Abstract
Immunosenescence involves modifications of humoral and cellular immunity. In a previous study, we have shown a locus-dependent reduction of HLA class-I cell surface expression on peripheral lymphocytes and monocytes with advancing age. Here we report the quantitative analysis of HLA-A and -B transcripts from PBL of 54 healthy subjects aged 21–90 years. Using a competitive RT-PCR method, we observed a significant decrease of HLA-A (P < 0·0001) and -B (P = 0·0025) mRNA contents with increasing age. Secondly, to investigate this locus-dependent alteration of HLA class-I transcription, we performed EMSA using nuclear extracts from PBL of five young (24–31-year-old) and 5 elderly (58–69 years old) donors with locus A and B sequences of the Enh-A as probes. No qualitative variation of EMSA profiles appeared between the two groups of donors with 6 and 4 bandshift for the locus A and B, respectively. Quantitatively, we observed a significant increase of B4 intensity in the elderly group compared to the young group (P < 0·05). These results suggest that the variation of DNA binding protein could contribute to the lower transcription of HLA-A and -B with ageing. These alterations of HLA class-I expression at the transcriptional level could lead to the unresponsiveness of CD8 T cells due to default of antigen presentation with ageing.
Keywords: competitive RT-PCR, HLA transcription, immunosenescence, mobility shift assay, peripheral blood leucocytes
Introduction
Advancing age is associated with a decline in cell-mediated and humoral immune responsiveness [1,2] which involves various mechanisms [3,4]. In spite of the key role of the major histocompatibility complex (MHC) in immune response, data concerning MHC implications in ageing remain scattered.
MHC molecules are highly polymorphic molecules, essential in the human response because they present peptide to T cells. MHC products may be implicated in ageing in two ways: their polymorphism and their capacity to present peptides to T cells. The implication of human leucocyte antigens (HLA) polymorphism on human longevity are controversial: Izaks et al. [5] did not observe any HLA alleles involved in mortality after the age of 85, whereas some data support the direct involvement of HLA class-II alleles in survival at a very old age [6].
MHC class-I-molecules present endogenous peptides to cytotoxic T cells, whereas MHC class-II molecules present exogenous peptides to helper T cells [7]. The recognition of T cell requires cell surface expressions of the T cell receptor and of the peptide-MHC molecules. Any alteration of the expression of one of those components can affect the T cell recognition and therefore the immune response. In senescent mice, the density of TCRαβ remains stable [8] while MHC class-I molecules expression increase on splenocytes [9]. In humans, we have shown a decrease of HLA class-I expression on peripheral T and B lymphocytes and on monocytes with increasing age [10]. The density of one specific peptide-MHC complex requires a minimum of 200 molecules to allow T cell recognition and lysis by cytotoxic T lymphocytes [11]. Therefore, the decreased expression of HLA class-I could contribute to immunological unresponsiveness, especially towards endogenous tumoral and viral antigens. In contrast, we did not observe any modification of HLA class-II (DR) expression on peripheral B cells and monocytes, which are involved in the presentation of antigens to helper-T cells [10]. The decrease of HLA class-I expression was not associated to a modification of expression of other cell differentiation markers such as CD3, CD19 and CD14. Therefore our data show that the loss of HLA class-I expression is a feature of senescence. This observation could be due to different mechanisms: (i) cell shedding, which may explain the increase of soluble HLA-I (sHLA-I) level in the elderly [12]; (ii) modification of assembling and transport; and (iii) alteration of synthesis and its regulation.
As a consequence of their different immunological functions, the control of MHC class-I and class-II gene expression shares some differences but also some similarities [13]. Schematically, MHC class-I and class-II promoters exhibit a common set of regulatory elements. In addition, MHC class-I promoter is distinguished by two cis-acting elements: the Enhancer A (Enh-A) and the interferon stimulating response element (ISRE). MHC class-I gene expression is controlled mainly at the transcriptional level in a locus-specific manner [14]. Notably, HLA-A and -B genes are not tightly co-ordinated in their transcription, especially regarding the Enh-A cis-trans regulation.
The aim of our study was to analyse potent mechanisms involved in the changes of HLA expression on peripheral blood leucocytes (PBL) with ageing: first, we performed a quantification of both HLA-A and -B transcripts from PBL using a competitive RT-PCR procedure. Secondly, we searched for the implication of transcription elements by electromobility shift assays (EMSA) of locus A and B Enh-A sequences using nuclear extracts of PBL from young and elderly donors.
Materials and methods
Quantitative RT-PCR
Blood donors and isolation of PBL
Fifty-four healthy volunteers were included in this study, recruited according to the SENIEUR recommendations [15]. Subjects were aged from 21 to 90 years, consisting of 31 females and 23 males, and were informed of the objectives of the study. Blood (10 ml) was drawn into sterile heparinized tubes and the mononuclear cells were isolated by centrifugation over Ficoll-Hipaque (Eurobio, Les Ulis, France).
RNA extraction and reverse transcription
Total RNA was extracted from PBL using RNA NOW (Ozyme, Montigny-le-Bretonneux, France). cDNA was reverse transcribed from 1 µg of total RNA using an oligo d(T) primer (Pharmacia, Uppsala, Sweden) and 200 U of reverse transcriptase (Life Technologies, Cergy-Pontoise, France) as described previously [16].
Production of internal standards
HLA-A and -B fragments differing from one another in length (257 and 259 pb, respectively) were produced by PCR amplifications of 1 µl of a cDNA sample in 50 µl of Taq 1× PCR buffer containing 200 µm of each dNTP, 100 ng of each primer and 1 U Taq polymerase (Pharmacia). The sense primer, corresponding to the α3 extracellular domain of HLA class-I molecules encoded by the fourth exon was shared by HLA-A and -B genes (P1 : 5′-CTACC CTGCGGAGATCAC-3′). The antisense primers, corresponding to the transmembrane part of HLA class-I molecules encoded by the fifth exon, were designed to be specific for HLA-A or -B genes (P2 : 5′-AGAGAACCAGGCCAGCAAT-3′ and P3 : 5′-TAGGACAGCCAGGCCAGCAACA-3′, respectively) [17] (Fig. 1). Amplifications were carried out using multistep programmes: 1 cycle at 94°C for 5 min, 35 cycles (94°C for 30 s, 54°C (HLA-A) or 56°C (HLA-B) for 30 s and 72°C for 30 s) and 1 cycle at 72°C for 7 min. HLA-A and -B internal standards were obtained by mutagene PCR amplifications of the 257 and 259 bp fragments using a sense primer (P1mut: 5′-CTACCCTGC GGAGATCACCCTGGCAGCGGGATG-3′) and the previous antisense primers (P2 or P3). These later amplifications were used to generate 252 and 254 bp fragments by the deletion of the first five nucleotides following P1. These HLA-A and -B mutated fragments were separated on a 1·2% agarose gel, eluted using a QIAquick Gel Extraction Kit (Qiagen, Hilden, Germany) and cloned into the pCRII-TOPO vector (Invitrogen, Leek, the Netherlands) according to the manufacturer's instructions, and sequenced to check the integrity of the sequence.
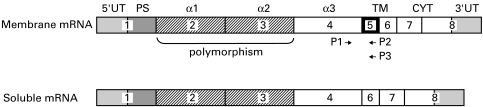
Fig. 1.
Membrane bound and soluble forms of HLA class-I mRNAs. The soluble form results from an alternative splicing of the exon 5 encoding the transmembrane region (TM). The sense primer P1 position is 696–713 in the exon 4 and antisense primers P2 and P3 positions are 934–952 and 633–954 in the exon 5, respectively.
Competitive PCR
Competitive PCR was realized as described previously by Vincent et al. [16]. Briefly, competitive PCR consisted of the co-amplification of 2 µl of cDNA sample and a known quantity of HLA-A or -B internal standard determined by O.D.260nm using a fluorescent P1 as sense primer and P2 or P3 as antisense primers for HLA-A or -B loci. Amplifications were carried out using previously described multistep programmes with a reduced number of amplification cycles (× 25).
Electrophoresis and quantification of the mRNAs
Each amplification was diluted in distilled water (1/50) and 1 µl was mixed with 25 µl of formamide solution and 0·5 µl of ROX-500 (Perkin-Elmer, Foster City, CA, USA) was added as size marker. The resulting mixture was heat denaturated at 95°C for 5 min and loaded on a 4% acrylamide gel and analysed by capillary electrophoresis on an ABI PRISM 310 DNA Sequencer (Perkin-Elmer). The two co-amplified PCR products were separated according to their size which differed by 5 bp. Using the Genescan software, peaks of fluorescence were displayed, allowing the determination of the fluorescence intensities of the PCR products corresponding to the calculated area of the peaks.
Statistical analysis
The correlations between age and HLA-A or HLA-B mRNA levels were calculated to obtain the linear regression curves, the value of the correlation coefficient and the statistical significance in keeping with the Fisher and Yates table.
Electrophoretic mobility shift assay
Blood donors and isolation of PBL
Blood samples (20 ml) were obtained from five young (from 24 to 31 years of age) and five elderly (from 59 to 68 years of age) healthy volunteers (two females and three males in the young group; three females and two males in the elderly group), recruited according to the SENIEUR recommendations [15], and PBL were isolated by centrifugation over Ficoll-Hipaque (Eurobio).
Preparation of nuclear extracts
Nuclear extracts were prepared by the method of Schreiber et al. [18]. Cells were washed with phosphate buffered saline (PBS), resuspended in four packed cell volumes of cold hypotonic buffer (10 mm HEPES pH 7·9; 10 mm KCl; 1·5 mm MgCl2; 0·1 mm EDTA; 0·1 mm EGTA; 1 mm DTT) and left on ice for 20 min. A complete protease inhibitor cocktail (Boehringer-Mannheim, Mannheim, Germany) was added in all the buffers. PBL were lysed with Nonidet P-40 (0·5%) by vigorous agitation for 10 s. Nuclei were pelleted by centrifugation at 1000 g and 4°C for 10 min and resuspended in 2 volumes of cold hypertonic buffer (20 mm HEPES pH 7·9; glycerol 20%; 400 mm NaCl; 1·5 mm MgCl2; 1 mm EDTA; 1 mm EGTA; 1 mm DTT; 0·5 mm PMSF) and rocked vigorously at 4°C for 30 min. Cellular remains were discarded and supernatant was dialysed by centrifugation on a mini-column (Nalgene, 4K MWCO) (Polylabo, Strasbourg, France) and frozen at −80°C. Protein concentration was measured by the Bradford method [19].
Preparation of probes
Hybridization products of single-strand synthetic oligonucleotides (Life Technologies) corresponding to the Enh-A sequence of locus-A (5′-ATGGATTGGGGAGTCCCA GCCTTGGGGATTCCCCAA-3′) and locus-B (5′-CTGCAATGG GGAGGCGCAGCGTT GGGGATTCCCCAC-3′) 5′ extended by a NotI site were used as probes. DNA fragments (200 ng) were end-labelled with [α32P]dCTP (Amersham, Orsay, France) and 4 U of the Klenow fragment of DNA polymerase I (Ozyme).
Gel mobility shift assay
In the binding reaction, labelled DNA (25 000 cpm) was incubated in the presence of 2 µg of the non-specific competitor DNA double-stranded poly[dI-dC] (Pharmacia) and nuclear extract (10 µg) in a total volume of 20 µl of binding buffer (HEPES 20 mm pH 7·9; KCl 100 mm; MgCl2 1 mm; EDTA 1 mm; ZnSO4 10 µm; DTT 1 mm; glycerol 25%). Reactions were carried out at 4°C for 30 min and followed by electrophoresis on a native 6% polyacrylamide gel (acrylamide: bisacrylamide 32 : 1) with buffer (Tris-base 90 mm; boric acid 90 mm; Na2EDTA 2·5 mm) at 25 mA for 1·5 h. The gel was dried, autoradiographed at −80°C and the bandshift intensities were assessed using an InstantImager (Packard Instrument, Meriden, CT, USA).
Statistical analysis
The variation of bandshift intensities with ageing were analysed using the anova test, a level of P < 0·05 was accepted as significant.
Results
Quantitative PCR sensitivity
Successive dilutions of cDNA sample were co-amplified with a known quantity of internal standard (data not shown): the initial quantity of cDNA sample is proportional to the ratio of peak areas (standard/sample) with a correlation coefficient R2 = 0·99. In addition, the intra- and interassays variations were checked for each locus and never exceeded 5%.
Quantification of HLA-A and -B transcripts from PBL during ageing
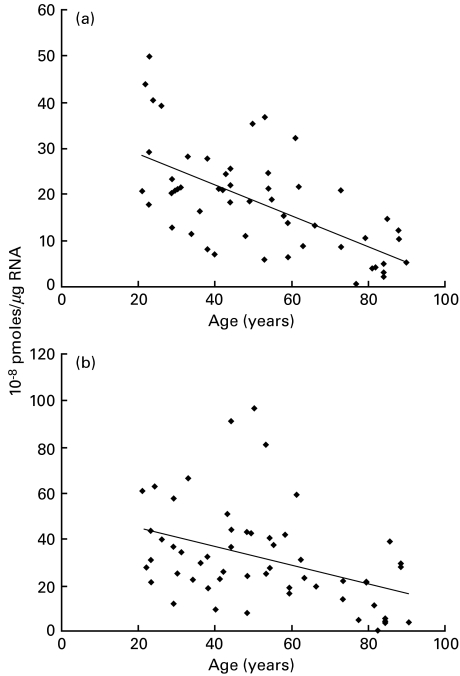
First, the HLA mRNA content is higher in PBL for the HLA-B locus (mean: 32·10−8 pmoles/µg of total RNA) than for the HLA-A locus (mean: 18·10−8 pmoles/µg of total RNA). Secondly, the amounts of HLA-A and -B transcripts are significantly decreased with increasing age (Fig. 2). The linear regression analysis shows variations of HLA-A mRNA (R2 = 0·38; P < 0·0001) (Fig. 2a) and HLA-B mRNA contents (R2 = 0·16; P = 0·0025) (Fig. 2b).
Fig. 2.
Quantification of HLA-A (a) and HLA-B (b) mRNAs from PBLs of 54 healthy subjects aged from 21 to 90 years using competitive RT-PCR procedure. Linear regression analysis shows that the correlation coefficients between age and transcription rate are R2 0·38 with P < 0·0001 (a) and R2 0·16 with P = 0·0025 (b).
Qualitative analysis of EMSA with nuclear extract of PBL from young and elderly
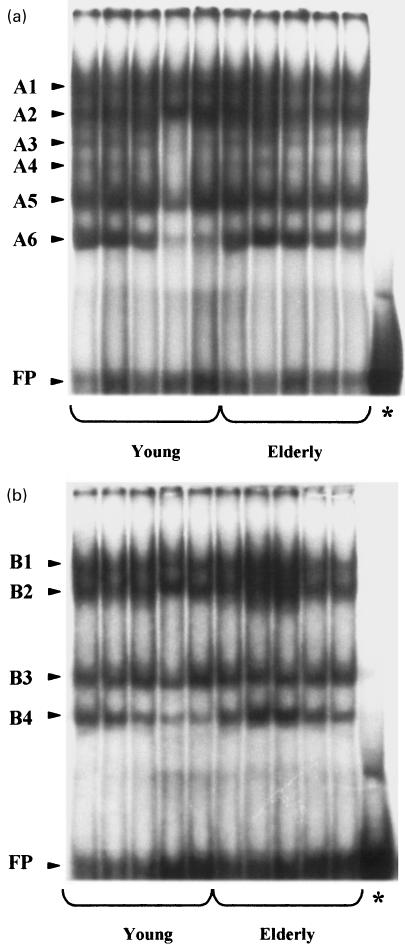
The binding activities of nuclear extracts from PBL on Enh-A probes are shown in Fig. 3. Six bands (four major and two minor) were observed with the nuclear extracts obtained from PBL using the locus A Enh-A sequence (Fig. 3a. These bands were designated as A1, A2, A3, A4, A5 and A6 (in the order of mobility). Concerning the locus B, four major bands were observed using the Enh-A sequence, namely B1, B2, B3 and B4 (Fig. 3b). Concerning the incidence of ageing upon EMSA profiles, no qualitative modification was observed between the young and the elderly groups of donors either with the locus A or the locus B Enh-A probes.
Fig. 3.
EMSA using Enh-A locus A (a) or locus B (b) probes and 10 µg of PBLs nuclear extracts from five young healthy donors aged from 24 to 31 years and five elderly healthy donors aged from 59 to 68 years. FP indicates free probe and (*) indicates unbound DNA probe without nuclear extract. A1–A6 and B1–B4 correspond to the bandshifts in the order of mobility for the locus A and locus B, respectively.
Quantitative analysis of EMSA with nuclear extract of PBL from young and elderly
The mean intensity (cpm) of the Enh-A bandshift is summarized in Table 1. Concerning the locus B bandshift, the B4 band was significantly lower (P < 0·05) in the young group (221·7 ± 44·6 cpm) than in the elderly group of donors (319·3 ± 51·0 cpm) (Table 1b). In contrast, no quantitative variation was noticed with increasing age for the locus A bandshift (Table 1a).
Table 1.
Quantification of the Enh-A locus-A (a) and locus-B (b) bandshifts by PBLs nuclear extracts from five young and five elderly healthy donors
| A1 | A2 | A3 | A4 | A5 | A6 | Total | ||
|---|---|---|---|---|---|---|---|---|
| (a) | ||||||||
| Young | 360·5 ± 23·1 | 342·5 ± 15·1 | 178·8 ± 16·6 | 159·7 ± 16·2 | 355·2 ± 23·6 | 236·2 ± 47·1 | 1633 ± 98 | |
| Elderly | 323·8 ± 45·4 | 324·8 ± 36·7 | 171·7 ± 18·9 | 154·4 ± 15·8 | 343·2 ± 19·6 | 288·7 ± 45·0 | 1607 ± 147 | |
| (b) | B1 | B2 | B3 | B4 | Total | |||
| Young | 450·6 ± 22·0 | 471·2 ± 10·4 | 346·5 ± 40·5 | 221·7 ± 44·6* | 1490 ± 36 | |||
| Elderly | 516·9 ± 99·6 | 539·7 ± 99·4 | 380·3 ± 16·8 | 319·3 ± 51·0* | 1756 ± 255 | |||
Mean cpm ± ESM significantly different by anova at
P < 0·05.
Discussion
The decrease of HLA class-I expression reported previously on lymphocytes and monocytes in the elderly [10] may be secondary to any step required for HLA synthesis, assembling, transport or expression at the cell surface. The fact that modifications of HLA class-I expression exist with locus-dependent specificity evokes the fact that transcriptional and/or post-transcriptional locus specific events can be involved [20]. First, we searched for putative alteration of the transcription rate of HLA-A and -B genes during ageing using a quantitative RT-PCR method [16]. For this purpose, we located the sense primer in the nonpolymorphic sequence of the fourth exon and the antisense primers in the fifth exon encoding the transmembrane region to discard sHLA-I transcripts produced by alternative splicing (Fig. 1).
To validate this quantitative approach, the specific RT-PCR conditions had to be determined. The reverse transcription was conducted as previously described by Vincent et al. [16], which shows no difference in the efficiency of the reaction, allowing correlation of the amounts of cDNAs with the corresponding mRNAs. The competitive PCR method can be used for the quantification only if both the sample sequence and the corresponding internal standard are co-amplified with the same efficiency. For this purpose, we chose to use internal standard not only amplified by the same set of primers as the cDNA sample, but also corresponding to the fragment of interest with a 5 nucleotides deletion. This difference in length is the minimum required to obtain a clear separation of PCR products according to their size. Using this method, we quantified the HLA-A and -B transcripts extracted from PBL of 54 healthy subjects aged from 21 to 90 years. Because the cell surface expression of HLA class I (A and B) decreases in the same way for both lymphocytes and monocytes, we performed our study on the whole PBL.
The first feature is that the HLA mRNA content is higher in PBL for the HLA-B than for the HLA-A. These results match our previous analysis of cell surface expression of HLA-A and -B molecules [10]. Collectively, these data suggest that locus specific HLA class-I molecules expressed on the cell surface correlates with the amount of locus specific mRNA. PBL mRNA profiles were similar to the one obtained on EBV transformed B cells of the donor but differed from the one obtained on various cell lines [21]. HLA class-I locus expression depend of cell type. This cellular differential expression of HLA class-I loci could act on the immune response by preferentially presenting distinct peptides to T cells.
The second feature is a significant decrease in the amounts of HLA-A and -B transcripts with increasing age. The alteration of HLA class-I transcription was slightly more pronounced for the HLA-A locus (− 64%) than HLA-B locus (− 50%) when elderly donors (> 60 years) were compared with young donors (< 30 years).
Lastly, HLA class-I phenotypes were determined by the standard NIH microlymphocytotoxicity method [22]. From this limited number of donors with a homogeneous distribution of HLA alleles during ageing, the modifications of HLA-A and -B transcription were not depending on HLA polymorphism.
Because of the decrease of HLA class-I transcription we report here, we can affirm that the lower HLA class-I surface expression observed previously on PBL during ageing is not due exclusively to cell shedding. Decrease of HLA class-I transcripts could result from mRNA instability and/or modification of the transcriptional control. To assess the incidence of ageing upon the regulation of HLA class-I genes transcription, we performed EMSA using nuclear extracts of PBL from young and elderly donors.
Regulation at the transcriptional level is mediated by several cis-acting regulatory elements in the promoter region of HLA class-I genes [23]. In particular, the enhancer-A presents locus-specific nucleotidic sequences and seems to be essential for the HLA class-I locus-specific transactivation [20,24–26]. We aimed to study the transcription factor pattern able to bind these enhancer-A in PBL from five elderly donors in comparison with five young healthy donors. For this purpose, we performed EMSA using locus A and B Enh-A probes exhibiting locus-specific conservation [27,28].
We have detected several binding activities; 6 and 4 bands were, respectively, observed using the locus-A and -B Enh-A sequences; the results are in agreement with data observed on B cell line and confirmed the fact that HLA class-I locus-A and -B are driven by different cis-trans regulatory mechanisms [14,29].
Concerning the incidence of ageing on EMSA profiles, no qualitative modification was observed between the young and the elderly groups of donors but a quantitative variation was noticed regarding the B4 band with a lower intensity in the young than in the elderly group of donors. Because the EMSA bandshift profile on lymphocytes is not yet clarified fully, we were not able to characterize the modification of the B4 band. Nevertheless, our results show that the Enh-A-mediated transregulation could participate to the down regulation of locus-B, whereas other cis-acting elements and their specific-binding proteins could account for the down-regulation in locus-A [13,30,31].
In conclusion, in present study we searched for modification of the amount of locus specific HLA class-I mRNA from PBL with ageing. With advancing age, we observed a decreased mRNA quantification affecting HLA-A products and to a lesser extent the HLA-B products. Therefore we can deduce that the decrease of transcription contributes to the lower HLA class-I surface expression on PBL with ageing. The mechanisms involved in the lower transcription may be complex: we observed a locus-dependent incidence of ageing upon the Enh-A transactivation, but this observation does not exclude a defect in assembling, transport and excess of cell shedding. Early reports indicated that the CD8+ T-cell repertoire was markedly altered during ageing, whereas the CD4+ T-cell repertoire was unchanged [32]. With regard to the different immunological functions of HLA class-I and class-II products and the differential incidence of age upon their expression, we speculate that the lower expression of HLA class-I molecules during ageing may alter the recognition of some endogenous peptides by the TCR of cytotoxic-T cells and participate to the alteration of the CD8+ cells repertoire. Because of the implication of HLA class-I molecules in viral and tumoral expression, we can propose that alteration of HLA class-I molecules could contribute to the increase of infectious and tumoral disease in the elderly.
Acknowledgments
We are grateful to the laboratory of Histocompatibility (CHRU Dupuytren, Limoges, France) for the determination of HLA phenotypes. This work was supported in part by grants from Région Limousin and Ligue Régionale du Limousin contre le Cancer. We thank Béatrice Desvergne (Lausanne, Switzerland) for her technical help concerning electromobility shift assay.
References
- 1.Segre D, Miller RA, Abraham GN, Weigle WO, Warner HR. Aging and the immune system. J Gerontol. 1989;44:164–8. doi: 10.1093/geronj/44.6.b164. [DOI] [PubMed] [Google Scholar]
- 2.Thoman ML, Weigle WO. The cellular and subcellular bases of immunosenescence. Adv Immunol. 1989;46:221–61. doi: 10.1016/s0065-2776(08)60655-0. [DOI] [PubMed] [Google Scholar]
- 3.Cossariza A, Ortolani C, Monti D, Franceschi D. Cytometric analysis of immunosenescence. Cytometry. 1997;27:297–313. doi: 10.1002/(sici)1097-0320(19970401)27:4<297::aid-cyto1>3.0.co;2-a. [DOI] [PubMed] [Google Scholar]
- 4.Solana R, Pawelec G. Molecular and cellular basis of immunosenescence. Mech Ageing Dev. 1998;102:115–29. doi: 10.1016/s0047-6374(98)00029-3. [DOI] [PubMed] [Google Scholar]
- 5.Izaks G, Van Houwelingen H, Schreuder G, Ligthard G. The association between human leucocyte antigens (HLA) and mortality in community residents aged 85 and older. J Am Geriatr Soc. 1997;45:56–60. doi: 10.1111/j.1532-5415.1997.tb00978.x. [DOI] [PubMed] [Google Scholar]
- 6.Ivanova R, Henon N, Lepage V, Charron D, Vicaut E, Schachter F. HLA-DR alleles display sex-dependent effects on survival and discriminate between individual and familial longevity. Hum Mol Genet. 1998;7:187–94. doi: 10.1093/hmg/7.2.187. [DOI] [PubMed] [Google Scholar]
- 7.Neefjes JJ, Momburg F. Cell biology of antigen presentation. Curr Opin Immunol. 1993;5:27–34. doi: 10.1016/0952-7915(93)90077-6. [DOI] [PubMed] [Google Scholar]
- 8.Wakikawa A, Utsuyama M, Hirokawa K. Altered expression of various receptors on T cells in young and old mice after mitogenic stimulation: a flow cytometric analysis. Mech Ageing Dev. 1997;94:113–22. doi: 10.1016/s0047-6374(97)01880-0. [DOI] [PubMed] [Google Scholar]
- 9.Sidman CL, Luther EA, Marshall JD, Nguyen KA, Roopenian DC, Worthen SM. Increased expression of major histocompatibility complex antigens on lymphocytes from aged mice. Proc Natl Acad Sci. 1987;84:7624–8. doi: 10.1073/pnas.84.21.7624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Le Morvan C, Cogné M, Troutaud D, Charmes JP, Sauvage P, Drouet M. Modification of HLA expression on peripheral lymphocytes and monocytes during ageing. Mech Ageing Dev. 1998;105:209–20. doi: 10.1016/s0047-6374(98)00096-7. [DOI] [PubMed] [Google Scholar]
- 11.Christinck E, Luscher M, Barber B, William D. Peptide binding to class-I MHC on living cells and quantitation of complexes required for CTL lysis. Nature. 1991;352:67–70. doi: 10.1038/352067a0. [DOI] [PubMed] [Google Scholar]
- 12.Le Morvan C, Cogné M, Drouet M. An elevation in the concentration of HLA class-I molecules in human blood due to ageing. Mech Ageing Dev. 2001;122:335–40. doi: 10.1016/s0047-6374(00)00250-5. [DOI] [PubMed] [Google Scholar]
- 13.van den Elsen PJ, Gobin SJP, van Eggermond MCJA, Peijnenburg A. Regulation of MHC class I and II gene transcription: differences and similarities. Immunogenetics. 1998;48:208–21. doi: 10.1007/s002510050425. [DOI] [PubMed] [Google Scholar]
- 14.Soong TW, Hui KM. Locus-specific transcriptional control of HLA genes. J Immunol. 1992;149:2008–20. [PubMed] [Google Scholar]
- 15.Ligthart GJ, Corberand JX, Fournier C, et al. Admission criteria for immuno-gerontological studies in man: the SENIEUR protocol. Mech Ageing Dev. 1984;28:47–55. doi: 10.1016/0047-6374(84)90152-0. [DOI] [PubMed] [Google Scholar]
- 16.Vincent R, Louis P, Gongora C, Papa I, Clot J, Eliaou JF. Quantitative analysis of the expression of the HLA-DRB genes at the transcriptional level by competitive polymerase chain reaction. J Immunol. 1996;156:603–10. [PubMed] [Google Scholar]
- 17.Arnett KL, Parham P. HLA class I nucleotide sequences. Tissue Antigens. 1995;45:217–57. doi: 10.1111/j.1399-0039.1995.tb03124.x. [DOI] [PubMed] [Google Scholar]
- 18.Schreiber E, Matthias P, Muller MM, Schaffner W. Rapid detection of octamer binding proteins with ‘mini-extracts’ prepared from a small number of cells. Nucl Acids Res. 1989;17:6419. doi: 10.1093/nar/17.15.6419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bradford MM. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem. 1976;72:248–54. doi: 10.1016/0003-2697(76)90527-3. [DOI] [PubMed] [Google Scholar]
- 20.Gobin SPJ, Keijsers V, Woltman AM, Peijnenburg A, Wilson L, van den Elsen PJ. Locus specific regulation of HLA class I gene expression. In: Charron D, editor. Genetic diversity of HLA functional and medical implication. II. Paris: Medical and Scientific International Publisher; 1997a. pp. 295–7. [Google Scholar]
- 21.Johnson DR. Differential expression of human major histocompatibility class I loci: HLA-A, -B, and -C. Hum Immunol. 2000;61:389–96. doi: 10.1016/s0198-8859(99)00186-x. [DOI] [PubMed] [Google Scholar]
- 22.Terasaki PI, Bernoco D, Park MS, Ozturk G, Iwaki Y. Microdroplet testing for HLA-A, B, C antigens. Am J Clin Pathol. 1978;69:103–20. doi: 10.1093/ajcp/69.2.103. [DOI] [PubMed] [Google Scholar]
- 23.Le Bouteiller P. HLA class-I chromosomal region, genes, and products: facts and questions. Crit Rev Immunol. 1994;14:89–109. doi: 10.1615/critrevimmunol.v14.i2.10. [DOI] [PubMed] [Google Scholar]
- 24.Mansky P, Brown WM, Park JH, Choi JW, Yang SY. The second κB element, κB2, of the HLA-A class I regulatory complex is an essential part of the promoter. J Immunol. 1994;153:5082–90. [PubMed] [Google Scholar]
- 25.Gobin SJP, Keijsers V, van Zutphen M, van den Elsen PJ. The role of enhancer A in the locus-specific transactivation of classical and nonclassical HLA class I genes by nuclear factor κB. J Immunol. 1998;161:2276–83. 1993. [PubMed] [Google Scholar]
- 26.Girdlestone J, Isamat M, Gewert D, Milstein C. Transcriptional regulation of HLA-A and -B: differential binding of members of the Rel and IRF families of transcription factors. Proc Natl Acad Sci. 1568–72;90:1. doi: 10.1073/pnas.90.24.11568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Cereb N, Yang SY. The regulatory complex of HLA class I promoters exhibits locus-specific conservation with limited allelic variation. J Immunol. 1994;152:3873–83. [PubMed] [Google Scholar]
- 28.Yao Z, Volgger A, Scholtz S, Albert ED. Sequence polymorphism in the HLA-B promoter region. Immunogenetics. 1995;41:343–53. doi: 10.1007/BF00163991. [DOI] [PubMed] [Google Scholar]
- 29.Park JH, Lee HW, Fleischhauer KL, Kim CG, Scheffery M, Yang SY. DNA-binding proteins for transcription enhancing region of HLA class I gene. Tissue Antigens. 1993;42:78–86. doi: 10.1111/j.1399-0039.1993.tb02241.x. [DOI] [PubMed] [Google Scholar]
- 30.Gobin SJP, Peijnenburg A, Keijsers V, van den Elsen PJ. Site α is crucial for two routes of IFNγ-induced MHC class I transactivation: the ISRE-mediated route and a novel pathway involving CIITA. Immunity. 1997b;6:601–11. doi: 10.1016/s1074-7613(00)80348-9. [DOI] [PubMed] [Google Scholar]
- 31.Girdlestone J. Synergistic induction of HLA class I expression by RelA and CIITA. Blood. 2000;95:3804–8. [PubMed] [Google Scholar]
- 32.Pawelec G, Effros RB, Caruso C, Remarque E, Barnett Y, Solana R. T‐cells and ageing (update February 1999) Front Biosci. 1999;4:216–69. doi: 10.2741/pawelec. [DOI] [PubMed] [Google Scholar]