K in PBS for 30 min at 37°C. After washing with PBS, the sections were refixed with 4% paraformaldehyde in PBS, placed in 0.2 M HCl for 10 min, acetylated by incubation in 0.1 M triethanolamine·HCl (pH 8.0)/0.25% acetic anhydride for 10 min, and dehydrated through a series of ethanol solutions. Probe for a 525-bp cDNA fragment was designated from positions 300 to 824 of Tret1 cDNA and labeled with digoxygenin RNA labeling kit (Roche Diagnostics). Hybridization was performed with the probe at a concentration of 100 ng/ml in the Probe Diluent (Genostaff) at 60°C for 16 h. After hybridization, the sections were washed in 5× HybriWash (Genostaff), equal to 5× SSC, at 60°C for 20 min, and then in 50% formamide/2× HybriWash at 60°C for 20 min, followed by RNase treatment in 50 μg/ml RNaseA in 10 mM Tris·HCl (pH 8.0)/1 M NaCl/1 mM EDTA. Then the sections were washed twice with 2× HybriWash at 60°C for 20 min, twice with 0.2× HybriWash at 60°C for 20 min, and once with TBS-T (0.1% Tween 20 in TBS). After treatment with 0.5% blocking reagent (Roche Diagnostics) in TBS-T for 30 min, the sections were incubated with antidigoxygenin alkaline phosphatase conjugate (Roche Diagnostics) diluted 1:1,000 with TBS-T for 2 h. The sections were washed twice with TBS-T and then incubated in 100 mM NaCl/50 mM MgCl2/0.1% Tween 20/100 mM Tris·HCl (pH 9.5). Coloring reactions were performed with BM purple alkaline phosphatase substrate (Roche Diagnostics) overnight and then washed with PBS. The sections were counterstained with Kernechtrot stain solution (Muto Pure Chemicals, Tokyo, Japan) and mounted with Malinol (Muto Pure Chemicals).
Tret1 Capped RNA (cRNA) Synthesis.
The cRNA expression vector pXbG-PvTRET1 was constructed as follows: ORF of Tret1 cDNA was amplified with PCR using specific primers containing BglII site for forward primers and EcoRI site for reverse primers at the 5′ end on each primer, respectively. The PCR products were digested with the restriction enzymes and cloned into the site of BglII/EcoRI in pXbG-2, a derivative of pXbG-ev1 (34), containing BglII/EcoRV/EcoRI sites between 28 bp of 5′- and 141 bp of 3′-untranslated region of Xenopus b-globin cDNA. ORF of hGLUT1 cDNA was obtained by digestion of pSPMM1 (from M. Mueckler, Washington University School of Medicine, St. Louis, MO) with BamHI and then subcloned into the site of BglII in pXbG-2. For expression of the TRET1::AcGFP1 fusion protein, TRET1 ORF lacking a stop codon was inserted into BglII/EcoRI sites in pT7XbG2-AcGFP1 vector (DNA Data Base in Japan accession no. AB255038). Template DNAs for cRNA synthesis were amplified from the corresponding cRNA expression vectors with a high-fidelity DNA polymerase, KOD Plus (Toyobo, Osaka, Japan), and primers such as SK (5′-CGGCCGCTCTAGAACTAGTGGATC-3′) and T7-XbG (5′-GGATCCTAATACGACTCACTATAGGGCTTGTTCTTTTTGCAGAAACTCAGA-3′) containing T7 promoter. The cRNAs were synthesized with the mMESSAGE mMACHINE T7 kit (Ambion) and then purified with the RNeasy MiniElute Cleanup kit (Qiagen, Hilden, Germany).
Expression of TRET1 in Xenopus Oocytes.
Stage V or VI oocytes were extirpated from Xenopus laevis females, and digested ovarian lobes with 0.2% (wt/vol) collagenase, type II (Sigma–Aldrich) in Ca2+-free modified Barth's saline [88.0 mM NaCl, 1.0 mM KCl, 2.4 mM NaHCO3, 15.0 mM Tris·HCl, 0.82 mM MgSO4, 10 μg/μl penicillin, and streptomycin (pH 7.6)] at 15°C for 3 h. The oocytes were microinjected with 40 nl of either 1 ng/nl TRET1::AcGFP1 or AcGFP1 cRNA, or nuclease-free water (Invitrogen) as a negative control. To express TRET1, the oocytes were incubated in modified Barth's saline (containing 0.41 mM CaCl2) for 3–4 days after injection at 15°C. Fluorescence images of the oocytes injected with the cRNA were observed and analyzed with a fluorescence microscope BZ-8000 (Keyence, Osaka, Japan).
Functional Zero-trans Assay of TRET1 in Xenopus Oocyte.
All uptake assays for zero-trans trehalose and/or other sugars were performed by using Xenopus oocytes expressing TRET1 at 15°C in modified Barth's saline containing appropriate concentrations of sugars. Sugar concentrations were determined by using HPLC (24). All assays were carried out in triplicate. No degradation of the sugars used in this study was detected in Xenopus oocytes extracts when incubated for 3 h at 37°C (data not shown).
Functional Assay of TRET1 in Mammalian Cells.
The TRET1 expression vector, pPvTRET1-IRES2-AcGFP1, was constructed as follows: ORF of Tret1 was obtained by digestion of pXbG-PvTRET1 with EcoRI and BglII and subcloned into the BglII/EcoRI site in pIRES2-AcGFP1 vector (Clontech). The cells were seeded on 35-mm culture dishes (Falcon 1008; BD Biosciences, Franklin Lakes, NJ) at 2 × 105 per 2 ml of medium per dish, incubated for 24 h, and transfected with pPvTRET1-IRES2-AcGFP1 using FuGene6 (Roche Diagnostics) according to the instruction manual. Control cells were transfected with the vector alone (pIRES2-AcGFP1). Two days after transfection, fresh medium containing 100 mM trehalose was added. After incubation for 3 h, the cells were rinsed with ice-cold Dulbecco's PBS (Sigma–Aldrich) three times and harvested, and trehalose was measured in the cells. All assays were carried out in triplicate. No degradation of trehalose was detected in extracts of these cells when incubated for 3 h at 37°C (data not shown). To examine transfection efficiency, we counted cells expressing AcGFP1 with a flow cytometer (EPICS Elite; Beckman Coulter, Fullerton, CA).
Statistical Analysis.
Results are reported as means ± SEM. Statistical differences were evaluated with Tukey's multiple-comparison tests after one-way ANOVA (Prism version 4; GraphPad Software, San Diego, CA).
Supplementary Material
Acknowledgments
We thank Drs. M. Mueckler and C. Makepeace (Washington University School of Medicine, St. Louis, MO) for providing hGLUT1 cDNA. This work was supported in part by Promotion of Basic Research Activities for Innovative Bioscience and a grant-in-aid (Bio Design Program) from the Ministry of Agriculture, Forestry, and Fisheries of Japan.
Abbreviations
- GLUT
facilitated glucose transporter
- TRET
facilitated trehalose transporter
- MFS
major facilitator superfamily
- cRNA
capped RNA.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: The sequences reported in this paper have been deposited in the DNA Data Base in Japan (accession no. AB272983).
This article contains supporting information online at www.pnas.org/cgi/content/full/0702538104/DC1.
References
- 1.Crowe JH, Crowe LM, Wolkers WF, Oliver AE, Ma X, Auh J-H, Tang M, Zhu S, Norris J, Tablin F. Integr Comp Biol. 2005;45:810–820. doi: 10.1093/icb/45.5.810. [DOI] [PubMed] [Google Scholar]
- 2.Crowe JH, Crowe LM, Carpenter JF, Aurell Wistrom C. Biochem J. 1987;242:1–10. doi: 10.1042/bj2420001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Crowe JH, Carpenter JF, Crowe LM. Annu Rev Physiol. 1998;60:73–103. doi: 10.1146/annurev.physiol.60.1.73. [DOI] [PubMed] [Google Scholar]
- 4.Elbein AD, Pan YT, Pastuszak I, Carroll D. Glycobiology. 2003;13:17R–27R. doi: 10.1093/glycob/cwg047. [DOI] [PubMed] [Google Scholar]
- 5.Benaroudj N, Lee DH, Goldberg AL. J Biol Chem. 2001;276:24261–24267. doi: 10.1074/jbc.M101487200. [DOI] [PubMed] [Google Scholar]
- 6.Nishizaki Y, Yoshizane C, Toshimori Y, Arai N, Akamatsu S, Hanaya T, Arai S, Ikeda M, Kurimoto M. Nutr Res. 2000;20:653. [Google Scholar]
- 7.Tanaka M, Machida Y, Niu S, Ikeda T, Jana NR, Doi H, Kurosawa M, Nekooki M, Nukina N. Nat Med. 2004;10:148–154. doi: 10.1038/nm985. [DOI] [PubMed] [Google Scholar]
- 8.Davies JE, Sarkar S, Rubinsztein DC. Hum Mol Genet. 2006;15:23–31. doi: 10.1093/hmg/ddi422. [DOI] [PubMed] [Google Scholar]
- 9.Sarkar S, Davies JE, Huang Z, Tunnacliffe A, Rubinsztein DC. J Biol Chem. 2007;282:5641–5652. doi: 10.1074/jbc.M609532200. [DOI] [PubMed] [Google Scholar]
- 10.Ramon M, Rolland F, Thevelein JM, Van Dijck P, Leyman B. Plant Mol Biol. 2007;63:195–206. doi: 10.1007/s11103-006-9082-2. [DOI] [PubMed] [Google Scholar]
- 11.Satoh-Nagasawa N, Nagasawa N, Malcomber S, Sakai H, Jackson D. Nature. 2006;441:227–230. doi: 10.1038/nature04725. [DOI] [PubMed] [Google Scholar]
- 12.Rolland F, Baena-Gonzalez E, Sheen J. Annu Rev Plant Biol. 2006;57:675–709. doi: 10.1146/annurev.arplant.57.032905.105441. [DOI] [PubMed] [Google Scholar]
- 13.Brumfiel G. Nature. 2004;428:14–15. doi: 10.1038/428014a. [DOI] [PubMed] [Google Scholar]
- 14.Wolkers WF, Walker NJ, Tablin F, Crowe JH. Cryobiology. 2001;42:79–87. doi: 10.1006/cryo.2001.2306. [DOI] [PubMed] [Google Scholar]
- 15.Boos W, Shuman H. Microbiol Mol Biol Rev. 1998;62:204–229. doi: 10.1128/mmbr.62.1.204-229.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Stambuk BU, De Araujo PS, Panek AD, Serrano R. Eur J Biochem. 1996;237:876–881. doi: 10.1111/j.1432-1033.1996.0876p.x. [DOI] [PubMed] [Google Scholar]
- 17.Stambuk BU, da Silva MA, Panek AD, de Araujo PS. FEMS Microbiol Lett. 1999;170:105–110. doi: 10.1111/j.1574-6968.1999.tb13361.x. [DOI] [PubMed] [Google Scholar]
- 18.Han EK, Cotty F, Sottas C, Jiang H, Michels CA. Mol Microbiol. 1995;17:1093–1107. doi: 10.1111/j.1365-2958.1995.mmi_17061093.x. [DOI] [PubMed] [Google Scholar]
- 19.Pessin JE, Bell GI. Annu Rev Physiol. 1992;54:911–930. doi: 10.1146/annurev.ph.54.030192.004403. [DOI] [PubMed] [Google Scholar]
- 20.Uldry M, Thorens B. Pflügers Arch. 2004;447:480–489. doi: 10.1007/s00424-003-1085-0. [DOI] [PubMed] [Google Scholar]
- 21.Wyatt GR. Adv Insect Physiol. 1967;4:287–360. [Google Scholar]
- 22.Keilin D. Proc R Soc London Ser B. 1959;150:149–191. doi: 10.1098/rspb.1959.0013. [DOI] [PubMed] [Google Scholar]
- 23.Kikawada T, Minakawa N, Watanabe M, Okuda T. Integr Comp Biol. 2005;45:710–714. doi: 10.1093/icb/45.5.710. [DOI] [PubMed] [Google Scholar]
- 24.Watanabe M, Kikawada T, Minagawa N, Yukuhiro F, Okuda T. J Exp Biol. 2002;205:2799–2802. doi: 10.1242/jeb.205.18.2799. [DOI] [PubMed] [Google Scholar]
- 25.Watanabe M, Kikawada T, Okuda T. J Exp Biol. 2003;206:2281–2286. doi: 10.1242/jeb.00418. [DOI] [PubMed] [Google Scholar]
- 26.Kikawada T, Nakahara Y, Kanamori Y, Iwata K, Watanabe M, McGee B, Tunnacliffe A, Okuda T. Biochem Biophys Res Commun. 2006;348:56–61. doi: 10.1016/j.bbrc.2006.07.003. [DOI] [PubMed] [Google Scholar]
- 27.Uldry M, Ibberson M, Hosokawa M, Thorens B. FEBS Lett. 2002;524:199–203. doi: 10.1016/s0014-5793(02)03058-2. [DOI] [PubMed] [Google Scholar]
- 28.Guo N, Puhlev I, Brown DR, Mansbridge J, Levine F. Nat Biotechnol. 2000;18:168–171. doi: 10.1038/72616. [DOI] [PubMed] [Google Scholar]
- 29.Singer MA, Lindquist S. Mol Cell. 1998;1:639–648. doi: 10.1016/s1097-2765(00)80064-7. [DOI] [PubMed] [Google Scholar]
- 30.Eroglu A, Russo MJ, Bieganski R, Fowler A, Cheley S, Bayley H, Toner M. Nat Biotechnol. 2000;18:163–167. doi: 10.1038/72608. [DOI] [PubMed] [Google Scholar]
- 31.Elliott GD, Liu XH, Cusick JL, Menze M, Vincent J, Witt T, Hand S, Toner M. Cryobiology. 2006;52:114–127. doi: 10.1016/j.cryobiol.2005.10.009. [DOI] [PubMed] [Google Scholar]
- 32.Lapinski J, Tunnacliffe A. FEBS Lett. 2003;553:387–390. doi: 10.1016/s0014-5793(03)01062-7. [DOI] [PubMed] [Google Scholar]
- 33.Ma X, Jamil K, Macrae TH, Clegg JS, Russell JM, Villeneuve TS, Euloth M, Sun Y, Crowe JH, Tablin F, et al. Cryobiology. 2005;51:15–28. doi: 10.1016/j.cryobiol.2005.04.007. [DOI] [PubMed] [Google Scholar]
- 34.Preston GM, Carroll TP, Guggino WB, Agre P. Science. 1992;256:385–387. doi: 10.1126/science.256.5055.385. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.