Abstract
The accumulation of amyloid plaques and amyloid congophilic angiopathy (ACA) in the brains of affected individuals is one of the main pathological features of Alzheimer's disease. Within these deposits, the βA4 (Aß) polypeptide represents a major component with the C-terminal 39–43 amino acid variants being most abundant. Using a mouse IgG1 MoAb produced by hybridoma βA4[35–43]-95.2 3B9, which reacts with the epitope is defined by the amino acid residues βA438[GVV]40, this study has identified a unique conformation within the carboxyl terminus of human βA4[1–42]. Although the βA438[GVV]40 sequence is present within the C-termini of human βA4[1–40] and βA4[1–43] and the βA4-containing region of human APP, the βA4[35–43]-95.2 3B9 MoAb (designated MoAb 3B9) does not bind these polypeptides, demonstrating a high degree of specificity for the βA438[GVV]40 epitope as presented within the βA4[1–42] sequence. The βA4[1–42] epitope bound by MoAb 3B9 is sensitive to heating (100°C for 5 min) and is denatured by SDS but not by oxidative radio-iodination of βA4 or by adsorption to plastic surfaces or nitrocellulose. The recognition of βA4 plaque deposits and ACA by MoAb 3B9 within formalin-fixed sections of human AD brain demonstrates the potential of these antibodies for investigating the role of the unique βA4[1–42] conformation in the development of Alzheimer's disease.
Keywords: βA4, amyloid, Alzheimer's disease, monoclonal antibody, polypeptide
Introduction
The pathological feature of Alzheimers's disease (AD) is the formation of neuritic and diffuse amyloid plaques within brain parenchyma, and amyloid congophilic angiopathy (ACA) associated with cerebral and leptomeningeal blood vessels [1]. The predominant component of these amyloid deposits is the approx. 4-kD βA4 (Aß) peptide which is a cleavage product of APP [2].
Although many N- and C-terminal truncated βA4 variants have been identified in the human AD brain, the principal variants associated with extracellular amyloid deposits are the 40–43 amino acid βA4 polypeptides [3–6].
The βA4 molecule comprises an amphipathic N-terminal region and a hydrophobic C-terminal region (residues 29–42) which adopts a structure consisting of a β-turn (residues 26–29) flanked by two strands of β-sheets [4,7]. Based on this predicted conformation of the βA4 monomer, it has been proposed that the β-pleated sheet structure of the insoluble amyloid fibrils arises from hydrogen bonding between the N-terminal β-strands of βA4 monomers and the C-terminal β-strands of other monomers [7]. Further, the association of βA4[1–42] with amyloid plaques is reflected in the variations in polypeptide solubility under physiological conditions such that βA4[1–42] more readily polymerizes into fibrils than βA4[1–40] or βA4[1–43] [6]. This tendency of βA4 to self aggregate at physiological conditions may explain the fibrillar deposits in plaques and blood vessels in AD.
It has been reported that the last three C-terminal residues of βA4[1–42] are crucial to amyloid deposition [8]. Using a newly developed monoclonal antibody, MoAb 3B9, this study identifies a unique conformation within the C-terminal region of βA4[1–42] that is absent from the other major amyloidogenic forms of βA4, namely βA4[1–40] and βA4[1–43].
Materials and methods
Chemicals used in this study were of analytical reagent grade. All buffers were prepared using Milli-Q purified distilled, deionized water (Millipore Corp., Bedford, MA) and were filtered through 0·22-μm filters (Millipore).
Animals
Inbred BALB/c mice 28 days of age were obtained from Monash University Central Animal Services and maintained within the Department of Pathology Animal House at the University of Melbourne.
Antibodies
The monoclonal IgG1 βA4[35–43]-95.2 3B9 antibody (designated MoAb 3B9) was generated from the spleen of an inbred BALB/c mouse immunized with βA4[35–43] peptide coupled to diphtheria toxoid using cell fusion techniques performed as previously described [9,10]. Isotype control antibodies included mouse IgG1 anti-human α-lactalbumin ET-1 MoAbs or anti-sheep erythrocyte MUI-1 MoAbs [11]. MoAb SKB1E8, specific for βA4, was kindly provided by Drs S. Holmes and C. Gray (SmithKline Beecham; Harlow, UK) and was used as a capture antibody for anti-βA4 double antibody capture ELISAs. MoAb WO-2, which reacts with residues 5–8 within the N-terminal region of the βA4 polypeptide (Ida et al., 1996 [21]), was produced in this laboratory from hybridoma cells kindly provided by Professor K. Beyreuther (Centre for Molecular Biology, University of Heidelberg). Polyclonal mouse antiserum specific for residues 34–40 of βA4 was produced in this laboratory following immunization of mice with βA4[34–40] peptide coupled to diphtheria toxoid.
Amyloid precursor protein
Amyloid precursor protein was purified from human AD brains in these laboratories using the method described by Moir et al. [12].
Peptides
βA4[35–43] peptides for the production of MoAbs were synthesized by Chiron Mimotopes Pty. Ltd. (Melbourne, Australia). The βA4[35–43] peptides were either coupled to diphtheria toxoid for immunization of mice or biotinylated for the screening of specific antibodies.
Binding of the antibodies to different carboxyl termini truncation variants of the βA4 polypeptide was assessed using synthetic βA4[1–40], βA4[1–42] and βA4[1–43]. The synthetic βA4[1–40], βA4[1–42] and βA4[1–43] polypeptides were synthesized by k-Biologicals Inc. (Rancho Cucamonga, CA) and peptide purity verified by mass spectrometry.
Mapping of the βA4 epitopes identified by the MoAb 3B9 was undertaken using an overlapping set of 15mer biotinylated peptides, incrementing by three amino acid residues, encompassing the human APP695 [589–652] sequence (Chiron Mimotopes).
Immunohistochemistry
To determine the reactivity of MoAb 3B9 to βA4 amyloid in the brains of AD patients immunoperoxidase immunohistochemistry was performed. Tissues from the brains of patients with AD and control individuals were used for immunohistochemistry. Briefly, paraffin-embedded, formalin-fixed 4-μm tissue sections were incubated in 80% formic acid for 5 min at room temperature and washed with ddH2O. Slides were then treated with 3% (v/v) H2O2 (BDH Chemicals, Sydney, Australia) for 5 min at room temperature to block endogenous peroxidase activity and washed once in ddH2O and once in TBS pH 7·4. The sections were then blocked with 20% (v/v) swine serum in TBS for 30 min at room temperature. Following blocking, 50 μl of test and isotype control MoAbs were incubated with the sections for 1 h at 37°C. The sections were then washed three times for 5 min per wash in TBS pH 7·4 and incubated with 50 μl of biotinylated rabbit anti-mouse immunoglobulins (1:500 in TBS containing 20% (v/v) swine serum; Dako Corp., Carpinteria, CA) for a further 30 min at room temperature. The sections were again washed three times in TBS pH 7·4 and 50 μl of streptavidin–hosreradish peroxidase (HRP) (1:100 in TBS containing 20% (v/v) swine serum; Dako) added to each section and incubated for 30 min at room temperature. The sections were then washed three times and incubated with 100 μl of HRP substrate solution (0·06% (w/v) DAB in 10 ml TBS pH 7·6 containing 0·015% (v/v) H2O2) for 5 min at room temperature. The slides were then washed in TBS pH 7·4 and ddH2O and counterstained with Mayer's haematoxylin for 1 min. The slides were then washed in ddH2O, dehydrated with absolute alcohol and mounted for viewing using DePeX mounting medium (BDH Chemicals).
ELISA
Indirect ELISA
Antibody titrations were performed by indirect ELISA. Fifty nanograms of each of the peptides or proteins, diluted in 0·05 m carbonate-bicarbonate buffer pH 9·6, were coated onto each well of 96-well ELISA plates (Cliniplate EB, Labsystems, Helsinki, Finland) for 4 h at room temperature. Peptide-coated plates were blocked for non-specific binding with 1% (w/v) bovine serum albumin (BSA; Sigma Chemical Co., St Louis, MO) overnight at 4°C. All primary antibodies were diluted with PBS−0·05% Tween 20 (PBS–T) containing 0·5% (w/v) BSA, added onto precoated ELISA plates and incubated for 30 min at room temperature. After nine washes with PBS–T, HRP-conjugated goat anti-mouse IgG (Zymed antibodies, San Francisco, CA) was added at 1:500 dilution and incubated for 30 min at room temperature. After another nine washing steps, 100 μl HRP substrate solution containing H2O2 and o-phenylenediamine were added to each well and the plates incubated, protected from the light, for 1 h. The reaction was stopped by the addition of 25 μl 8 m H2SO4 to each well. Optical density (OD) values were then determined at 490 nm for each well using a computer-controlled BioRad (Richmond, CA) Model 450 microplate reader.
A different coating method was used for the biotinylated βA4[35–43] peptide. Cliniplate EB 96-well ELISA plates were coated with 50 μl of streptavidin (500 ng; Sigma) in ddH2O and air-dried at 37°C overnight. Streptavidin-coated plates were blocked for non-specific binding with 1% (w/v) BSA for 1 h at room temperature. βA4[35–43] biotinylated peptide was added at 50 ng per well in PBS–T and incubated for 1 h. These plates were then used as above to determine the antibody reactivity.
Double antibody capture ELISA
βA4-specific SKB1E8 capture antibody was coated onto Cliniplate EB ELISA plates at 400 ng per well in 50 μl 0·05 m carbonate-bicarbonate buffer pH 9·6 for 4 h at room temperature. Coated plates were blocked for non-specific binding with 1% (w/v) BSA overnight at 4°C. All peptides were diluted in 0·5% (w/v) BSA in PBS–T and incubated in the antibody-coated plates for 30 min at room temperature. After nine washes with PBS–T, biotinylated MoAb 3B9 or isotype control MoAb were added at 2 μg/ml to appropriate wells and the plates incubated for a further 30 min. After nine washes with PBS–T, HRP-coupled streptavidin (Southern Biotechnology Associates, Birmingham, AL) was added at 1:2000 dilution and incubated for 30 min at room temperature. After another nine washing steps, 100 μl HRP substrate solution containing H2O2 and o-phenylenediamine were added to each well and the plates incubated, protected from the light, for 1 h. The reaction was stopped by the addition of 25 μl 8 m H2SO4 to each well. OD values were then determined at 490 nm for each well using a computer-controlled BioRad Model 450 microplate reader.
Dot blot analysis
βA4 peptides were dot blotted onto nitrocellulose using a Bio-Dot microfiltration apparatus (BioRad). βA4[1–40], βA4[1–42] and βA4[1–43] peptides were added onto nitrocellulose at 1 μg per dot and incubated for 1 h at room temperature. Nitrocellulose was then washed three times with PBS–T, removed from the microfiltration apparatus and blocked with 1% (w/v) BSA for 1 h at room temperature. After three 5-min washes with PBS–T, the nitrocellulose membrane was cut into strips and incubated with MoAb 3B9 or isotype control MoAbs (1 μg/ml diluted in PBS–T containing 0·5% (w/v) BSA) for 30 min at room temperature. Following another three 5-min washes with PBS–T, the nitrocellulose strips were incubated with HRP-conjugated rabbit anti-mouse immunoglobulin (1:500 dilution in PBS–T containing 0·5% (w/v) BSA; Dako) for 30 min at room temperature. Following another three washes, HRP enzyme substrate solution containing 0·5 mg/ml 4-chloro-1-naphthol and 0·015% (v/v) H2O2 dissolved in PBS was added and the nitrocellulose strips incubated for 20 min at room temperature. Colour development was stopped after 20 min incubation by washing the nitrocellulose strips three times in ddH2O for 15 min for each wash.
Immunoprecipitation
Immunoprecipitations were performed with the MoAb 3B9 which was used to capture iodinated βA4[1–40] and βA4[1–42] synthetic peptides from solution. Five micrograms of test and isotype control antibodies (diluted in PBS containing 1% (w/v) BSA) were incubated with 1 × 106 ct/min of iodinated βA4 (diluted in PBS containing 1% (w/v) BSA) for 12 h on a rotating mixer at 4°C. Fifty micrograms of affinity-purified rabbit anti-mouse IgG (diluted in PBS containing 1% (w/v) BSA) were added to each of the test and control tubes and tubes were incubated for 2 h on a rotating mixer at room temperature. Then, 50 μl of Staphylococcus aureus (Pansorbin ® cells; Calbiochem-Novabiochem Corp., La Jolla, CA) were added to each tube and the tubes incubated for a further 30 min with mixing at room temperature. Following five washes with PBS–T, the final pellets was counted for bound radioactivity on a computer-controlled LKB-Wallac 1261 Multigamma counter. The pellets were then re-suspended in SDS-reducing sample buffer, boiled for 5 min, centrifuged to remove S. aureus cells, and the immunoprecipitated proteins in the supernatants separated on a 20% SDS–polyacrylamide gel and identified by autoradiography.
Epitope mapping analysis
To confirm further the specificity of the MoAb 3B9, binding was examined using a PepSet comprising an overlapping set of linear 15mer peptides, incrementing by three amino acid residues per peptide, encompassing the human APP695[589–652] sequence in a similar manner to that described elsewhere [10]. The peptides comprising the PepSet were synthesized by Chiron Mimotopes and designed to enhance uniformity of ELISA plate-coating of the dissimilar peptides by biotinylation via an amino terminal-SGSG-linker (i.e. Biotin-SGSG-NH2-Peptide). For epitope mapping, 50-μl aliquots of streptavidin (500 ng) dissolved in ddH2O were added to wells of 96-well ELISA plates (Cliniplate EB, Labsystems). The streptavidin was allowed to evaporate to dryness at 37°C overnight. The ELISA plate wells were then blocked with 100 μl/well of 1% (w/v) BSA for 1 h at room temperature and washed three times with PBS–T. Peptide concentrations were adjusted to 10 μg/ml and 50 μl of each of the 17, 15mer biotinylated peptides were added, in duplicate, to appropriate wells of the ELISA plate and incubated for 1 h at room temperature. Following three washes with PBS–T, 50 μl of the test and control antibodies, diluted in PBS–T containing 0·5% (w/v) BSA, were added to each well of the ELISA plate and incubated for 30 min at room temperature. ELISA plates were then washed nine times with PBS–T and 50 μl HRP-conjugated rabbit anti-mouse immunoglobulins (diluted 1:500 in PBS–T containing 0·5% (w/v) BSA; Dako) added to each well. After 30 min incubation, the plates were washed nine times with PBS–T and 100 μl of HRP substrate solution containing H2O2 and o-phenylenediamine were added to each well and the reaction allowed to proceed for 1 h at room temperature protected from the light. The reaction was stopped by the addition of 25 μl 8 m H2SO4 to each well. The presence of bound mouse immunoglobulins was detected spectrophotometrically at 490 nm using a computer-controlled BioRad 450 ELISA plate reader.
Control ELISAs were performed simultaneously to determine levels of background binding of the antibodies to streptavidin and BSA in the absence of biotinylated peptides.
Results
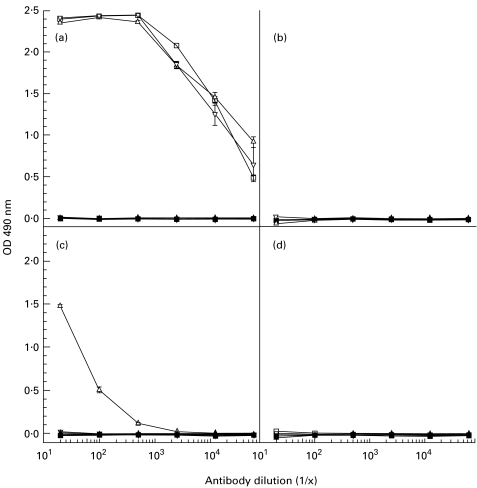
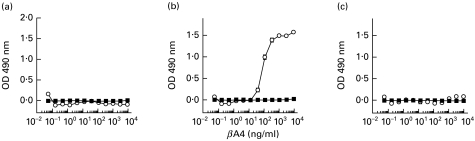
All mice immunized with βA4[35–43] peptide conjugated to diphtheria toxoid demonstrated high-titre anti-peptide antibody responses. As shown in Fig. 1, only one of these mice produced antiserum that cross-reacted with βA4[1–42] but not with βA4[1–40] or βA4[1–43].
Fig. 1.
ELISA reactivity of immune and preimmune sera from mice immunized with βA4[35–43]-DT. (a) Responses of mice to βA4[35–43] peptide used as the immunogen (mouse 1 (□), mouse 2 (Δ), mouse 3 (∇)). (b,c,d) Reactivity of these antisera to βA4[1–40], βA4[1–42] and βA4[1–43], respectively. Preimmune sera are delineated by the following symbols: mouse 1 (▪); mouse 2 (▴); and mouse 3 (▾).
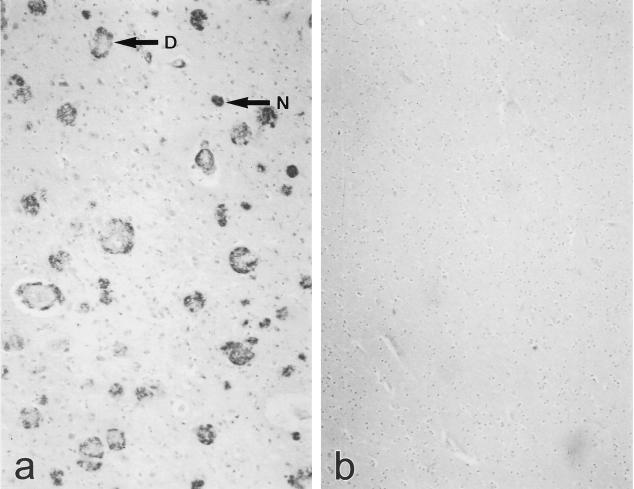
Fusion of the spleen cells from this mouse with NS-1 myeloma cells resulted in the production of a limit dilution-cloned hybridoma, designated βA4[35–43]-95.2 3B9, which secreted an IgG1 MoAb (MoAb 3B9) reactive with neuritic and diffuse plaques in 4-μm sections of paraffin-embedded, formalin-fixed brain tissue from AD patients (Fig. 2).
Fig. 2.
Immunohistochemical staining of 4-μm sections of paraffin-embedded, formalin-fixed human Alzheimer's disease (AD) and control temporal lobe using MoAb 3B9. Staining of neuritic plaques (N) and diffuse plaques (D) is apparent in tissue from an AD patient (a) but not in tissue from a non-demented control individual (b).
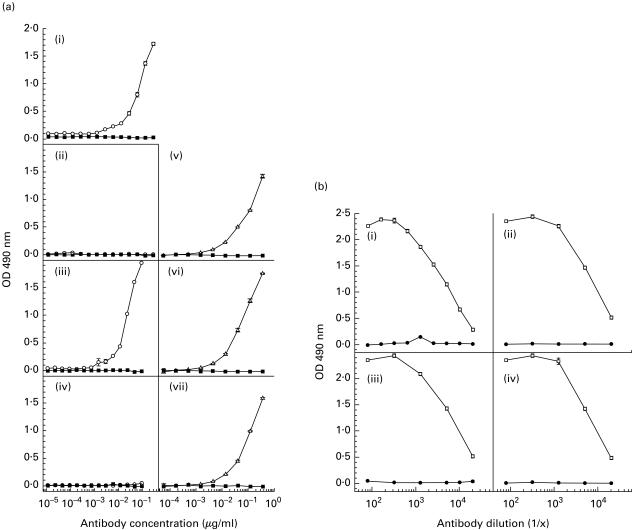
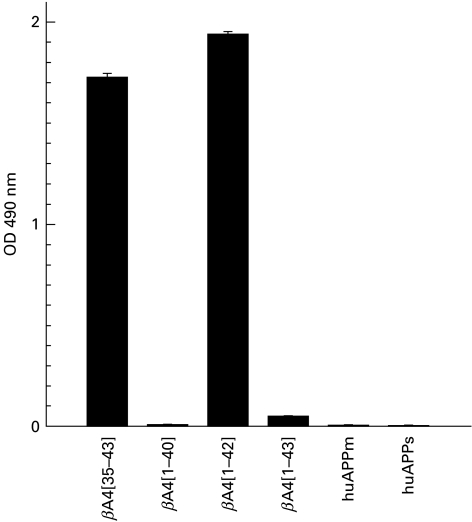
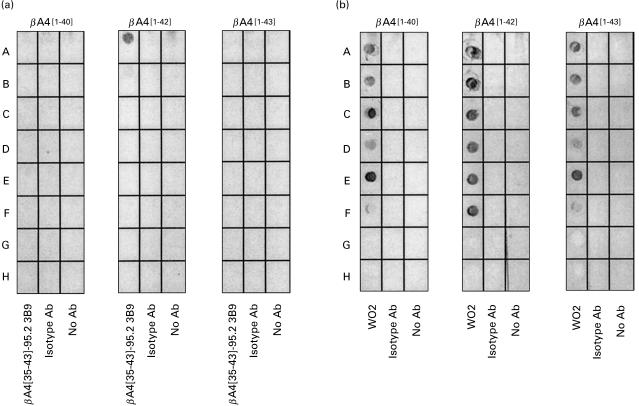
MoAb 3B9 also demonstrated high-titre reactivity to the βA4[35–43] peptide immunogen and βA4[1–42] but no reactivity to βA4[1–40] or βA4[1–43] by indirect ELISA (Fig. 3a). Lack of reactivity of the MoAb 3B9 to βA4[1–40] and βA4[1–43] was not due to obscuring of the epitope present within the C-termini of these peptides following adsorption to the ELISA plates, as mouse anti-βA4[34–40] antibodies exhibited high-titre binding to βA4[1–40], βA4[1–42] and βA4[1–43] when these variants were adsorbed to the ELISA plates (Fig. 3b). Reactivity to purified secreted and full length forms of human APP was not detected by indirect ELISA (Fig. 4).
Fig. 3. (a).
ELISA titrations showing the reactivity of purified MoAb 3B9 (○) and isotype control MUI-1 MoAb (▪) to βA4[35–43] peptide (i), βA4[1–40] (ii), βA4[1–42] (iii) and βA4[1–43] (iv). In these experiments the WO2 MoAb was used as a positive control for peptide binding [21]. (v,vi,vii) Reactivity of MoAb WO2 (Δ) and the isotype control MUI-1 MoAb (▪) to βA4[1–40], βA4[1–42] and βA4[1–43], respectively. (b) ELISA titrations showing the reactivity of polyclonal mouse anti-βA4[34–40] antiserum (○) and preimmune serum (•) to βA4[34–40] peptide (i), βA4[1–40] (ii), βA4[1–42] (iii) and βA4[1–43] (iv).
Fig. 4.
Reactivity of MoAb 3B9 to βA4[35–43], C-terminal βA4 variants, soluble (huAPPs) and full length (huAPPm) purified human brain amyloid precursor protein.
Characterization of the specificity of the MoAb 3B9 using a double antibody capture ELISA confirmed the observations from the indirect ELISAs. The double antibody capture ELISA was performed by coating ELISA plates with 400 ng/well of anti-βA4 SKB1E8 MoAbs (epitope βA4[19 – 22]). Bound βA4 peptide was then identified using biotinylated MoAb 3B9 and streptavidin–HRP (Fig. 5).
Fig. 5.
Double antibody capture ELISA demonstrating the specific binding of biotinylated MoAb 3B9 (○) to βA4[1–42] (b) but not βA4[1–40] (a) or βA4[1–43] (c). Biotinylated isotype control ET-1 MoAb (▪) exhibited no binding.
The specific binding of MoAb 3B9 to 125I-labelled βA4[1–42] was also observed following immunoprecipitation (Fig. 6). These data demonstrate the binding of this MoAb to βA4[1–42] peptide in solution and indicate that the carboxyl terminal βA4[35–43] epitope is not sensitive to the oxidative radioiodination. Under the conditions examined, both the double antibody capture ELISA and the immunoprecipitation confirmed the βA4[1–42] specificity of MoAb 3B9.
Fig. 6.
Immunoprecipitation of 125I-labelled βA4 using MoAb 3B9 (lane 1) showing recognition of βA4[1–42] but not βA4[1–40]. Samples include: lane 1, 125I-βA4[1–40] or [1–42], MoAb 3B9, RAM-IgG, Pansorbin cells; lane 2, 125I-βA4[1–40] or [1–42], isotype control ET-1 MoAb, RAM-IgG, Pansorbin cells; lane 3, 125I-βA4[1–40] or [1–42] and Pansorbin cells; lane 4, 125I-βA4[1–40] or [1–42], RAM-IgG and Pansorbin cells. Immunoprecipitation performed as described above.
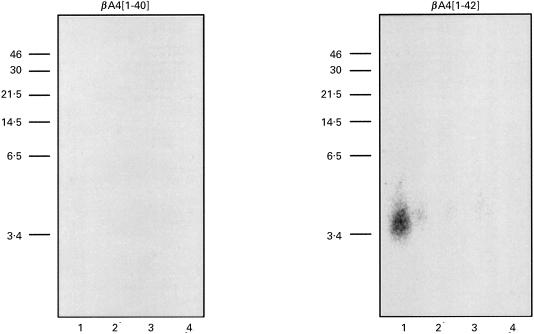
As shown in Fig. 7a, the conformation of the epitope identified by MoAb 3B9 was profoundly altered following boiling or in the presence of SDS detergent, so that this MoAb was unable to bind to βA4[1–42] peptide treated in this manner. Weak binding of MoAb 3B9 to the βA4[1–42] peptide which had been boiled for 5 min was detected, suggesting differential rates of epitope denaturation possibly associated with the extent of βA4[1–42] aggregation within the sample. These experiments further confirmed the anti-βA4[1–42] specificity of the MoAb 3B9, with detectable binding to βA4[1–42] peptide but not βA4[1–40] or βA4[1–43] following adsorption of these peptides to nitrocellulose at 1·0 μg/dot and probing with high concentrations of the MoAb (1·0 μg MoAb/ml). The sensitivity of the epitopes identified by the MoAb 3B9 to SDS detergent and boiling precludes their use in conventional immunoblotting.
Fig. 7.
(a) Investigation of the stability of the βA4 38[GVV]40 epitope identified by MoAb 3B9 showed recognition of native βA4[1–42] but loss of this antigenic activity following heating and solubilization in 1% (w/v) SDS detergent. In this experiment, nitrocellulose was dotted with 1·0 μg of βA4[1–40], [1–42] or [1–43] and treated as follows: A, peptide dissolved in PBS; B, peptide dissolved in PBS and boiled for 5 min; C, peptide dissolved in 1% (w/v) SDS-non-reducing sample buffer; D, peptide dissolved in 1% (w/v) SDS-non-reducing sample buffer and boiled for 5 min; E, peptide dissolved in 1% (w/v) SDS-reducing sample buffer containing 25 mm DTT; F, peptide dissolved in 1% (w/v) SDS-reducing sample buffer containing 25 mm DTT and boiled for 5 min; rows G and H were not dotted with βA4 samples. These strips of nitrocellulose were then reacted with MoAb 3B9 (lane 1), isotype control ET-1 MoAb (lane 2) or PBS (lane 3) and bound mouse immunoglobulin detected as described for dot blotting above. (b) Reactivity of the WO2 MoAb [21] to each of the βA4 variants under identical conditions to those described for MoAb 3B9.
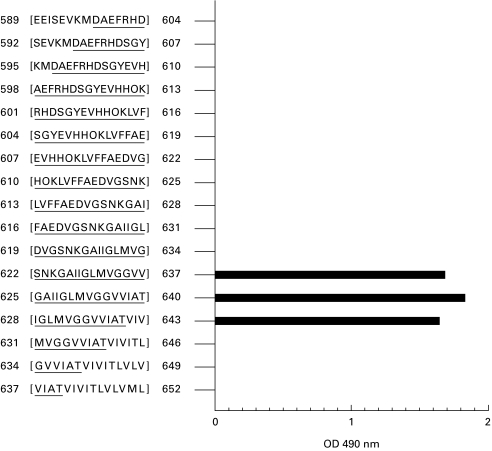
Epitope mapping analysis was undertaken using an overlapping set of 15mer linear peptides incrementing by three amino acids per peptide and encompassing the region APP695[589–652]. High levels of binding of the MoAb 3B9 to the linear peptide APP695 625[GAIIGLMVGGVVIAT]640 containing the βA4 35[MVGGVVIAT]43 peptide immunogen sequence were observed as expected (Fig. 8). Significantly, the MoAb 3B9 bound the APP695 622[SNKGAIIGLMVGGVV]637 linear peptide which contains the GVV terminal sequence of βA4[1–40]. Recognition of an epitope within the carboxy terminal 15 amino acid residues of βA4[1–40], when represented as a linear peptide, in contrast to the lack of binding to intact βA4[1–40] by the MoAb 3B9, indicates conformational variations between βA4[1–40], βA4[1–42] and βA4[1–43]. Lack of MoAb 3B9 binding to the PepSet sequences APP 631[NH2MVGGVVIATVIVITL-COOH]646 and APP 634[NH2GVVIATVIVITLVLV-COOH]649 which contain the GVV epitope is most probably due to steric hindrance, as the biotin linker is covalently coupled to the NH2 terminus of the peptides.
Fig. 8.
The binding profile of MoAb 3B9 defined by ELISA using an overlapping set of 17 peptides encompassing the APP695[589–652] sequence. Antibody binding to the 15mer peptides containing the βA4 38[GVV]40 epitope in an accessible conformation is shown.
Discussion
Monoclonal antibodies secreted by hybridoma βA4[35–43]-95.2 3B9, isolated following fusion of splenocytes from mice immunized with βA4[35–43], react with βA4[1–42] but not with βA4[1–40] or βA4[1–43].
Indirect ELISAs in which the polypeptides were adsorbed to ELISA plate well surfaces produced high-titre responses to βA4[1–42], but no responses to βA4[1–40] or βA4[1–43]. As neoepitopes on polypeptides have been previously reported following adsorption to plastics [13], the demonstration of the binding of MoAb 3B9 to epitopes on native βA4 was investigated. Using a double antibody capture ELISA, MoAb 3B9 bound βA4[1–42] peptide captured from free solution by the βA4[19–22]-specific SKB1E8 MoAb. Further, direct binding of these MoAbs to the βA4[1–42] peptide in free solution was confirmed by immunoprecipitation demonstrating that the reactive epitope was not a neoepitope created by binding to ELISA plate well surfaces.
Epitope mapping using a linear array of 17, 15mer peptides encompassing the sequence APP695[589–652] and incrementing by three residues per peptide revealed that the MoAb 3B9 bound the GVV epitope within the carboxyl terminal region of βA4. Within this peptide series, antibody binding was only detected in those sequences in which this GVV epitope was accessible at the solvent-exposed interface of the peptide array. Thus, MoAb 3B9 bound the GVV epitope in peptide arrays created by the sequences: APP695 622[SNKGAIIGLMVGGVV]637, the carboxyl terminal of βA4[1–40]; APP695 625[GAIIGLMVGGVVIAT]640, the carboxyl terminal of βA4[1–43] and APP695 628[IGLMVGGVVIATVIV]643.
Although this MoAb bound the GVV epitope within peptides which constitute the carboxyl terminal regions of βA4[1–40] (βA4 [35MVGGVV40]) and βA4[1–43] (βA4[35MVGGVVIAT43]), MoAb 3B9 did not bind this epitope in when presented within the entire polypeptide sequences of βA4[1–40] or βA4[1–43]. These data suggest that the amino acid residues of βA4 which are absent in the 15mer PepSet peptides impose conformational constraints on the C-terminal regions of the βA4[1–40], [1–42] and [1–43] polypeptides and consequently influence accessibility of the C-terminal GVV epitope to MoAb 3B9 binding. Thus, the GVV epitope was only recognized by MoAb 3B9 in the context of the βA4[1–42] sequence (βA4[35MVGGVVIA42]), indicating that significant conformational variation exists between the carboxyl termini of βA4[1–40], βA4[1–42] and βA4[1–43].
The lack of reactivity of MoAb 3B9 to highly purified human brain-derived APP further supports the proposal that this antibody recognizes the βA438[GVV]40 epitope when presented in the unique conformation of the βA4[1–42] C-terminal sequence.
Structural variations between C-terminal variants of βA4 are indicated by variations in peptide solubility. Fibril formation by the polymerization of βA4[1–42] is significantly greater than that of βA4[1–40] in water and is further enhanced in physiological solutions [8,17–20]. Using CD spectroscopy, analysis of βA4 has demonstrated that the N-terminal 1–28 region may exist as an α-helix in organic solvents. This structure is modified with increasing temperature and pH so that at pH 1–4 or above 7 the peptides adopt a random coil structure but aggregate in a β-sheet conformation between pH 4 and 7. In contrast, the region encompassing βA4[29–42] has been shown to adopt a β-strand conformation regardless of temperature and pH, and as such has been reported to direct the protein folding of the entire βA4 sequence to produce the β-pleated sheet structure found in amyloid deposits [7,8,15,16]. Reported differences in the solution conformations of βA4[1–39] and βA4[1–42] from CD spectroscopy studies focus attention on βA440[VIA]42 as being the region critical for the formation of amyloid fibrils.
In light of these observations, this study has demonstrated a unique C-terminal conformation within βA4[1–42], as defined using MoAb 3B9 which binds the GVV epitope. These results support the hypothesis that key residues within the C-terminal region of βA4[1–42] generate a unique peptide conformation which may be responsible for initiating the βA4 polymerization and amyloid fibril formation observed in the neuritic and diffuse plaques of AD.
In conclusion, the antibodies generated in this study recognize the βA438[GVV]40 sequence when presented within the C-terminal region of βA4[1–42], the most predominant form of βA4 detected in plaques within Alzheimer's disease brain. The unique βA4 conformation formed by the C-terminal of βA4[1–42] which contains the βA438[GVV]40 sequence may be the key to understanding the molecular basis of βA4 aggregation that results in potentially neurotoxic amyloid plaques which are believed to lead ultimately to cognitive decline. The MoAb described in this study exhibits specific recognition of βA4[1–42] and as such provides a powerful tool for more detailed immunohistochemical examination of AD pathology and the precise definition of the levels of βA4[1–42] in the brains and body fluids of AD sufferers and age- and sex-matched control individuals.
Acknowledgments
The authors would like to thank Mr Michael Lidman for supplying the purified human brain APP used in this study, Ms Jo Merriner and Ms Tina Cardamone for performing immunohistochemical staining, and Mr Ben Kreunen for preparation of photography. The authors acknowledge and thank SmithKline Beecham for supporting this work.
References
- 1.Glenner G. Amyloid deposits and amyloidosis; the β-fibrilloses. New Engl J Med. 1980;302:1283–2. doi: 10.1056/NEJM198006053022305. [DOI] [PubMed] [Google Scholar]
- 2.Kang J, Lemarie H, Unterbeck A, et al. The precursor of Alzheimer's disease amyloid A4 protein resembles a cell surface receptor. Nature. 1987;325:733–6. doi: 10.1038/325733a0. [DOI] [PubMed] [Google Scholar]
- 3.Bush AI, Beyreuther K, Masters Cl. βA4 amyloid protein and its precursor in Alzheimer's disease. Pharmac Ther. 1992;56:97–117. doi: 10.1016/0163-7258(92)90039-3. [DOI] [PubMed] [Google Scholar]
- 4.Hilbich C, Kisters-Woike B, Masters CL, Beyreuther K. Amyloid protein precursor in development, aging and Alzheimer's disease. Berlin: Springer-Verlag; 1994. Amyloid βA4 of Alzheimer's disease: structural requirements for folding and aggregation; pp. 21–35. [Google Scholar]
- 5.Younkin Sg. Evidence that Aß42 is the real culprit in Alzheimer's disease. Ann Neurol. 1995;37:287–8. doi: 10.1002/ana.410370303. [DOI] [PubMed] [Google Scholar]
- 6.Greenberg Bd. The COOH-terminus of the Alzheimer amyloid Aß peptide: differences in length influence the process of amyloid deposition in Alzheimer brain, and tell us something about relationships among parenchymal and vessel-associated amyloid deposits. Amyloid: Int J Exp Clin Invest. 1995;2:195–203. [Google Scholar]
- 7.Soto C, Branes MC, Alvarez J, Inestrosa Nc. Structural determinants of Alzheimer's amyloid β-peptide. J Neurochem. 1994;63:1191–8. doi: 10.1046/j.1471-4159.1994.63041191.x. [DOI] [PubMed] [Google Scholar]
- 8.Barrow C, Yasuda A, Kenny P, Zagorski Mg. Solution conformations and aggregational properties synthetic amyloid β-peptides of Alzheimer's disease: analysis of circular dichroism spectra. J Mol Biol. 1992;225:1075–93. doi: 10.1016/0022-2836(92)90106-t. [DOI] [PubMed] [Google Scholar]
- 9.Underwood JR, Pederson JS, Chalmers PJ, Toh Bh. Hybrids from normal, germ free, nude and neonatal mice produce monoclonal autoantibodies to eight different intracellular structures. Clin Exp Immunol. 1985;60:417–26. [PMC free article] [PubMed] [Google Scholar]
- 10.Underwood JR, Cartwright GA, McCall AM, Tribbick G, Geysen MH, Hearn Mtw. Monoclonal anti-H1 histone autoantibodies from unimmunized Balb/c mice. Specificity and VH and VL domain sequences. J Autoimmun. 1994;7:291–320. doi: 10.1006/jaut.1994.1022. [DOI] [PubMed] [Google Scholar]
- 11.Underwood JR, McCall AM, Csar Xf. Naturally-occurring anti-thymocyte autoantibody which identifies a restricted CD4+CD8+CD3−/lo/int thymocyte subpopulation exhibits extensive polyspecificity. Thymus. 1996;24:61–8. [PubMed] [Google Scholar]
- 12.Moir RD, Martins RN, Bush AI, et al. Human brain βA4 amyloid protein precursor of Alzheimer's disease: purification and partial characterization. J Neurochem. 1992;59:1490–8. doi: 10.1111/j.1471-4159.1992.tb08465.x. [DOI] [PubMed] [Google Scholar]
- 13.Hollander Z, Katchalski-Katzir E. Use of monoclonal antibodies to detect conformational alterations in lactate dehydrogenase isoenzyme 5 on heat denaturation and on adsorption to polystyrene plates. Mol Immunol. 1986;23:927–33. doi: 10.1016/0161-5890(86)90122-7. [DOI] [PubMed] [Google Scholar]
- 14.Halverson K, Fraser PE, Kirschner DA, Lansbury P. Molecular determinants of amyloid deposition in Alzheimer's disease: conformational studies of synthetic β-protein fragments. Biochemistry. 1990;29:2639–44. doi: 10.1021/bi00463a003. [DOI] [PubMed] [Google Scholar]
- 15.Barrow CJ, Zagorski Mg. Solution structures of β peptide and its constituent fragments: relation to amyloid deposition. Science. 1991;253:179–82. doi: 10.1126/science.1853202. [DOI] [PubMed] [Google Scholar]
- 16.Zagorski MG, Barrow Cj. NMR studies of amyloid β-peptides: proton assignments, secondary structure and mechanism of an α-helix (r) β-sheet conversion for a homologous, 28-residue, N-terminal fragment. Biochem. 1992;31:5621–31. doi: 10.1021/bi00139a028. [DOI] [PubMed] [Google Scholar]
- 17.Burdick D, Soreghan B, Kwon M, et al. Assembly and aggregation properties of synthetic Alzheimer's A4/β amyloid peptide analogs. J Biol Chem. 1992;267:546–54. [PubMed] [Google Scholar]
- 18.Jarret JT, Lansbury P. T Seeding ‘one-dimensional crystallization’ of amyloid: a pathogenic mechanism in Alzheimer's disease and scrapie? Cell. 1993;73:1055–8. doi: 10.1016/0092-8674(93)90635-4. [DOI] [PubMed] [Google Scholar]
- 19.Jarret JT, Costa PR, Griffin RG, Lansbury Pt. Model of the β protein C-terminus: differences in amyloid structure may lead to segregation of ‘long’ and ‘short’ fibrils. J Am Chem Soc. 1993;116:9741–2. [Google Scholar]
- 20.Weinreb PH, Jarret JT, Lansbury Pt. Peptide models of hydrophobic cluster at the C-terminus of the β-amyloid protein. J Am Chem Soc. 1994;116:10835–6. [Google Scholar]
- 21.Ida N, Hartmann T, Pantel J, et al. Analysis of heterogeneous βA4 peptides in human cerebrospinal fluid and blood by a newly developed sensitive Western blot assay. J Biol Chem. 1996;271:22908–14. doi: 10.1074/jbc.271.37.22908. [DOI] [PubMed] [Google Scholar]