Abstract
The synthetic immunomodulator murabutide (MB) presents multiple biological activities with minimal toxicity in animals and in man. Although MB is known to target cells of the reticuloendothelial system and to regulate cytokine synthesis, the molecular mechanisms underlying several of its biological effects are still largely unknown. In an effort to define cellular factors implicated in the immunomodulatory and HIV-suppressive activities of MB, we have undertaken profiling the regulated expression of genes in human monocyte-derived macrophages (MDM) following a 6-h stimulation with this synthetic glycopeptide. Oligonucleotide microarray analysis was performed on RNA samples of differentiated MDM from four separate donors, using probe sets corresponding to 1081 genes. We have identified, in a reproducible fashion, the enhanced expression of 40 genes and the inhibition of 16 others in MB-treated MDM. These regulated genes belonged to different families of immune mediators or their receptors, transcription factors and kinases, matrix proteins and their inhibitors, ion channels and transporters, and proteins involved in cell metabolic pathways. Additional verification of the regulated expression of selected genes was carried out using Northern blots or the quantification of released proteins in MDM cultures. The profile of MB-regulated genes in MDM provides a molecular basis for some of its previously reported biological activities, and reveals new set of genes targeted by the immunomodulator suggesting potential application in novel therapeutic indications.
Keywords: immunomodulators, monocytes/macrophages, microarray
INTRODUCTION
The advent of gene microarrays has permitted the analysis of the expression pattern of a large number of genes in different disease stages or in response to stimuli. Thus, it has become a valuable tool in deciphering complex regulatory pathways [1, 2]. Large-scale gene expression studies also allows a better understanding of the mode of action of pharmacological compounds as well as the identification of new drug targets [3]. As an example, IFNs belong to a family of clinically important compounds with antiviral, antitumour and immunomodulatory activities [4]. Expression profiling using oligonucleotide arrays following IFN- α, β and γ treatment of the human fibrosarcoma cell line HT 1080 provided not only potential explanations for their differential activities, but also suggested some possible new therapeutic applications [5]. On the other hand, systematic analysis of gene expression has allowed a better understanding of the mechanism of LPS toxicity and revealed new targets for the treatment of septic shock or inflammatory diseases [6, 7]. These studies have highlighted the pleiotropic effects of LPS and IFNs, and provided a molecular basis for the mode of action of these immunomodulatory compounds.
Murabutide (MB) is a safe synthetic immunomodulator derived from muramyl dipeptide (MDP), the minimal bioactive structure of the bacterial cell wall peptidoglycan. Unlike the parent molecule MDP, MB is apyrogenic [8], and has been shown to enhance non-specific resistance to bacterial and viral infections [9], as well as to decrease the lethal effect of LPS in mice [10]. MB was also found to synergize with cytokines of therapeutic interest. For instance, MB enhances the antiviral and anti-inflammatory effects of IFN-α[11, 12] and potentiates the antitumour activity of both IFN-α and IL-2 in murine models [11, 13]. Based on its immunomodulatory activity in the absence of prohibiting side-effects [14], MB was chosen to be clinically evaluated and was found to modulate the release of cytokines without significant production of proinflammatory mediators [15]. Mononuclear phagocytes have been reported to be the major target for muramyl peptides as demonstrated by the capacity of these compounds to regulate macrophage function and the release of monokines [16]. Indeed, in vitro, MB has been shown to activate specific signalling pathways in macrophages [17]. Furthermore, a recent study has demonstrated that MB can inhibit HIV-1 replication dramatically in acutely infected monocyte-derived macrophages (MDM) and dendritic cells [18]. The HIV-suppressive activity was not linked to a direct effect on virus entry or viral enzymes but was associated with the regulation of cellular factors needed at different steps of the virus life cycle [18]. For instance, MB has been reported to induce the activation of the truncated C/EBPβ isoform [17], which is known to inhibit HIV transcription [19]. Additional support for the capacity of MB to regulate the expression of cellular genes, and consequently to inhibit viral replication, has been provided by a recent study using endogeneously infected lymphocytes. Thus, MB-treated CD4+ cells from HIV-infected patients were found to down-regulate the expression of c-myc, a host factor known to be essential for the nuclear transport of preintegration complexes in activated T-lymphocytes [20].
Despite a well-defined immunopharmacological profile, little is known about the molecular mechanisms by which MB promotes its various effects. Therefore, we analysed the modulation of over 1000 genes in human MDM stimulated with MB. The validity of this approach was confirmed by successfully iden-tifying several transcripts known to be regulated by this immunomodulator. The transcript profile obtained using microarrays provided a comprehensive framework for studying the response of MDM to MB. We also identified the regulation of several transcripts which could either explain certain biological effects or suggest new potential therapeutic applications of this compound.
MATERIALS AND METHODS
Preparation of MDM
Monocytes were isolated from buffy coats obtained from healthy donors (Etablissement de transfusion sanguine, Lille, France). PBMC were prepared by Ficoll Hypaque density gradient centrifugation (Amersham Pharmacia Biotech, Orsay, France) and were resuspended in RPMI 1640 medium (Life Technologies, Cergy Pontoise, France) supplemented with 10% heat-inactivated human AB+ serum (obtained from Etablissement de transfusion sanguine, Lille, France) to allow their differentiation to MDM. After overnight adherence to tissue culture flasks (ATGC Biotechnologie, Marne la Vallee, France), monocytes were recovered by gentle scraping and were cultured in six-well plates at 106 cells/ml for 7 days in RPMI 1640 medium containing 10% AB+ serum. MB (N-acetyl-muramyl-l-alanyl-d-glutamine n-butyl ester), provided by ISTAC Biotechnology, Lille, France, was dissolved in PBS at 10 mg/ml stock solution. The absence of endotoxin contaminant (< 6 × 10–2 EU/ml) was verified by the Limulus amoebocyte lysate assay (BioWhittaker France, Fontenay-sous-bois, France). MDM were stimulated with MB at 10 μg/ml for up to 24 h. This concentration was previously found to induce, in vitro, maximal biological effects which could be reproduced in experimental models at a dose of 5–10 mg/kg of body weight [8–13].
Cytokines assays
Cell supernatants from unstimulated and MB-treated MDM were collected and stored at –70°C. Levels of TNF-α, IL-1β, IL-6, IL-8, macrophage inflammatory protein-1 beta (MIP-1β), and RANTES were determined in supernatants of MDM cultures using ELISA kits purchased from R&D Systems (Abingdon, UK).
Preparation of samples for microarray hybridizations
Six hours after stimulation with MB or after culture in fresh medium alone, total RNA was prepared from MDM using RNA plus (Bioprobe Systems, Montreuil, France). RNA samples obtained from cells of four different donors were labelled fluorescently using the two-step procedure according to the supplier’s instructions (fluorescent labelling kit, Clontech, Palo Alto, CA, USA). Briefly, 20 μg of total RNA (treated with 1 U RNase-free DNase, Promega, Charbonniere, France) were reverse transcribed at 48°C for 30 min using the specific cDNA synthesis primer set provided in the kit, 500 U Moloney murine leukaemia virus reverse transcriptase, in presence of dATP, dCTP, dGTP, dTTP and aminoallyl-dUTP in total volume of 50 μl. To monitor the cDNA synthesis reaction, samples were spiked with a synthetic RNA, derived from phage lambda provided with the kit (Clontech). Then, the RNA was digested with 5 U RNase h at 37°C for 15 min, and the reaction mix was treated with QuickClean resin provided in the kit. The cDNA was precipitated by addition of 5·5 μl 3 m sodium acetate and 137·5 μl 100% ethanol at – 20°C for 1 h. Then in two experiments, the monofunctional N-hydroxysuccinimide- (NHS)-activated fluorescent dye Cy3 was added to the cDNA synthesized from RNA isolated from untreated MDM, whereas the NHS-activated fluorescent dye Cy5 was added to the cDNA synthesized from RNA isolated from MB-treated MDM. To ensure that the differences observed were not due to differences in the intrinsic fluorescence intensity of the fluorochromes or to variability in NHS-Cy5 or NHS-Cy3 incorporation into cDNA, the dyes were interchanged in two other experiments. NHS-Cy5 or NHS-Cy3 were both from Amersham Pharmacia Biotech, Orsay, France. In addition, a synthetic aminoallyl-modified oligonucleotide provided in the kit (Clontech) was used to monitor the labelling reaction. The purified labelled samples along with appropriate controls were hybridized to a Human Glass Array 1·0 (Clontech) for 16 h at 50°C. After washes, the dried microarray slides were scanned using Affymetrix 418 Array Scanner (Affymetrix, Santa Clara, CA, USA).
Data analysis
Data were collected after laser scanning and pixel levels of fluorescence were analysed using ScanAlyse 2·44 software (developed by M. Eisen, Stanford University, CA, USA). Typically, the observed fluorescence ranged between 0 and 55 000 arbitrary fluorescence units. Raw data were imported into a Microsoft Excel database for analysis. The corrected mean pixel intensities from Cy3 (Ig) and Cy5 (I’g) were used to calculate ratio of gene expression (g). The mean background fluorescent level for Cy3 (BG) and Cy5 (BG′) were determined on empty spots. These mean background values were subtracted from the fluorescence level obtained for each gene spot ((Ig-BG) and (I′g-BG′) for Cy3 and Cy5, respectively). A threshold of 20 arbitrary fluorescence units was assigned to any gene with a calculated fluorescence intensity below 20. The fluorescence level of five different housekeeping genes (hg) coding for GAPDH, hypoxanthine-guanine phosphoribosyltranspherase (HPRT), phospholipase A2 (PLA2) and the 60S and 40S ribosomal proteins was calculated similarly ((Ihg-BG) and (I′hg-BG′) for Cy3 and Cy5, respectively). The ratio of Cy3/Cy5 intensities was calculated for each gene represented on the array with respect to the fluorescence level of the five housekeeping genes according to the formulae: Cy3 = (Ig-BG)/(Ihg-BG)] and Cy5 = (I′g-BG′)/(I′hg-BG′)]. Then, the mean of the five ratios was used for subsequent comparisons. Arbitrarily, we considered any gene with a mean ratio >1·5 to be positively regulated following MB stimulation, and to be negatively regulated with a mean ratio < 0·7 in at least two different MDM preparations.
Northern blot analysis
Total RNA isolated from MDM, treated or not with MB, were electrophoresed under denaturing conditions in 1% agarose/formaldehyde gel and transferred onto a Hybond N+ nylon membrane (Amersham Pharmacia Biotech). RNA was UV cross-linked and prehybridized in a hybridization oven for 4h at 60°C in prehybridization solution and processed as described previously [21]. Hybridization was performed with 32P-labelled oligonucleotide probes (synthesized by MWG-Biotech, Courtaboeuf, France) specific for ninjurin-1 (NINJ-1), glucose transporter 1 (GLUT-1) or heat shock protein 70 kDa (HSP70). The sequences of the probes used were: NINJ-1: 5′-CGTCCAGCATGCTCTCCGCCACGCTCTTCTTG-3′; GLUT-1: 5′-CATCTGCCGACTCTCTTCCTTCATCTCCTG-3′ and HSP70: 5′-GTAGAATGCCTTGGTCTCCCCCTTGTAG-3′. Oligonucleotides were labelled at the 3′ ends with α-32P] dCTP (Amersham Pharmacia Biotech) using terminal transferase (Roche, Meylan, France). Following hybridization, membranes were exposed for 24–72 h at – 70°C to X-Omat Kodak film using intensifying screens. Membranes were stripped off in 0·1% SDS at 75°C for 1 h and were rehybridized with a GAPDH probe to control for equal loading of RNA. The signal intensity of each band was quantified using Kodak Digital Software and was normalized with respect to GAPDH band intensity.
Statistical analysis
Statistical analyses of the differences in cytokine and chemokine levels observed following treatment with MB were carried out using Wilcoxon matched-pair test. P-values < 0·05 were considered statistically significant.
RESULTS
Microarray analysis of MB-regulated genes
To identify changes in gene expression triggered by stimulation with the immunomodulator, we prepared fluorescence-labelled cDNA samples from unstimulated and MB-treated MDM. We performed microarrays experiments on RNA isolated from MDM prepared from four different donors, and exchanged the dyes in two experiments. The reproducibility of the detection of gene expression by microarrays was additionally confirmed in two experiments in which samples from the same donor were reanalysed after having interchanged the dyes. An identical profile of regulated genes was observed in these experiments demonstrating the independent nature of the results from the type of fluorochrome used to label the cDNA. Results derived from the four independent donors are summarized in Tables 1 and 2. We considered that a gene was up- or down-regulated following MB stimulation if a ratio > 1·5 or < 0·7, respectively, was observed in cells from at least two of the four donors tested. Thus, comparison of gene expression patterns between untreated and MB-treated MDM revealed that 56 transcripts were modulated, of which 40 showed increased expression in MB-treated MDM (Table 1). Conversely, 16 transcripts were down-regulated by MB (Table 2). Seven of the 56 transcripts were found to be modulated in all samples; 38 in three of the four samples, and 11 in only two samples. As expected, transcripts modulated by MB consisted of genes encoding cytokines, chemokines and their receptors. Among these were transcripts for genes that have been shown previously to be up-regulated following MB treatment such as IL-1β and IL-8 [22]. These results further demonstrated the validity of the micoarrays approach. Genes coding for MIP-1β and IL-8 were among those which showed the strongest increase in expression following MB stimulation, and this phenomenon was observed in the four experiments performed. MB treatment also up-regulated the expression of genes coding for cytokine receptors such as IL-1RII, while the expression of the chemokine receptor CXCR4 was down-regulated.
Table 1.
Microarray analysis of genes up-regulated in MDM following a 6-h stimulation with MB
| Regulated gene expression | |||||
|---|---|---|---|---|---|
| Ratio | |||||
| Genes | GenBank accession number | Abbreviation | Mean | Range | Frequency (out of 4) |
| Cytokines, chemokines and growth factors | |||||
| Macrophage inflammatory protein-1beta | J04130 | MIP-1β | 18·2 | 3·1–37·4 | 4 |
| Interleukin-8 | Y00787 | IL-8 | 12·9 | 2·7–23·6 | 4 |
| RANTES, small inducible cytokine A5 | M21121 | SCYA5 | 12·1 | 6·5–17·8 | 2 |
| Glial activating factor, fibroblast growth factor-9 | D14838 | FGF-9 | 10·5 | 1·9–20·7 | 3 |
| Bone morphogenetic protein-8, osteogenic protein 2 | M97016 | BMP-8 | 7·2 | 3·8–12·1 | 3 |
| Fibroblast growth factor-6 | X6345 | FGF-6 | 3·8 | 3·0–4·9 | 3 |
| IL-1 receptor accessory protein | AF029213 | IL-1RAcP | 3·1 | 2·3–3·8 | 2 |
| Interleukin-1beta | K02770 | IL-1β | 2·5 | 2·3–2·8 | 3 |
| Receptors | |||||
| Purinoreceptor P2 × 1, ATP receptor P2 × 1 | X83688 | P2 × 1 | 12·3 | 11·1–13·4 | 2 |
| Leukosialyn, sialophorin | J04536 | CD43 | 4·7 | 2·2–9·2 | 3 |
| Purinergic receptor P2Y5 | AF000546 | P2Y5 | 4·5 | 2·1–7·2 | 3 |
| IL-1 receptor type II | X59770 | IL-1RII | 4·3 | 3·2–4·9 | 3 |
| IL-6 receptor-alpha | X12830 | IL-6Rα | 3·4 | 2·8–4·0 | 2 |
| Gamma-aminobuturic acid A receptor epsilon subunit | U66661 | GABRE | 2·8 | 2·1–3·7 | 3 |
| Transcription factors/DNA binding | |||||
| NF-κB p-100 subunit | X61498 | NF-kB p100 | 6·7 | 3·1–10·4 | 2 |
| Interferon regulatory factor-2 | X15949 | IRF-2 | 2·5 | 2·2–2·9 | 3 |
| Cyclins/kinases and inhibitors | |||||
| cdc 27 | U00001 | CDC27HS | 15·2 | 6·2–22·7 | 3 |
| Extracellular regulated kinase-2 | M84489 | ERK-2 | 3·8 | 2·4–5·9 | 3 |
| GAP-associated protein | U17032 | ARHGAP5 | 3·3 | 2·3–4·7 | 3 |
| cAMP-dependent protein kinase beta catalytic subunit | M34181 | PKAC-β | 2·1 | 1·9–2·4 | 3 |
| Matrix proteins/enzymes and inhibitors | |||||
| Matrix metalloproteinase 3, stromelysin 1 | X05232 | MMP3 | 11·3 | 9·1–13·6 | 2 |
| Collagen 4 alpha 3 subunit | M92993 | COL4A3 | 7·7 | 2·8–14·8 | 3 |
| Tissue inhibitor of metalloproteinase 4 | U76456 | TIMP4 | 3·2 | 1·8–5·2 | 3 |
| Cytoskeletal 14 keratin, cytokeratin 14 | J00124 | CK14 | 2·8 | 1·7–5·1 | 3 |
| Matrix metalloproteinase 13, collagenase 3 | X75308 | MMP13 | 2·2 | 1·9–2·5 | 2 |
| Ion channels and transporters | |||||
| Na+/K+ transporting ATPase beta3 subunit | U51478 | ATPB3 | 7·8 | 4·7–9·9 | 3 |
| Organic cation transporter 1 | U77086 | OCT-1 | 7·2 | 1·9–17·4 | 3 |
| Glucose transporter 1, solute carrier 2 member 1 | K03195 | GLUT-1 | 2·7 | 2·6–2·9 | 2 |
| Adhesion molecules | |||||
| Ninjurin-1 | U72661 | NINJ-1 | 13·7 | 2·1–35·2 | 4 |
| Neural cell adhesion molecule L1 | M74387 | L1CAM | 4·1 | 2·5–6·2 | 3 |
| Apoptosis and proteolytic systems | |||||
| Plasminogen | X05199 | PLG | 12·9 | 11·4–14·3 | 2 |
| Casper | AF010127 | CASH-α,β | 2·7 | 2·2–3·7 | 3 |
| Proteasome component C9 | D00763 | PSMA4 | 2·6 | 1·8–3·1 | 3 |
| Metabolic pathways | |||||
| Alpha 1-acid glycoprotein, orosomucoid 1 | X02544 | ORM1 | 6·8 | 2·4–15·4 | 3 |
| Heat shock protein 70 kDa | M11717 | HSP70 | 4·8 | 3·1–5·7 | 3 |
| Ferrochelatase, heme synthetase | D00726 | 4·1 | 1·8–8·5 | 3 | |
| Cytochrome P450 2F1 | J02906 | CYP2F1 | 3·3 | 2·1–4·9 | 3 |
| Quinone oxidoreductase, beta-crystallin | L13278 | CRYZ | 3·2 | 2·5–4·1 | 3 |
| Other proteins | |||||
| Insulin-like growth factor-binding protein 5 | M65062 | IGFBP-5 | 4·6 | 1·9–9·6 | 3 |
| B94 protein | M92357 | TNFAI | 3·8 | 2·7–4·8 | 3 |
Table 2.
Microarray analysis of genes down-regulated in MDM following a 6-h stimulation with MB
| Regulated gene expression | |||||
|---|---|---|---|---|---|
| Ratio | |||||
| Genes | GenBank accession number | Abbreviation | Mean | Range | Frequency (out of 4) |
| Receptors | |||||
| Low density lipoprotein receptor related protein 1 | X13916 | LRP-1 | 0·2 | 0·1–0·3 | 3 |
| Serotonin receptor, 5-hydroxytryptamine 1D receptor | M89955 | HTR1D | 0·2 | 0·1–0·3 | 3 |
| Vascular endothelial growth factor type II receptor | X61656 | VEGFR2 | 0·3 | 0·1–0·4 | 2 |
| CXC chemokine receptor type 4 | D10924 | CXCR4 | 0·5 | 0·4–0·6 | 3 |
| Transcription factors/DNA binding | |||||
| PAX3/FORKHEAD | U02368 | FKHR | 0·3 | 0·2–0·4 | 4 |
| Nuclear factor 90 | U10324 | NF90 | 0·4 | 0·3–0·5 | 4 |
| Cyclins/kinases and inhibitors | |||||
| Cyclin-dependent kinase 4 inhibitor 2, p16-INK4 | L27211 | CDKN2 | 0·2 | 0·1–0·3 | 3 |
| c-Jun N-terminal kinase 2 | L31951 | JNK-2 | 0·3 | 0·1–0·6 | 3 |
| Cyclin dependent kinase 5 activator | X80343 | CDK-5A | 0·3 | 0·1–0·6 | 3 |
| Calmodulin-1 (phosphorylase kinase, delta) | D45887 | CALM | 0·6 | 0·5–0·6 | 3 |
| Apoptosis and proteolytic systems | |||||
| Cysteine protease | D55696 | 0·2 | 0·2–0·3 | 3 | |
| Caspase 2, ICH-1 L protease + ICH-1S protease | U13021 + U13022 | CASP-2 | 0·3 | 0·1–0·4 | 3 |
| Other proteins | |||||
| Delta-like protein | U15979 | DLK | 0·2 | 0·1–0·4 | 4 |
| DNA damage/repair/recombination | U12134 | RAD52 | 0·3 | 0·1–0·5 | 4 |
| Apolipoprotein E | M12529 | APOE | 0·4 | 0·3–0·5 | 2 |
| Plexin-related protein | U52111 | PLXR | 0·5 | 0·4–0·5 | 3 |
Importantly, analysis of the microarray experiments showed that MB also modulated the levels of transcripts encoding proteins with diverse functions. In particular, MB treament exerted a positive effect on the levels of mRNAs coding for proteins involved in the regulation of transcription such as that of interferon regulatory factor (IRF)-2 (Table 1). MB treatment also led to the activation of genes coding for proteins involved in intracellular signalling including the extracellular signal regulated kinase (ERK)-2. The transcription of certain genes involved in matrix modelling was affected positively following MB stimulation. In this group, the gene modulated most consistently was the collagen 4 alpha subunit 3. Among the genes coding for adhesion molecules, the expression of NINJ-1 was consistently and strongly activated by MB treatment. Also, in three of four experiments MB treatment increased the expression of genes coding for growth factors, including fibroblast growth factor (FGF)-9, bone morphogenetic protein (BMP)-8 and insulin like growth factor-binding protein (IGFBP)-5. Up-regulation of genes coding for membraneous transporters such as organic cation transporter (OCT)-1 and glucose transporter (GLUT)-1 also occurred following MB treatment. Conversely, exposure of MDM to MB led to a decrease in the expression of a set of genes (Table 2). Some of the consistently down-regulated genes included those encoding nuclear factors such as NF90 or RAD52, the latter being a protein involved in DNA repair. In addition, genes encoding kinases such as c-Jun N-terminal kinase (JNK)-2, cyclin dependent kinase-5 activator (CDK-5 A), were down-regulated in MB-treated MDM. Several receptors, such as the low density lipoprotein receptor related protein (LRP)-1 and the serotonin receptor (HTRD1), were also found to be down-regulated following MB treatment. In addition, stimulation of MDM with MB led to a decrease in the expression of transcripts encoding proteins involved in proteolysis such as cysteine protease and caspase-2. Thus, treatment of MDM with the immunomodulator led to changes in the transcription of a wide array of genes, many of which have never been previously reported to be either regulated by MB or linked to a specific biological activity of the molecule.
Northern blot analysis of MB-modulated genes observed by microarray
We further confirmed by Northern blot analysis the regulation of a panel of genes following MB stimulation of MDM over a 24-h period (Fig. 1). We selected genes which have been shown to be up-regulated in two (GLUT-1), three (HSP70) or four (NINJ-1) of the four microarray experiments, and which had not been reported previously to be regulated by MB. MDM from three additional donors were stimulated for different periods of time with MB before RNA was extracted. As shown in Fig. 1, NINJ-1, HSP70 and GLUT-1 transcript levels increased following MB stimulation. NINJ-1 transcript level peaked at 4 h, whereas the level of the other transcripts peaked later, at 6 h. Twenty-four hours after treatment, the levels of the transcripts analysed returned to unstimulated levels except for that of HSP70 mRNA which remained elevated (data not shown).
Fig. 1.
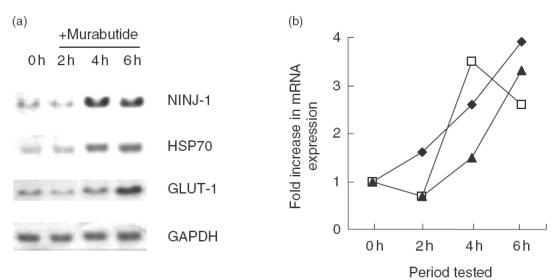
Kinetics of gene induction in MDM following stimulation with MB. (a) RNA samples (20 μg) prepared from MDM that were either left untreated (0 h) or stimulated with MB (2, 4 and 6 h) have been analysed by Northern blot using 32P-labelled oligonucleotide probes specific for NINJ-1, HSP70 or GLUT-1. The same membrane was hybridized with a GAPDH cDNA probe. (b) graphical representation of the changes in NINJ-1, HSP70 and GLUT-1 gene expresssion following MB stimulation. Autoradiographs were analysed by densitometry and the relative mRNA levels of each gene were normalized to those of GAPDH (results shown are of one representative experiment of three independent ones). ✦, HSP70; □, NINJ-1; ▴, GLUT-1.
Profile of secreted cytokines and chemokines in MDM culture
One of the caveats of the microarray experiments and indeed of any technique measuring transcript levels is that the level of a particular mRNA may not necessarily correspond to protein level. To gain further insight into the response of MDM to MB, we determined the level of some cytokines released in the culture medium of MDM treated for 24 h with MB, using cell cultures obtained from six separate donors. As shown in Fig. 2, secreted levels of the proinflammatory cytokines IL-6 and IL-8, as well as of the chemokine MIP-1β were found to be elevated significantly (P < 0·05) following MB treatment. A good correlation was obtained between the increase in cytokine transcript levels observed in the microarray experiments and the subsequent events leading to the secretion of the corresponding proteins in the medium. Only cytokines secreted several folds over background levels, such as MIP-1β and IL-8 were effectively those whose transcript levels were found to be increased in the microarray experiments. In addition, the variability between donors observed in the microarray analysis was also observed in the detection of cytokines secreted by MB-treated MDM. As an example, RANTES was found to be up-regulated at the transcript level in 2 out of 4 microarray experiments (Table 1), and at the level of protein secretion in 3 out of 6 experiments (Fig. 2).
Fig. 2.
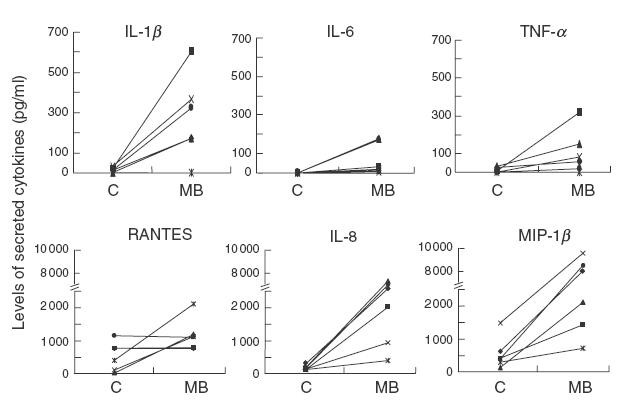
Profile of cytokines released in culture supernatants of MDM left untreated (c) or stimulated with 10 μg/ml MB for a 24-h period. The levels of cytokines and chemokines in the culture supernatants of MDM derived from six different donors were determined using commercially avalaible ELISA kits.
DISCUSSION
The synthetic immunomodulator MB has been selected for clinical development from over 200 MDP derivatives, based on a favourable activity to toxicity ratio [15]. Despite its interesting biological activities, the mode of action of this compound is not yet fully understood. Recently, some of the early intracellular events following stimulation of MDM with MB have been described and include the selective activation of specific kinases and of transcription factors [17]. However, the consequences of the activations of these cellular factors have not been studied in detail. Therefore, gene array technology was used to examine the differential gene expression in MDM stimulated with MB, with the aim to explain some of the biological effects of MB and to gain insight into new potential therapeutic applications.
In our gene array approach, expression of all individual genes, within every experiment, relative to each of the five housekeeping genes was highly concordant with less than 1·5-fold variation. This indicated that there was no need for further normalization of data against a set of most expressed genes. To further strengthen the confidence in the observed profile of regulated genes, a mean change in relative expression of > 1·5- or < 0·7-fold was accepted if such a change could be attained in at least four of the five calculated ratios against the five different housekeeping genes. In addition, a significant down-regulation (<0·7-fold) in gene expression was considered to be valid when the baseline expression level, in the absence of MB, could be detected at a minimum net value of 50 fluorescence units. Moreover, data were only considered significant, and subsequently reported in Tables 1 and 2, when the set threshold for regulated expression (>1·5- or < 0·7-fold) was achieved reproducibly in cells from at least two of the four tested donors. This stringency in our selection of significant variations was aimed to reduce potential artefacts which are frequently attributed to common variations between experiments or between cells from different donors.
As expected for a compound with immunomodulatory activity, we found that MB modulates the expression of a set of cytokine-related genes in vitro. We have recently reported the induction of MIP-1β and IL-1 receptor antagonist (IL-1Ra) and the lack of detection of both IL-1β and IL-8 following administration of MB to healthy volunteers [23]. Interestingly, MDM released higher levels of IL-8 and IL-1β following MB treatment whereas these two cytokines were not detectable in sera of healthy volunteers who received this immunomodulator [23]. This difference cannot be accounted for by the lack of responsiveness of monocytes as in vitro these cells do secrete large amounts of IL-8 following stimulation with MB [22]. Thus, at present, we cannot provide an explanation for this difference which will be the subject of future studies. With respect to IL-1β, which is one of the most potent endogenous pyrogens [24], although we cannot rule out the secretion in vivo of minute amounts of this cytokine, undetectable with the kits used, the absence of pyrogenicity of MB [15] argues against this possibility.
We also found that a cluster of genes coding for proteins involved in signal transduction pathways was modulated following MB stimulation. In particular, the expression of JNK-2 was down-regulated whereas ERK-2 and the catalytic subunit of the cAMP-dependent protein kinase (PKA) transcript levels were increased. Of note, none of these genes have been reported to be modulated at transcriptional level by LPS [25]. It is generally considered that JNK activation sends pro-apoptotic signals and that ERK activation leads to cell survival [26]. In addition, PKA activates ERK and is linked to G-protein coupled receptors [27], a large family of receptors which comprises chemokine receptors. Thus, following MB stimulation, the change in JNK-2, PKA and ERK-2 gene transcription may influence the response of MDM to chemokines induced by MB treatment itself.
Recently, it has been demonstrated that MB possesses the ability to suppress HIV replication in vitro and that this activity was mediated, in part, through the modulation of the expression of cellular genes involved in pro-viral DNA integration and/or virus transcription [18]. In the microarray experiments, we observed an increase in the transcript levels of p100. p100 is a member of the NF-κB family of transcription factors which plays an important role in HIV-1 transcription [28]. It has been decribed that p100 potently inhibits the transactivation of the HIV-1 long-terminal repeat mediated by the viral gene product Tat [29]. Thus, the modulation of p100 expression may participate in the inhibitory effect of MB on HIV infection and replication.
One of the important biological properties of MB is the ability of this compound to synergize with cytokines of therapeutic interest, and particularly with type I IFN. IFNs exert multiple biological effects through the induction of many genes, of which over 30 are encoding proteins with antiviral and immunomodulatory functions [5]. The mechanism of the synergy between MB and type I IFN has not yet been elucidated fully. A previous study has shown an increase in STAT-1 DNA binding activity in MB-treated MDM [17], a transcription factor involved in the response to IFN. However, this synergy may also be linked to an increased expression of some of the IFN-inducible genes triggered by MB. Interestingly, we found an increased expression of the transcription factor IRF-2. In response to IFN stimulation, IRF-2 exerts both positive and negative effects. It has been shown recently that IRF-2 is required for the generation of normal Th1 response in vivo[30]. Moreover, IRF-2 was found to be required for IFN-α-induced antiviral protection in vitro[31]. Thus, induction of IRF-2, along with STAT-1 activation by MB, could explain partly the synergistic effects observed when this compound is associated with type I IFN.
Results of the microarray experiments highlighted also the ability of MB to up-regulate the expression of certain genes, including HSP70, presenting some immunomodulatory effects. Complexes of HSP70 and peptides derived from either tumour cells or virus-infected cells trigger a strong immune response directed against tumour or infected cells [32, 33]. Therefore, the increase in HSP70 levels triggered by MB may lead to improved immune responses in the context of cancer or viral diseases and may also be involved in the adjuvant effect of the immunomodulator.
Interestingly, MB treatment resulted in the modulation of a set of transcripts encoding various transporters, which may have an impact on the functions of MDM. As an example, stimulation with MB leads to an increase in GLUT-1 transcripts, the only glucose transporter expressed in MDM [34]. It is well known that the activity of different cell types, including phagocytic cells, is regulated by glucose supply [35]. Indeed, depletion of glucose level leads to an impaired cytokine secretion by LPS-stimulated macrophages [36]. Thus, the putative increase in GLUT-1 molecules at the cell surface following MB treatment would lead to an increase in glucose uptake, thereby improving the capacity of MDM to mount a vigorous response.
One of the important applications of the microarray analysis is the identification of new pathways for a candidate drug, leading to novel therapeutic strategies. Unexpectedly, transcripts coding for growth factors involved in bone formation, such as BMP-8 and insulin-like growth factor-binding protein 5 (IGFBP-5) were found to be up-regulated by MB. IGFBP-5 is a member of the insulin-like growth factor (IGF) system which controls growth in many tissues and has been shown to activate the proliferation of osteoblasts in vitro[37]. Accordingly, systemic cutaneous injection of IGFBP-5 has been reported to stimulate bone formation [37]. Importantly, a reduced bone mineral content was found among HIV-infected patients, along with lower serum levels of osteocalcin, highlighting a reduced rate of bone formation. The severity of these defects were also correlated with lower CD4+ T cell counts, thus suggesting that the decrease in bone formation is mainly due to mechanisms of the impaired immune defence in HIV-infected patients [38]. Therefore, the increase in IGFBP-5 following MB stimulation could potentially be beneficial in diseases in which the balance between bone formation/resorption is impaired, particularly in chronic HIV-1 infection.
Among the genes modulated by MB, we observed a cluster of genes known to be involved in functions of the nervous system, including FGF-9, NINJ-1, HTR1D and the neural cell adhesion molecule L1 (L1CAM). Effects of muramyl peptides on the functions of the central nervous system have been observed, including pyrogenic and somnogenic activities [39]. However, in contrast to most muramyl peptides, MB is devoid of both activities [8]. Thus, this is the first observation suggesting a potential interaction between MB and the nervous system. NINJ-1 is a cell-membrane associated protein whose expression is up-regulated after nerve injury in dorsal root ganglion neurone and in Schwann cells [40]. Macrophage activation and migration at the site of nerve injury in the peripheral nervous system or in the central nervous system appear to play an important role in promoting nerve regeneration [41]. In addition, LPS administration to rats that underwent transection of dorsal roots resulted in activation of macrophage/microglia and in an enhanced response to degenerating axon in the central nervous system [42]. As NINJ-1 and L1CAM are up-regulated by MB in macrophage, this may potentially be of relevance to the overall mechanism leading to nerve regeneration.
In conclusion, microarray analysis has shed light on the mode of action of the safe immunomodulator MB. In addition, these experiments have allowed the generation of new hypotheses aimed at explaining the antiviral activity of MB and the molecular events underlying the synergy between MB anf IFN. Importantly, this work has also suggested new therapeutic indications for this compound that warrant further investigation. Most interestingly, the observed profile of regulated genes by the non-specific immunomodulator, known to act mainly on elements of innate immunity, provides a new framework for understanding and evaluating the approach of non-specific immunotherapy for the treatment of chronic viral infections and tumours. Finally, analysis by microarrays could be highly efficient in explaining the differences in immunopharmacological and biological properties between MDP and the safe derivative MB. Such differences may well be linked to a different profile of gene regulation in target cells and further studies will be needed to dissect the structure–function relationship among these analogues.
Acknowledgments
This work was supported by a grant from the Association Stop SIDA (Lille, France) and by the Director’s initiative fund at the Pasteur Institute in Lille. We are grateful to Y. Lemoine, D. Hot and L. Huot for their valuable help in the use of the laser fluorescence scanner and in the analysis of results.
References
- 1.Perou CM, Sorlie T, Eisen MB, et al. Molecular portraits of human breast tumours. Nature. 2000;406:747–52. doi: 10.1038/35021093. [DOI] [PubMed] [Google Scholar]
- 2.Gasch AP, Spellman PT, Kao CM, et al. Genomic expression programs in the response of yeast cells to environmental changes. Mol Biol Cell. 2000;11:4241–57. doi: 10.1091/mbc.11.12.4241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Marton MJ, DeRisi JL, Bennett HA, et al. Drug target validation and identification of secondary drug target effects using DNA microarrays. Nat Med. 1998;4:1293–301. doi: 10.1038/3282. [DOI] [PubMed] [Google Scholar]
- 4.Baron S, Tyring SK, Fleischmann WR, Jr, et al. The interferons. Mechanisms of action and clinical applications. JAMA. 1991;266:1375–83. doi: 10.1001/jama.266.10.1375. [DOI] [PubMed] [Google Scholar]
- 5.Der SD, Zhou A, Williams BR, Silverman RH. Identification of genes differentially regulated by interferon alpha, beta, or gamma using oligonucleotide arrays. Proc Natl Acad Sci USA. 1998;95:15623–8. doi: 10.1073/pnas.95.26.15623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hancock RE, Scott MG. The role of antimicrobial peptides in animal defenses. Proc Natl Acad Sci USA. 2000;97:8856–61. doi: 10.1073/pnas.97.16.8856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hashimoto S, Suzuki T, Dong HY, Yamazaki N, Matsushima K. Serial analysis of gene expression in human monocytes and macrophages. Blood. 1999;94:837–44. [PubMed] [Google Scholar]
- 8.Chedid LA, Parant MA, Audibert FM, et al. Biological activity of a new synthetic muramyl peptide adjuvant devoid of pyrogenicity. Infect Immun. 1982;35:417–24. doi: 10.1128/iai.35.2.417-424.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Parant M, Chedid L. Stimulation of non-specific resistance to infections by synthetic immunoregulatory agents. Infection. 1984;12:230–4. doi: 10.1007/BF01640913. [DOI] [PubMed] [Google Scholar]
- 10.Parant MA, Pouillart P, Le Contel C, Parant FJ, Chedid LA, Bahr GM. Selective modulation of lipopolysaccharide-induced death and cytokine production by various muramyl peptides. Infect Immun. 1995;63:110–5. doi: 10.1128/iai.63.1.110-115.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bahr GM, Pouillart PR, Chedid LA. Enhancement in vivo of the antiinflammatory and antitumor activities of type I interferon by association with the synthetic immunomodulator murabutide. J Interferon. 1996;16:297–306. doi: 10.1089/jir.1996.16.297. [DOI] [PubMed] [Google Scholar]
- 12.Pouillart PR, Audibert FM, Chedid LA, Lefrancier PL, Bahr GM. Enhancement by muramyl peptides of the protective response of interferon-alpha/beta against encephalomyocarditis virus infection. Int J Immunopharmacol. 1996;18:183–92. [Google Scholar]
- 13.Bahr GM, Darcissac E, Pouillart PR, Chedid LA. Synergistic effects between recombinant interleukin-2 and the synthetic immunomodulator murabutide: selective enhancement of cytokine release and potentiation of antitumor activity. J Interferon Cytokine Res. 1996;16:169–78. doi: 10.1089/jir.1996.16.169. [DOI] [PubMed] [Google Scholar]
- 14.Telzak E, Wolff SM, Dinarello CA, et al. Clinical evaluation of the immunoadjuvant murabutide, a derivative of MDP, administered with a tetanus toxoid vaccine. J Infect Dis. 1986;153:628–33. doi: 10.1093/infdis/153.3.628. [DOI] [PubMed] [Google Scholar]
- 15.Bahr GM, Darcissac E, Bevec D, Dukor P, Chedid L. Immunopharmacological activities and clinical development of muramyl peptides with particular emphasis on murabutide. Int J Immunopharmacol. 1995;17:117–31. doi: 10.1016/0192-0561(94)00094-5. [DOI] [PubMed] [Google Scholar]
- 16.Bahr GM, Chedid L. Immunological activities of muramyl peptides. Fed Proc. 1986;45:2541–4. [PubMed] [Google Scholar]
- 17.Vidal VF, Castéran N, Riendeau CJ, et al. Macrophage stimulation with Murabutide, an HIV-suppressive muramyl peptide derivative, selectively activates extracellular signal-regulated kinases 1 and 2, C/EBPβ and STAT1: role of CD14 and Toll-like receptors 2 and 4. Eur J Immunol. 2001;31:1962–71. doi: 10.1002/1521-4141(200107)31:7<1962::aid-immu1962>3.0.co;2-v. [DOI] [PubMed] [Google Scholar]
- 18.Darcissac EC, Truong MJ, Dewulf J, Mouton Y, Capron A, Bahr GM. The synthetic immunomodulator murabutide controls human immunodeficiency virus type 1 replication at multiple levels in macrophages and dendritic cells. J Virol. 2000;74:7794–802. doi: 10.1128/jvi.74.17.7794-7802.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Henderson AJ, Connor RI, Calame KL. C/EBP activators are required for HIV-1 replication and proviral induction in monocytic cell lines. Immunity. 1996;5:91–101. doi: 10.1016/s1074-7613(00)80313-1. [DOI] [PubMed] [Google Scholar]
- 20.Bahr GM, Darcissac EC, Casteran N, et al. Selective regulation of human immunodeficiency virus-infected CD4 (+) lymphocytes by a synthetic immunomodulator leads to potent virus suppression in vitro and in hu-PBL-SCID mice. J Virol. 2001;75:6941–52. doi: 10.1128/JVI.75.15.6941-6952.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Schreck R, Bevec D, Dukor P, Baeuerle PA, Chedid L, Bahr GM. Selection of a muramyl peptide based on its lack of activation of nuclear factor-kappa B as a potential adjuvant for AIDS vaccines. Clin Exp Immunol. 1992;90:188–93. doi: 10.1111/j.1365-2249.1992.tb07926.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Darcissac EC, Bahr GM, Pouillart PR, Riveau GJ, Parant MA. Selective potentiation of cytokine expression in human whole blood by murabutide, a muramyl dipeptide analogue. Cytokine. 1996;8:658–66. doi: 10.1006/cyto.1996.0088. 10.1006/cyto.1996.0088. [DOI] [PubMed] [Google Scholar]
- 23.Darcissac ECA, Vidal V, Guillaume M, Thebault J-J, Bahr GM. Clinical tolerance and profile of cytokine induction in healthy volunteers following the simultaneous administration of IFN-alpha and the synthetic immunomodulator murabutide. J Interferon Cytokine Res. 2001;21:655–61. doi: 10.1089/107999001753124381. 10.1089/107999001753124381. [DOI] [PubMed] [Google Scholar]
- 24.Dinarello CA, Gatti S, Bartfai T. Fever: links with an ancient receptor. Curr Biol. 1999;9:R147–50. doi: 10.1016/s0960-9822(99)80085-2. [DOI] [PubMed] [Google Scholar]
- 25.Rosenberger CM, Scott MG, Gold MR, Hancock RE, Finlay BB. Salmonella typhimurium infection and lipopolysaccharide stimulation induce similar changes in macrophage gene expression. J Immunol. 2000;164:5894–904. doi: 10.4049/jimmunol.164.11.5894. [DOI] [PubMed] [Google Scholar]
- 26.Chang L, Karin M. Mammalian MAP kinase signalling cascades. Nature. 2001;410:37–40. doi: 10.1038/35065000. [DOI] [PubMed] [Google Scholar]
- 27.Mitchell RA, Metz CN, Peng T, Bucala R. Sustained mitogen-activated protein kinase (MAPK) and cytoplasmic phospholipase A2 activation by macrophage migration inhibitory factor (MIF). Regulatory role in cell proliferation and glucocorticoid action. J Biol Chem. 1999;274:18100–6. doi: 10.1074/jbc.274.25.18100. [DOI] [PubMed] [Google Scholar]
- 28.Cullen BR. Regulation of HIV-1 gene expression. FASEB J. 1991;5:2361–8. doi: 10.1096/fasebj.5.10.1712325. [DOI] [PubMed] [Google Scholar]
- 29.Harhaj E, Blaney J, Millhouse S, Sun SC. Differential effects of I kappa B molecules on Tat-mediated transactivation of HIV-1 LTR. Virology. 1996;216:284–7. doi: 10.1006/viro.1996.0062. 10.1006/viro.1996.0062. [DOI] [PubMed] [Google Scholar]
- 30.Lohoff M, Duncan GS, Ferrick D, et al. Deficiency in the transcription factor interferon regulatory factor (IRF)-2 leads to severely compromised development of natural killer and T helper type 1 cells. J Exp Med. 2000;192:325–36. doi: 10.1084/jem.192.3.325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Rubinstein YR, Pontzer CH. Loss of interferon alpha and interferon tau-induced antiviral protection in interferon regulatory factor-2 DNA-binding domain dominant negative mutants. Antiviral Res. 2000;46:207–13. doi: 10.1016/s0166-3542(00)00087-5. [DOI] [PubMed] [Google Scholar]
- 32.Przepiorka D, Srivastava PK. Heat shock protein–peptide complexes as immunotherapy for human cancer. Mol Med Today. 1998;4:478–84. doi: 10.1016/s1357-4310(98)01345-8. [DOI] [PubMed] [Google Scholar]
- 33.Moroi Y, Mayhew M, Trcka J, et al. Induction of cellular immunity by immunization with novel hybrid peptides complexed to heat shock protein 70. Proc Natl Acad Sci USA. 2000;97:3485–90. doi: 10.1073/pnas.070550797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Fukuzumi M, Shinomiya H, Shimizu Y, Ohishi K, Utsumi S. Endotoxin-induced enhancement of glucose influx into murine peritoneal macrophages via GLUT1. Infect Immun. 1996;64:108–12. doi: 10.1128/iai.64.1.108-112.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Spolarics Z, Bagby GJ, Lang CH, Spitzer JJ. Up-regulation of glucose metabolism in Kupffer cells following infusion of tumour necrosis factor. Biochem J. 1991;278:515–9. doi: 10.1042/bj2780515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Orlinska U, Newton RC. Role of glucose in interleukin-1 beta production by lipopolysaccharide-activated human monocytes. J Cell Physiol. 1993;157:201–8. doi: 10.1002/jcp.1041570126. [DOI] [PubMed] [Google Scholar]
- 37.Richman C, Baylink DJ, Lang K, Dony C, Mohan S. Recombinant human insulin-like growth factor-binding protein-5 stimulates bone formation parameters in vitro and in vivo. Endocrinology. 1999;140:4699–705. doi: 10.1210/endo.140.10.7081. [DOI] [PubMed] [Google Scholar]
- 38.Teichmann J, Stephan E, Discher T, et al. Changes in calciotropic hormones and biochemical markers of bone metabolism in patients with human immunodeficiency virus infection. Metabolism. 2000;49:1134–9. doi: 10.1053/meta.2000.8609. 10.1053/meta.2000.8609. [DOI] [PubMed] [Google Scholar]
- 39.Polanski M, Johnson TS, Karnovsky ML. Muramyl peptides as paradigms in neuroimmunomodulation. Ann NY Acad Sci. 1992;650:218–20. doi: 10.1111/j.1749-6632.1992.tb49125.x. [DOI] [PubMed] [Google Scholar]
- 40.Araki T, Milbrandt J. Ninjurin, a novel adhesion molecule, is induced by nerve injury and promotes axonal growth. Neuron. 1996;17:353–61. doi: 10.1016/s0896-6273(00)80166-x. [DOI] [PubMed] [Google Scholar]
- 41.Rapalino O, Lazarov-Spiegler O, Agranov E, et al. Implantation of stimulated homologous macrophages results in partial recovery of paraplegic rats. Nat Med. 1998;4:814–21. doi: 10.1038/nm0798-814. [DOI] [PubMed] [Google Scholar]
- 42.Lazar DA, Ellegala DB, Avellino AM, Dailey AT, Andrus K, Kliot M. Modulation of macrophage and microglial responses to axonal injury in the peripheral and central nervous systems. Neurosurgery. 1999;45:593–600. doi: 10.1097/00006123-199909000-00030. [DOI] [PubMed] [Google Scholar]


