Abstract
Transgenic and knockout mouse models have been invaluable for the elucidation of basic mechanisms in autoimmunity and have contributed new experimental models of human autoimmune diseases. Transgenic models of self tolerance have helped to change our view of this state from a process mediated purely by thymic deletion to a more complex process encompassing deletion, peripheral anergy, down-regulation of receptors and modulation by regulatory cells. Experiments in which the genes for the candidate target antigens in autoimmune disease are over-expressed or under-expressed have helped to clarify the targets of attack. Several examples of T cell receptor transgenic mice have been described in which T cells carry the receptor derived from a human or mouse autoimmune T cell clone. Such mice allow the characterization of T cell specificities contributing to disease and of the additional factors and checkpoints influencing disease development. In addition, the expression of disease associated HLA alleles in ‘humanised’ transgenic lines allows the mapping of HLA-restricted T cell epitopes and investigation of the mechanisms underlying these genetic associations. These approaches are leading to the generation of new disease models, offering hope for the design and testing of novel immunotherapeutic strategies.
Keywords: autoimmunity, HLA, transgenic mouse
The field of autoimmune disease has benefited from a long and profitable interaction between basic science and clinical practice. Much of the impetus in the field has come from rat and mouse models. In some cases, this has involved the identification of autoimmune lesions during the breeding of inbred mouse strains, such as nonobese diabetic (NOD) mice or lupus-prone MRL, BXSB, and (NZB × NZW)f1 mice. An alternative approach that has proved valuable has been the experimental induction of disease in susceptible strains. These include immunization with myelin antigens to induce experimental allergic encephalomyelits (EAE) as a model for multiple sclerosis (MS), with acetylcholine receptor to model myasthenia, retinal antigens in uveitis and renal basement membrane antigens for Goodpasture’s disease. From these basic, disease-induction experiments have come most of our current concepts and therapeutic hopes for autoimmune disease, including views on effector mechanisms and modalities for their inhibition.
With the advent of transgenic mouse technology in the early 1980s came many new opportunities for investigating autoimmune mechanisms. These have been used to address questions ranging from basic mechanisms in the maintenance and breakdown of self tolerance to the role of specific molecules implicated in human autoimmune disease. Some 10 years later came the impact of many studies looking at the effect of knocking out specific molecules in gene targeting experiments. The aim of this review is to highlight briefly some of the key developments in this field. Transgenic and knockout strains have become major tools for addressing the central questions in autoimmune disease. What are the normal mechanisms through which the immune system can eliminate or control self-reactive T and B cells? Which are the specific T cells mediating disease and which receptors do they express? What is the contribution to disease of known or unknown susceptibility genes, including MHC genes? Which of the array of candidate target antigens for a given disease, account for the pattern of pathology? Which cytokines and chemokines are implicated? Which costimulatory molecules are involved in the up-regulation or down-regulation of disease? In addition, experiments in transgenic models can illuminate the mechanisms underlying clinical observations from autoimmune diseases, for example, the fact that there is clearly a genetic predisposition, overlaid with environmental/infectious risk factors. The ultimate test of these models is whether or not they can offer new insights into disease aetiology that lead to the design of new therapies, and whether the models can themselves be valuable in testing therapies.
Transgenic and knockout models have been used both as models to test firmly established hypothesese of autoimmunity, for example, that particular MHC alleles may be protective in type I diabetes, and as a source of unpredicted phenotypes leading to new ideas of disease aetiology. An example of the latter type is the fortuitous development of spontaneous colitis in a number of immune deficient knockout strains, including TCRα, IL-2 and IL-2Rα knockouts [1–3]. These observations have triggered research into T cell mechanisms in human inflammatory bowel disease (IBD) as well as offering experimental systems in which therapies may be tested. Rather than describing and highlighting the models arranged according to the human diseases they mimic, models will be discussed according to the types of genes introduced and the aspect of the immune response influenced.
TRANSGENIC MICE FOR THE ANALYSIS OF MECHANISMS OF CENTRAL AND PERIPHERAL SELF-TOLERANCE
It was once believed that all self-reactive T cells must be negatively selected during thymic development, so that any self-reactivity would be regarded as an aberrant breakdown of this process. It then became clear that healthy individuals readily show T cells responses to some self antigens [4], so that the prevention of disease must also reside at other levels, including the induction of peripheral anergy and the effect of regulatory cells. Some of the early transgenic experiments asked how the immune system of a mouse would cope if all receptors (either B or T cell) were self reactive for some self antigen which was then also expressed in the mouse line, potentially creating an entirely autoimmune immune system. One experiment of this type, conducted in von Boehmer’s laboratory, asked what would happen if most T cells express a transgenic, class I restricted TCR specific for the antigen H-Y and then meet their target antigen, more or less ubiquitously expressed on tissues of male (but not female) mice [5]. Since these experiments were reported, the H-Y antigen has been characterized as several male cell derived peptides, bound to class I and II molecules, and originating from the sequences of a number of Y chromosome encoded genes that have differential sequence from the X chromosome copy [6]. The initial studies showed that most of the autoreactive, TCR transgenic T cells engaged H-Y peptides in the thymus and were deleted at the CD4+CD8+ double positive stage of differentiation [5]. However, many of the transgenic T cells were able to mature and were exported to the periphery, where they were found to have much reduced levels of CD8 or TCR, which rendered them anergic. In another set of experiments, mice were made to carry only the H-Y reactive TCRβ chain. It was found that T cells expressing this chain in the context of a self-reactive TCR heterodimer were able to mature and leave the thymus, but their use of the paired α chains had been edited such that the surviving cells tended to use a subset of α chains probably associated with much reduced affinity [7]. Thus, despite carrying an immune system dominated by self-recognizing cells, no mouse was described to develop any form of disease pathology.
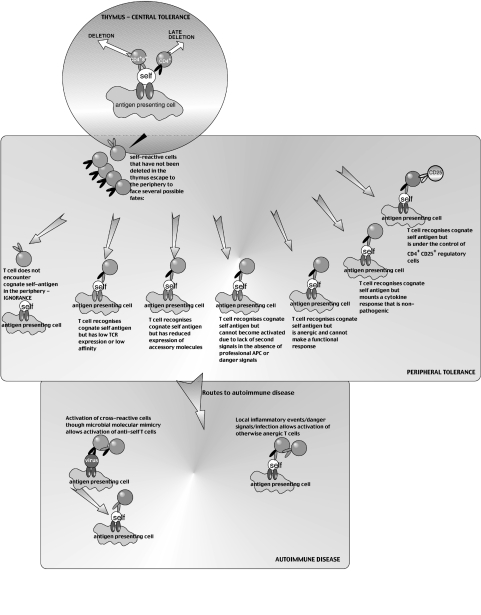
A related experiment investigated the induction of tolerance in TCR transgenic mice where a transgenically expressed cognate, self antigen is directed to be expressed exclusively in the pancreas through use of an insulin promoter. These mice generally do not show evidence of deletion; they either show complete ignorance of the target antigen, or are anergic, the T cells having down-regulated receptors or accessory molecules, so rendering them functionally inert in vivo [8]. Since experiments of this type were first reported from Miller’s laboratory in the late 1980s, there have been many examples studied. Many different mechanisms have been described, including down-regulation of receptors, immune deviation to a Th2 response and control by regulatory cells. In each case, the lesson is that the immune system is remarkably resistant to causing autoimmune disease, with many different safeguards (Fig. 1). In many or most cases, the mere existence of self-reactive cells is not sufficient to cause disease – some additional, inflammatory insult is generally required. An example of this comes from the experiments showing that coexpression in transgenic mice of an islet cell expressed lymphochoriomeningitis virus (LCMV) epitope and a corresponding TCR does not lead to disease, unless the mice are infected with live virus, in which case the T cells can become activated to cause full blown diabetes [9]. Experiments of this type are in line with the general view from clinical studies that humans may harbour autoreactive cells without developing autoimmune disease, some other environmental components being required. While the additional components may come from other susceptibility genes, the factors clearly cannot be only genetic, since autoimmune diseases show concordance in monozygotic twins of 35% or less.
Fig. 1.
Mechanisms leading to central tolerance, peripheral tolerance or autoimmune tolerance
TRANSGENIC MICE EXPRESSING T CELL RECEPTORS DERIVED FROM AUTOIMMUNE T CELL CLONES
The experiments described above are, on the whole, testament to how difficult it is to overcome the checks and balances of the immune system and create an autoimmune mouse. Nevertheless, several laboratories have been able to generate models of autoimmunity by making transgenic lines that express TCRs derived from T cell clones previously proven to transfer autoimmune disease. In mice expressing a TCR derived from an encephalitogenic, MBP specific CD4 clone, there is a much enhanced susceptibility to the induction of CNS disease following injection of MBP peptide and B. pertussis toxin [10]. The survival of self-reactive T cell clones in this and similar models is believed to depend on the very low affinity of the MBP peptide (MBP 1–11) for MHC and TCR [11]. Interestingly, a proportion of the mice spontaneously develop CNS disease in early adulthood, but this is dependent on the microbial status of the mouse facility in which they are housed [10,12]. Mice maintained under specific pathogen free conditions rarely develop the disease. In the MBP TCR mice, as in mice carrying a TCR from a NOD mouse diabetogenic clone (BDC2·5) specific for a beta cell granule antigen, spontaneous disease is much enhanced in mice crossed onto a knockout strain for either recombinase activating gene (RAG) or TCRα[13,14]. The relevance of these experiments is that allelic exclusion at the TCRα locus is relatively leaky, such that a sizeable proportion of cells may additionally manage to rearrange and express endogenous TCRα chains, giving rise to additional TCR specificities, through pairing of the encephalitogenic TCRβ chain with the repertoire of endogenous α chains. Within this population reside regulatory cells, possibly the CD4+CD25+ cells described in other systems, that have the ability to suppress the response to MBP 1–11, so inhibiting the disease process [15].
In general, the fact that TCR transgenic mice do not develop autoimmune disease at birth and that, even with the delayed onset of disease in adulthood, penetrance can be poor, has provided many other opportunities for investigating the checkpoints that may also control the onset of human autoimmune disease. Work in transgenic models of diabetes, particularly the BDC2·5 diabetogenic TCR transgenic derived in Mathis and Benoist’s laboratory, have helped to elucidate some of these checkpoints [16]. The first checkpoint relates to the fact that the TCR transgenic mice possess a full repertoire of fully functional, islet beta cell specific T cells from birth, yet these do not appear as islet infiltrates until 3 weeks or later. T cells in the BDC2·5 TCR model change their behaviour in early adulthood and start to migrate to the pancreas generating an insulitis, this correlating with changes in the expression of addressins and other mediators involved in lymphocyte homing. In other words, autoreactive T cells may remain nonpathogenic prior to disease onset because they lack the necessary signals to home to the tissue involved. In this and many other NOD transgenic models, there is a second checkpoint that must be passed, from a peri-islet infiltrate during which T cells seem to queue up outside the islet waiting to cause damage, to the full-blown invasion and destruction of beta cells. Studies in the NOD backcrosses to identify diabetes loci identified separate genetic controls for the two checkpoints. In various models, a range of different events may contribute to the progression from insulitis to diabetes. These may include the recruitment of other cell types, changes in patterns of Th1 and Th2 cytokine release, and changes in populations of regulatory cells.
Experiments with murine TCR expression from autoimmune clones have been used to look at receptors in a number of mouse models including EAE, type I diabetes and collagen induced arthritis. It was long appreciated that there would be great advantages in being able to study receptors from human patients’ T cell clones in vivo using transgenic mouse models. One problem when studying patients’ T cell clones recognizing a particular self peptide or expressing a particular TCR is that one cannot conclude with any certainty that the cells are causally involved in the disease process. Provided the T cells studied are specific for self peptides with sequence conservation between mouse and human, the demonstration that a given human T cell clone can cause disease when its TCR sequence is transposed into a mouse by transgenesis, is an important step forward in the argument. This would also help to jump an important therapeutic hurdle: many of the treatments that have been successful in the treatment of murine models are specific to the rat or mouse T cell models from which they were derived. These include T cell receptor DNA vaccinations and TCR-derived peptides. Hu-TCR transgenic models offer the chance of moving directly to tests of the equivalent human reagents. These experiments of course also require that the restricting human HLA molecule is also expressed transgenically in the model (see below). One group have described induction of an EAE-like disease in hu-TCR transgenic mice expressing a TCR directed against an immunodominant epitope of MBP [17]. We have also generated hu-TCR transgenic mice and find spontaneous autoimmunity associated with an overtly Th1 response to MBP peptide (manuscript in preparation). One caveat to these experiments is that the behaviour of the TCR transgene does not always reiterate faithfully its history in the T cell clones from which it was derived. One of the most aggressive spontaneous disease phenotypes so far observed in a TCR transgenic model uses a receptor derived from a clone that was originally nonpathogenic [18].
TRANSGENIC EXPRESSION OF CANDIDATE AUTOANTIGENS
An enduring problem in the T cell mediated autoimmune diseases is that, unlike the antibody mediated diseases where characterization of patient autoantibodies itself provides the means for purifying the target antigen, there is often little agreement as to the target recognized by T cells. For each disease, a number of possible antigens have been cited. In multiple sclerosis the primary target may be MBP, proteolipoprotein, myelin oligodendrocyte glycoprotein or αB-crystallin. In type I diabetes, the candidates are glutamic acid decarboxylase 65 (GAD65), insulin, the IA-2 phosphatase and Hsp60. For rheumatoid arthritis, collagens, proteoglycans, human cartilage gp39 (HCgp39) and heat shock proteins have been considered. One possible test of candidacy would be to look for antigens or peptides that are preferentially recognized from T cells of patients but not controls. However, as will be clear from the preceding sections, the existence of cells that can be activated in vitro to respond to a particular peptide may be a poor correlate of the induction of autoimmune pathology. Spontaneous animal models such as the NOD mouse offer the chance of studies to determine which of the antiself responses arises first in the disease process. Another possibility is to induce tolerance to the candidate self peptide, for example by oral or intranasal tolerization, and look for disease protection. These experiments tend to show that several different antigens can confer protection. Thus, either the path to autoimmune disease encompasses recognition of several different antigens in the same target tissue, or tolerance induction to various antigens can each turn off disease through a form of local, linked suppression.
An alternative approach is to look at the consequence of hyper-expression of the candidate self-antigen in transgenics with the aim of driving deletional tolerance of self-reactive cells, so protecting from disease. The logic for this approach came from the early transgeic experiments of Skowronski et al. [19], looking at the effect of simian virus 40 (SV40) large T antigen expression as a neoantigen, expressed in islet cells by controlling DNA expression from an insulin promoter. Some transgenic lines expressed the transgene from birth and showed full immunological tolerance to the antigen, while others showed delayed expression, and a failure to develop tolerance, this leading to insulitis.
We looked in NOD mice at Hsp60, one of the candidate diabetes autoantigens, generating Hsp60 hyperexpression in the APC of the thymus by driving expression of the murine Hsp60 cDNA from an H2-E class II promoter [20]. Interestingly, we found that very high levels of expression were achieved in the thymus, without producing an absolute loss in responsiveness to self Hsp60. However, responses to some specific Hsp60 epitopes were lost and though many mice showed lymphocytic peri-islet infiltrates, there was protection from full-blown diabetes. A similar approach has since been attempted for other candidate antigens in the disease, generally showing protection from disease. Diabetes was prevented by transgenic expression of proinsulin [21]. There have been various examples of transgenic hyperexpression of GAD65, some leading to protection and others to disease exacerbation [22,23]. However, a compelling argument for the role of GAD65 as a target antigen comes from the elegant experiments of Yoon et al. [24], who made antisense transgenics in which GAD65 was ablated to varying degrees in the pancreas, but not in the CNS. Lines in which GAD65 message was reduced to undetectable levels showed protection from diabetes and were resistant to transfer of disease by pathogenic cells from diabetic donors. An obvious caveat to the transgenic over- or under-expression approach is that the target tissue may be affected by this change in expression for nonimmunological reasons. For example, we do not know what are the physiological consequences of GAD65 deletion for the overall function of a beta cell.
HLA TRANSGENICS MODELS FOR DISEASE STUDIES AND EPITOPE MAPPING
Another transgene that can be incorporated into the experimental model is of course the predisposing HLA allele, since, for many autoimmune diseases, HLA genes are the strongest markers of genetic susceptibility [25,26]. Although these diseases clearly involve multiple genes as well as environmental factors, several groups have taken the view that since the HLA gene products confer a significant component of the genetic risk, one should express the disease related genes in transgenic mice and thus investigate mechanisms underlying susceptibility [27,28]. Since the HLA genes confer enhanced risk rather than acting as disease genes per se, it would not be anticipated that HLA transgenic mice would spontaneously develop a disease associated with that allele. The goal of HLA transgenic experiments is to make laboratory strains that can better mimic the molecular interaction giving rise to the human disease and create a mouse showing enhanced susceptibility to the induction of disease compared with a nontransgenic or an HLA transgenic carrying an irrelevant allele. HLA transgenics can be used to map the epitopes of candidate target antigens from self tissues recognized in the context of particular HLA alleles. An important, if often overlooked, starting point for transgenic studies to model the role of HLA genes in disease is to have a high degree of confidence as to the identity of the specific, implicated genes whose role is to be investigated. For some HLA-associated diseases there is considerable confidence that a gene has been identified which has a causal role in susceptibility. Examples are DRB1*1501 in Goodpasture’s disease and DQB1*0302 in type I diabetes. In some other cases, data point to a number of possible candidates in the HLA region. Early experiments with HLA class I and class II transgenic mice aimed to establish their usefulness for studying ‘humanized’ responses: whether the human molecules could function to present peptides to murine T cells, whether this would depend on a species matched interaction with CD4 or CD8 and whether the murine TCR repertoire and its plasticity would allow responses of similar specificity to the human HLA-restricted responses [29–40]. Where experiment have been done to investigate whether the specificity of murine, HLA-restricted responses show similarities to the responses of humans carrying those HLA alleles, similarities have been found 34–36, see Table 1].
Table 1.
Examples of HLA class II transgenic models of autoimmune responses
| HLA allele | Disease modelled | Antigen studied | Findings | Additional comments | Ref |
|---|---|---|---|---|---|
| HLA-DRB1*0401 | Rheumatoid arthritis | Type II collagen | 261–273 immunodominant epitope | Also found in RA patients’ responses | [42,54] |
| HLA-DRB1*0401, HLA-DRB1*0101 | Rheumatoid arthritis | Type II collagen | Immunization can cause HLA dependent polyarthritis | Induce with whole CII or 261–273. Similar findings in HLA-DR1 and HLA-DR4. | [55–57] |
| Epitope overlaps mouse H2-Aq epitope. | |||||
| HLA-DRB1*0401 and DRB1*0402 | Rheumatoid arthritis | HCgp39 | 3 immunodominant epitopes identified. | Epitopes confirmed in studies of humanpatient T cell responses. | [45] |
| Differential epitopes between susceptibility (0401)and control (0402) alleles | Patient T cell responses | ||||
| HLA-DRB1*0401 | Type I diabetes | GAD65 | Identification of GAD65 274–286, and 115–127as DR4 restricted epitopes | [44] | |
| HLA-DRB1*0401 | Type I diabetes | GAD65 | 6 immunodominant epitopes identified | Epitopes confirmed in DR4 patients. | [41] |
| HLA-DQB1*0302 | Type I diabetes | GAD65 | GAD521-535 implicated as epitope associatedwith susceptibility | Comparisons on susceptible and non susceptiblemice and affected and unaffected twins. | [47] |
| HLA-DQB1*0302 | Type I diabetes | GAD65 | Adoptive transfer of disease with GAD247–266 and 509–528 specific lines | Recipients needed pretreatment withstreptozotocin to generate islet insult | [58] |
| HLA-DQB1*0302 | Type I diabetes | GAD65 | GAD65 121–140, 201–220, 231–250,and 471–490 identified as DQ8 epitopes. | [48] | |
| HLA-DQB1*0302 | Type I diabetes | GAD65 | Three immunodominant regions identified,51–120, 111–180, 521–585, as well asdefinition of common core motif | Epitopes confirmed in DQ8 patients. | [46] |
| HLA-DQB1*0302 | Type I diabetes | Proinsulin | Differences in proinsulin epitope recognitionbetween DQ8 and DQ6 mice | [59] | |
| HLA-DRB1*0401 | Type I diabetes | Preproinsulin andproinsulin | Immunodominance of 73–90 peptide | Sequence spans region that is proteolyticallydestroyed during maturation of the insulinmolecule. | [43] |
| HLA-DQB1*0302 | Type I diabetes | endogenous | Spontaneous diabetes if mice are crossedwith RIP-B7-1 line | Study showed HLA specificity since miceexpressing the diabetes protectiveDQB1*0601 allele did not develop disease. | [50] |
| HLA-DRB1*0101 | Multiple sclerosis | MBP | Responses to MBP 139–154 reiterate thoseof DR1 patients | Responses were unaffected by the presence orabsence of a human CD4 transgene. | [38] |
| HLA-DRB1*0401 | Multiple sclerosis | MBP | EAE like symptoms resulting from immunizationwith PLP 175–192 | MBP 87–106 induced little response or disease. | [60] |
| Evidence for impact of HLA expression ingeneral rather than a disease associated allele. | |||||
| HLA-DRB1*1501 | Multiple sclerosis | MBP | Used transgenics as a tool for generation of a1501/MBP85-99 specific monoclonal | [61] | |
| HLA-DRB1*1502 | Multiple sclerosis | PLP | DR15-restricted PLP 95–116 specificmouse cells transfer CNS disease | Mimics 95–116 response of patients. Modelutilizes the 1502 allele showing a possible association in Japanese patients rather thanthe 1501 allele most commonly associatedwith disease. | [49] |
Responses to peptides spanning the amino acid sequence of self-antigens have been investigated for antigens including insulin and GAD65, type II collagen and human cartilage glycorpotein-39 (HCgp-39) 41–48, Table 1]. Generally, the mere induction of these responses is insufficient to induce disease. However, some groups have achieved disease transfer with HLA-restricted, transgenic mouse T cells [49]. Furthermore, evidence for a role of predisposing alleles comes from experiments where mice expressing HLA-DQ8 (the allele strongly implicated in diabetes susceptibility) do not develop spontaneous disease and nor do mice with transgenic expression of B7-1 targeted to beta cells. However, when the two lines are crossed, most mice develop diabetes [50].
Because of the polygenic, complex aetiology of autoimmune diseases, HLA transgenics are not predicted to develop spontaneously autoimmune disease in the absence of other disease factors. However, when rats carrying a high copy number of HLA-B27 were generated in an effort to model ankylosing spondylitis, they were found to develop pathology including inflammatory peripheral arthritis, inflammatory and fibrotic spinal lesions and gastrointestinal inflammation [51]. Various high-copy number B27 transgenics develop disease while HLA-B7 controls do not [52]. The subsequent application of the HLA-B27 transgenic rats to investigate the factors contributing to disease demonstrates some of the potential benefits of a transgenic approach for elucidating disease mechanisms [53].
CONCLUDING REMARKS
Transgenic mouse models have been invaluable for investigating many aspects of autoimmune disease, ranging from the mechanisms by which self-tolerance may be maintained or bypassed, to the nature of the target antigens and the role of HLA genes. In the postgenomic age, as more is learnt about the relative contributions of genetic and environmental factors to these complex diseases, it will be possible to build more faithful experimental models to further elucidate pathogenic mechanisms. This approach is already leading to the design of new therapeutic agents, able to target specific disease effector pathways without the risks associated with blanket immunosuppression.
References
- 1.Mombaerts P, Mizoguchi E, Grusby MJ, Glimcher LH, Bhan AK, Tonegawa S. Spontaneous development of inflammatory bowel disease in T cell receptor mutant mice. Cell. 1993;75:274–82. doi: 10.1016/0092-8674(93)80069-q. [DOI] [PubMed] [Google Scholar]
- 2.Bhan AK, Mizoguch E, Smith RN, Mizoguchi A. Colitis in transgenic and knockout models of human inflammatory bowel disease. Immunol Rev. 1999;169:195–207. doi: 10.1111/j.1600-065x.1999.tb01316.x. [DOI] [PubMed] [Google Scholar]
- 3.Blumberg RS, Saubermann LJ, Strober W. Animal models of mucosal inflammation and their relation to human inflammatory bowel disease. Curr Opin Immunol. 1999;11:648–56. doi: 10.1016/s0952-7915(99)00032-1. [DOI] [PubMed] [Google Scholar]
- 4.Burns J, Rosenzweig A, Zweiman B, Lisak RP. Isolation of myelin basic protein-reactive T-cell lines from normal human blood. Cell Immunol. 1983;81:435–40. doi: 10.1016/0008-8749(83)90250-2. [DOI] [PubMed] [Google Scholar]
- 5.Kisielow P, Bluthmann H, Staerz UD, Steinmetz M, von Boehmer H. Tolerance in T-cell-receptor transgenic mice involves deletion of nonmature CD4+8+ thymocytes. Nature. 1988;33:742–6. doi: 10.1038/333742a0. [DOI] [PubMed] [Google Scholar]
- 6.Scott D, Addey C, Ellis P, James E, Mitchell MJ, Saut N, Jurcevic S, Simpson E. Dendritic cells permit identification of genes encoding MHC class II-restricted epitopes of transplantation antigens. Immunity. 2000;12:711–20. doi: 10.1016/s1074-7613(00)80221-6. [DOI] [PubMed] [Google Scholar]
- 7.Bouneaud C, Kourilsky P, Bousso P. Impact of negative selection on the T cell repertoire reactive to a self-peptide: a large fraction of T cell clones escapes clonal deletion. Immunity. 2000;13:829–40. doi: 10.1016/s1074-7613(00)00080-7. [DOI] [PubMed] [Google Scholar]
- 8.Miller JF, Flavell RA. T-cell tolerance and autoimmunity in transgenic models of central and peripheral tolerance. Curr Opin Immunol. 1994;6:892–9. doi: 10.1016/0952-7915(94)90010-8. [DOI] [PubMed] [Google Scholar]
- 9.Ohashi PS, Oehen S, Burki K, et al. Ablation of ‘tolerance’ and induction of diabetes by virus infection in viral antigen transgenic mice. Cell. 1991;65:305–17. doi: 10.1016/0092-8674(91)90164-t. [DOI] [PubMed] [Google Scholar]
- 10.Goverman J, Woods A, Larson L, Weiner LP, Hood L, Zaller DM. Transgenic mice that express a myelin basic protein specific T cell receptor develop spontaneous autoimmunity. Cell. 1993;72:551–60. doi: 10.1016/0092-8674(93)90074-z. [DOI] [PubMed] [Google Scholar]
- 11.Liu GY, Fairchild P, Smith RM, Prowle PR, Kioussis D, Wraith DC. Low avidity recognition of self antigen by T cells permits escape from self tolerance. Immunity. 1995;3:407–15. doi: 10.1016/1074-7613(95)90170-1. [DOI] [PubMed] [Google Scholar]
- 12.Goverman J. Tolerance and autoimmunity in TCR transgenic mice specific for myelin basic protein. Immunol Rev. 1999;169:147–59. doi: 10.1111/j.1600-065X.1999.tb01313.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lafaillle J, Nagashima K, Katsuki M, Tonegawa S. High incidence of spontaneous autoimmune encephalomyelitis in immunodeficient anti-myelin basic protein T cell receptor transgenic mice. Cell. 1994;78:399–408. doi: 10.1016/0092-8674(94)90419-7. [DOI] [PubMed] [Google Scholar]
- 14.Ji H, Korganow AS, Mangialaio S, et al. Different modes of pathogenesis in T-cell-dependent autoimmunity: clues from two TCR transgenic systems. Immunol Rev. 1999;169:139–46. doi: 10.1111/j.1600-065x.1999.tb01312.x. [DOI] [PubMed] [Google Scholar]
- 15.Olivares-Villagomez D, Wang Y, Lafaille JJ. Regulatory CD4+ T cells expressing endogenous T cell receptor chains protect myelin basic protein specific transgenic mice from spontaneous autoimmune encephalomyelitis. J Exp Med. 1998;188:1883–94. doi: 10.1084/jem.188.10.1883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Andre I, Gonzalez A, Wang B, Katz J, Benoist C, Mathis D. Checkpoints in the progression of autoimmune disease: lessons from diabetes models. Proc Natl Acad Sci USA. 1996;93:2260–3. doi: 10.1073/pnas.93.6.2260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Madsen LS, Andersson EC, Jansson L, et al. A humanized model for multiple sclerosis using HLA-DR2 and a human T-cell receptor. Nat Genet. 1999;23:343–7. doi: 10.1038/15525. [DOI] [PubMed] [Google Scholar]
- 18.Waldner H, Whitters MJ, Sobel RA, Collins M, Kuchroo VK. Fulminant spontaneous autoimmunity of the central nervous system in mice transgenic for the myelin proteolipid protein-specific T cell receptor. Proc Natl Acad Sci USA. 2000;97:3412–7. doi: 10.1073/pnas.97.7.3412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Skowronski J, Jolicoeur C, Alpert S, Hanahan D. Determinants of the B-cell response against a transgenic autoantigen. Proc Natl Acad Sci USA. 1990;87:7487–91. doi: 10.1073/pnas.87.19.7487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Birk OS, Douek DC, Elias D, et al. A role of Hsp60 in autoimmune diabetes: analysis in a transgenic model. Proc Natl Acad Sci USA. 1996;93:1032–7. doi: 10.1073/pnas.93.3.1032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.French MB, Allison J, Cram DS, et al. Transgenic expression of mouse proinsulin II prevents diabetes in nonobese diabetic mice. Diabetes. 1997;46:34–9. doi: 10.2337/diab.46.1.34. [DOI] [PubMed] [Google Scholar]
- 22.Geng L, Solimena M, Flavell RA, Sherwin RS, Hayday AC. Widespread expression of an autoantigen-GAD65 transgene does not tolerize non-obese diabetic mice and can exacerbate disease. Proc Natl Acad Sci USA. 1998;95:10055–60. doi: 10.1073/pnas.95.17.10055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bridgett M, Cetkovic-Cvrlje M, O’Rourke R, et al. Differential protection in two transgenic lines of NOD/Lt mice hyperexpressing the autoantigen GAD65 in pancreatic beta-cells. Diabetes. 1998;47:1848–56. doi: 10.2337/diabetes.47.12.1848. [DOI] [PubMed] [Google Scholar]
- 24.Yoon JW, Yoon CS, Lim HW, et al. Control of autoimmune diabetes in NOD mice by GAD expression or suppression in beta cells. Science. 1999;284:1183–7. doi: 10.1126/science.284.5417.1183. [DOI] [PubMed] [Google Scholar]
- 25.Thorsby E. Invited anniversary review: HLA associated diseases. Hum Immunol. 1997;53:1–11. doi: 10.1016/S0198-8859(97)00024-4. [DOI] [PubMed] [Google Scholar]
- 26.Haines JL, Ter-Minassian M, Bazyk A, et al. A complete genomic screen for multiple sclerosis underscores a role for the major histocompatibility complex. The Multiple Sclerosis Genet Group. Nat Genet. 1996;13:469–71. doi: 10.1038/ng0896-469. [DOI] [PubMed] [Google Scholar]
- 27.Altmann DM, Ellmerich S, Boyton RJ. HLA Transgenic Mice for the Analysis of Autoimmune Disease. In: Friedland J, Lightstone E, editors. Infection and Immunity. Reading: Harwood Academic Publishers; 2001. pp. 000–000. [Google Scholar]
- 28.Fugger L. Human autoimmunity genes in mice. Curr Opin Immunol. 2000;12:698–703. doi: 10.1016/s0952-7915(00)00165-5. [DOI] [PubMed] [Google Scholar]
- 29.Kalinke U, Arnold B, Hammerling GJ. Strong xenogeneic HLA response in transgenic mice after introducing an alpha 3 domain into HLA B27. Nature. 1990;348:642–4. doi: 10.1038/348642a0. [DOI] [PubMed] [Google Scholar]
- 30.Engelhard VH, Lacy E, Ridge JP. Influenza A-specific, HLA-A2.1-restricted cytotoxic T lymphocytes from HLA-A2.1 transgenic mice recognize fragments of the M1 protein. J Immunol. 1991;146:1226–32. [PubMed] [Google Scholar]
- 31.Vitiello A, Marchesini D, Furze J, Sherman LA, Chesnut RW. Analysis of the HLA-restricted influenza-specific cytotoxic T lymphocyte response in transgenic mice carrying a chimeric human-mouse class I major histocompatibility complex. J Exp Med. 1991;173:1007–15. doi: 10.1084/jem.173.4.1007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kievits F, Ivanyi P, Krimpenfort P, Berns A, Ploegh HL. HLA-restricted recognition of viral antigens in HLA transgenic mice. Nature. 1987;329:447–9. doi: 10.1038/329447a0. [DOI] [PubMed] [Google Scholar]
- 33.Man S, Ridge JP, Engelhard VH. Diversity and dominance among TCR recognizing HLA-A2.1+ influenza matrix peptide in human MHC class I transgenic mice. J Immunol. 1994;153:4458–6. [PubMed] [Google Scholar]
- 34.Shirai M, Arichi T, Nishioka M, et al. CTL responses of HLA-A2.1-transgenic mice specific for hepatitis C viral peptides predict epitopes for CTL of humans carrying HLA-A2. 1 J Immunol. 1995;154:2733–42. [PubMed] [Google Scholar]
- 35.Blum-Tirouvanziam U, Servis C, Habluetzel A, et al. Localization of HLA-A2.1-restricted T cell epitopes in the circumsporozoite protein of Plasmodium falciparum. J Immunol. 1995;154:3922–31. [PubMed] [Google Scholar]
- 36.Wentworth PA, Vitiello A, Sidney J, Keogh E, Chesnut RW, Grey H, Sette A. Differences and similarities in the A2.1-restricted cytotoxic T cell repertoire in humans and human leukocyte antigen-transgenic mice. Eur J Immunol. 1996;26:97–101. doi: 10.1002/eji.1830260115. [DOI] [PubMed] [Google Scholar]
- 37.Yamamoto K, Fukui Y, Esaki Y, et al. Functional interaction between human histocompatibility leukocyte antigen (HLA) class II and mouse CD4 molecule in antigen recognition by T cells in HLA-DR and DQ transgenic mice. J Exp Med. 1994;180:165–71. doi: 10.1084/jem.180.1.165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Altmann DM, Douek DC, Frater AJ, Hetherington CM, Inoko H, Elliott JI. The T cell response of HLA-DR transgenic mice to human myelin basic protein and other antigens in the presence and absence of human CD4. J Exp Med. 1995;181:867–75. doi: 10.1084/jem.181.3.867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Woods A, Chen HY, Trumbauer ME, Sirotina A, Cummings R, Zaller DM. Human major histocompatibility complex class II-restricted T cell responses in transgenic mice. J Exp Med. 1994;180:173–81. doi: 10.1084/jem.180.1.173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Tishon A, LaFace DM, Lewicki H, van Binnendijk RS, Osterhaus A, Oldstone MB. Transgenic mice expressing human HLA and CD8 molecules generate HLA-restricted measles virus cytotoxic T lymphocytes of the same specificity as humans with natural measles virus infection. Virology. 2000;275:286–93. doi: 10.1006/viro.2000.0517. 10.1006/viro.2000.0517. [DOI] [PubMed] [Google Scholar]
- 41.Patel SD, Cope AP, Congia M, Chen TT, Kim E, Fugger L, Wherrett D, Sonderstrup-McDevitt G. Identification of immunodominant T cell epitopes of human glutamic acid decarboxylase 65 by using HLA-DR (alpha1*0101,beta1*0401) transgenic mice. Proc Natl Acad Sci USA. 1997;94:8082–7. doi: 10.1073/pnas.94.15.8082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Andersson EC, Hansen BE, Jacobsen H, et al. Definition of MHC and T cell receptor contacts in the HLA-DR4restricted immunodominant epitope in type II collagen and characterization of collagen-induced arthritis in HLA-DR4 and human CD4 transgenic mice. Proc Natl Acad Sci USA. 1998;95:7574–9. doi: 10.1073/pnas.95.13.7574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Congia M, Patel S, Cope AP, De Virgiliis S, Sonderstrup G. T cell epitopes of insulin defined in HLA-DR4 transgenic mice are derived from preproinsulin and proinsulin. Proc Natl Acad Sci USA. 1998;95:3833–8. doi: 10.1073/pnas.95.7.3833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Wicker LS, Chen SL, Nepom GT, et al. Naturally processed T cell epitopes from human glutamic acid decarboxylase identified using mice transgenic for the type 1 diabetes-associated human MHC class II allele, DRB1*0401. J Clin Invest. 1996;98:2597–603. doi: 10.1172/JCI119079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Cope AP, Patel SD, Hall F, et al. T cell responses to a human cartilage autoantigen in the context of rheumatoid arthritis-associated and nonassociated HLA-DR4 alleles. Arthritis Rheum. 1999;42:1497–507. doi: 10.1002/1529-0131(199907)42:7<1497::AID-ANR25>3.0.CO;2-#. [DOI] [PubMed] [Google Scholar]
- 46.Herman AE, Tisch RM, Patel SD, et al. Determination of glutamic acid decarboxylase 65 peptides presented by the type I diabetes-associated HLA-DQ8 class II molecule identifies an immunogenic peptide motif. J Immunol. 1999;163:6275–82. [PubMed] [Google Scholar]
- 47.Boyton RJ, Lohmann T, Londei M, et al. Glutamic acid decarboxylase T lymphocyte responses associated with susceptibility or resistance to type I diabetes: analysis in disease discordant human twins, non-obese diabetic mice and HLA-DQ transgenic mice. Int Immunol. 1998;10:1765–76. doi: 10.1093/intimm/10.12.1765. [DOI] [PubMed] [Google Scholar]
- 48.Liu J, Purdy LE, Rabinovitch S, Jevnikar AM, Elliott JF. Major DQ8-restricted T-cell epitopes for human GAD65 mapped using human CD4, DQA1*0301, DQB1*0302 transgenic IA (null) NOD mice. Diabetes. 1999;48:469–77. doi: 10.2337/diabetes.48.3.469. [DOI] [PubMed] [Google Scholar]
- 49.Kawamura K, Yamamura T, Yokoyama K, et al. HLA-DR2-restricted responses to proteolipid protein 95–116 peptide cause autoimmune encephalitis in transgenic mice. J Clin Invest. 2000;105:977–84. doi: 10.1172/JCI8407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Wen L, Wong FS, Tang J, Chen NY, Altieri M, David C, Flavell R, Sherwin R. In vivo evidence for the contribution of human histocompatibility leukocyte antigen (HLA) -DQ molecules to the development of diabetes. J Exp Med. 2000;191:97–104. doi: 10.1084/jem.191.1.97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Hammer RE, Maika SD, Richardson JA, Tang JP, Taurog JD. Spontaneous inflammatory disease in transgenic rats expressing HLA-B27 and human beta 2m: an animal model of HLA-B27-associated human disorders. Cell. 1990;63:1099–112. doi: 10.1016/0092-8674(90)90512-d. [DOI] [PubMed] [Google Scholar]
- 52.Taurog JD, Maika SD, Satumtira N, et al. Inflammatory disease in HLA-B27 transgenic rats. Immunol Rev. 1999;169:209–23. doi: 10.1111/j.1600-065x.1999.tb01317.x. [DOI] [PubMed] [Google Scholar]
- 53.Taurog JD, Richardson JA, Croft JT, Simmons WA, Zhou M, Fernandez-Sueiro JL, Balish E, Hammer RE. The germfree state prevents development of gut and joint inflammatory disease in HLA-B27 transgenic rats. J Exp Med. 1994;180:2359–64. doi: 10.1084/jem.180.6.2359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Fugger L, Rothbard JB, Sonderstrup-McDevitt G. Specificity of an HLA-DRB1*0401-restricted T cell response to type II collagen. Eur J Immunol. 1996;26:928–33. doi: 10.1002/eji.1830260431. [DOI] [PubMed] [Google Scholar]
- 55.Rosloniec EF, Brand DD, Myers LK, et al. An HLA-DR1 transgene confers susceptibility to collagen-induced arthritis elicited with human type II collagen. J Exp Med. 1997;185:1113–22. doi: 10.1084/jem.185.6.1113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Rosloniec EF, Brand DD, Myers LK, et al. Induction of autoimmune arthritis in HLA-DR4 (DRB1*0401) transgenic mice by immunization with human and bovine type II collagen. J Immunol. 1998;160:2573–8. [PubMed] [Google Scholar]
- 57.Holmdahl R, Andersson EC, Andersen CB, Svejgaard A, Fugger L. Transgenic mouse models of rheumatoid arthritis. Immunol Rev. 1999;169:161–73. doi: 10.1111/j.1600-065x.1999.tb01314.x. [DOI] [PubMed] [Google Scholar]
- 58.Wen L, Wong FS, Burkly L, Altieri M, Mamalaki C, Kioussis D, Flavell RA, Sherwin RS. Induction of insulitis by glutamic acid decarboxylase peptide-specific and HLA-DQ8-restricted CD4 (+) T cells from human DQ transgenic mice. J Clin Invest. 1998;102:947–57. doi: 10.1172/JCI2723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Raju R, Munn SR, David CS. T cell recognition of human preproinsulin peptides depends on the polymorphism at HLA DQ locus: a study using HLA DQ8 and DQ6 transgenic mice. Hum Immunol. 1997;58:21–9. doi: 10.1016/s0198-8859(97)00212-7. [DOI] [PubMed] [Google Scholar]
- 60.Ito K, Bian HJ, Molina M, et al. HLA-DR4-IE chimeric class II transgenic, murine class II-deficient mice are susceptible to experimental allergic encephalomyelitis. J Exp Med. 1996;183:2635–44. doi: 10.1084/jem.183.6.2635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Krogsgaard M, Wucherpfennig KW, Canella B, et al. Visualization of myelin basic protein (MBP) T cell epitopes in multiple sclerosis lesions using a monoclonal antibody specific for the human histocompatibility leukocyte antigen (HLA) -DR2-MBP 85–99 complex. J Exp Med. 2000;191:1395–412. doi: 10.1084/jem.191.8.1395. [DOI] [PMC free article] [PubMed] [Google Scholar]