Abstract
Phenylalanine hydroxylase stimulator (PHS) is a component of the phenylalanine hydroxylation system that is involved in the regeneration of the cofactor tetrahydrobiopterin. It is also identical to the dimerization cofactor of hepatocyte nuclear factor 1 (HNF1) (DCoH) that is able to enhance the transcriptional activity of HNF1. Moreover, it has the structural potential for binding macromolecules such as proteins and nucleic acids, consistent with its involvement in gene expression. We investigated whether PHS/DCoH could enhance the expression of phenylalanine hydroxylase (PAH). Cotransfection assays showed that DCoH itself could not transactivate the 9-kb human PAH 5′ flanking fragment. However, this 9-kb fragment was transactivated by HNF1 in a dose-dependent manner with a maximum of nearly 8-fold activation; DCoH potentiated this transactivation by another 1.6-fold. The HNF1 binding sites were located at −3.5 kb in a region that is 77.5% identical to the mouse liver-specific hormone-inducible PAH gene enhancer. This study suggests a possible dual function of PHS in vivo in the human phenylalanine hydroxylation system: it is involved in the regeneration of the cofactor tetrahydrobiopterin and can also enhance the expression of the human PAH gene.
Phenylalanine hydroxylase stimulator (PHS) is a component of the phenylalanine hydroxylation system (1) that can stimulate phenylalanine hydroxylase (PAH) activity in vitro at higher concentrations of PAH and at higher pH (2). Subsequently, it was demonstrated that PHS is a 4α-carbinolamine dehydratase (3, 4) that participates in the regeneration of tetrahydrobiopterin, the cofactor for PAH (5) and other aromatic amino acid hydroxylases. Tetrahydrobiopterin is converted to a 4α-carbinolamine during the hydroxylation of phenylalanine to tyrosine by PAH. The 4α-carbinolamine then undergoes a PHS-catalyzed dehydration reaction to form quinonoid dihydrobiopterin, which is finally reduced back to tetrahydrobiopterin by the NADH-dependent dihydropteridine reductase. This cycle enables tetrahydrobiopterin to function catalytically during phenylalanine hydroxylation. The importance of PHS in the regeneration of tetrahydrobiopterin is supported by the observations that certain hyperphenylalaninemic patients with a deficiency of this enzyme excrete large amounts of a chemically rearranged form of biopterin, 7-biopterin (6, 7), which is an uncoupling cofactor (8) and a potent inhibitor of PAH (6, 9). In addition, in vitro studies confirmed that the nonenzyme rearrangement of 6-biopterin to 7-biopterin occurs in the absence of PHS and that PHS can prevent this side reaction (10, 11).
In addition to its catalytic function in the regeneration of tetrahydrobiopterin, PHS was shown to be identical to the dimerization cofactor of hepatocyte nuclear factor 1 (HNF1) (DCoH) (12, 13). DCoH was initially copurified from rat liver nuclear extract with HNF1 (14), a liver-specific transcription factor involved in the liver-specific expression of a number of genes such as α- and β-fibrinogen, transthyretin, albumin, α-fetoprotein, and α1-antitrypsin gene (for a minireview, see ref. 15). In vitro studies showed that HNF1 dimers are stabilized by DCoH by forming HNF1–DCoH heterotetramers (14). Moreover, DCoH is able to activate the transcriptional activity of HNF1 through a still poorly defined mechanism (14, 16). Similar to HNF1, DCoH is also highly expressed in liver and kidney, consistent with it having a role in the regulation of HNF1-regulated genes.
Interestingly, a homologue of human DCoH has been identified in Pseudomonas aeruginosa (17). Its gene, phhB, is located in an operon including phhA, which encodes a homologue of mammalian PAH (17). Moreover, truncation of the phhB gene significantly decreased the expression of phhA, suggesting a regulatory function of phhB (17). In addition, the crystal structure of DCoH reveals that this protein is a tetramer containing two saddles that have the potential to bind macromolecules such as proteins and nucleic acids (18, 19). The pattern of DCoH folding is similar to that seen in the TATA-binding protein, a general transcription factor. These structural characteristics suggest that DCoH has the potential to regulate gene expression in varied ways. In this context, we are interested in whether DCoH regulates the expression of the human PAH gene. In this article, we report that DCoH enhances the expression of the human PAH gene in the presence of HNF1 and identifies the HNF1 binding sites in the 5′ flanking region of the human PAH gene.
MATERIALS AND METHODS
Plasmid Constructs.
The DCoH- and HNF1-expressing plasmid pBJ5-DCoH, pBJ5-HNF1, and the vector pBJ5 were generous gifts from G. Crabtree (14). phPAH9-CAT (20) and phPAH1.5-CAT (21), in which the reporter gene for chloramphenicol acetyltransferase (CAT) is driven by a 9- or 1.5-kb 5′ flanking human PAH gene fragment, were provided by Savio Woo. The restriction endonucleases used to construct the deletion plasmids and to determine the direction of the subclones are shown in Fig. 3. The plasmids pXbaI1.5 and pXbaI1.7 were constructed by digesting phPAH9-CAT with XbaI and then religating the largest fragment with the purified 1.5-kb or 1.7-kb XbaI–XbaI fragment, respectively. KpnI and BamHI were used to determine the insertion orientation. pBglII was constructed by completely digesting phPAH9-CAT with BglII and then religating the largest fragment. pTK-Luc-HNF1 was constructed by inserting the 0.5-kb BamHI–XbaI fragment, which is highly identical to the mouse PAH enhancer, upstream of the thymidine kinase promoter of the reporter vector pTK-Luc.
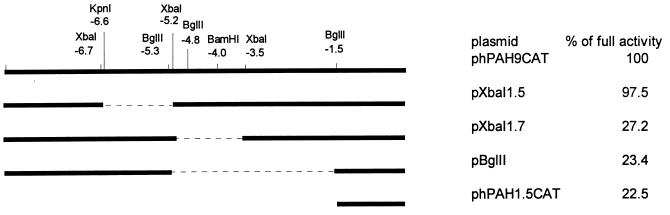
Figure 3.
Deletion analysis of the 9-kb fragment. The positions (in kb) of the restriction endonucleases used to make deletion constructs are shown above; the name of each is given at the right. Each construct was analyzed by the transactivation assay in the absence or presence of pBJ5-HNF1. The activity of the complete 9-kb fragment transactivated by HNF1 was set to 100%. The activity of other constructs was expressed as percent of this activity.
Cell Culture and Electroporation.
Chinese hamster ovary (CHO) cells were grown in Ham’s F-12 medium supplied with 10% fetal calf serum (GIBCO/BRL) at 37°C under 5% CO2. Twenty-four hours before transfection, cells were seeded at a density of 10–15 × 106 cells per T180 flask. Then, cells were harvested and resuspended in growth medium. Between 0.25 ml and 0.3 ml of a suspension of CHO cells (0.8 to 1× 107 cells) was mixed with plasmid DNA in a 0.4-cm electroporation cuvette by shaking. The total amount of plasmid DNA transfected in each experiment was held constant by adjusting the amount of pBJ5 vector. pSV-β-Galactosidase (Promega; 2 μg) was included in every electroporation as an internal control for normalization of transfection efficiency. The amounts of other plasmids used in the electroporations are indicated in the text. CHO cells were transfected by electroporation at 270 V and 960 μF with a Bio-Rad Gene Pulser. After electroporation, cells were seeded into a T75 cell flask containing 13 ml of growth medium and incubated under 5% CO2 at 37°C for 48 h.
CAT, β-Galactosidase, and Luciferase Assay.
CAT activity was measured according to the protocol of Seed and Sheen (22). Briefly, cells were harvested by trypsinization. After washing with PBS (Life Technologies), cells were lysed in 1× Reporter lysis buffer (Promega). Crude cell extracts were used for CAT, β-galactosidase, and luciferase activity assays. For CAT assays, 58.5 or 117 μl of cell extract was heated for 10 min at 60°C to inactivate endogenous deacetylase activity. Subsequently, [14C]chloramphenicol (50 μCi/ml; Dupont/NEN; 3 μl) and n-butyryl CoA (Promega; 5 mg/ml) were added, and then 1× Reporter lysis buffer was used to adjust the final volume to 125 μl. The reactions were incubated at 37°C for 6–16 h. Mixed xylenes (Aldrich; 300 μl) were added to terminate the reactions and extract the products. The upper phase, 200 μl, was back-extracted with 100 μl of 0.25 M Tris⋅HCl (pH 8.0). Radioactivity was measured in exactly 150 μl of the resulting upper phase by scintillation counting. For β-galactosidase activity assay, 2 μl of crude cell extract was diluted to 150 μl with 1× Reporter lysis buffer, and then 150 μl of Assay 2× Buffer (Promega) containing the substrate o-nitrophenyl β-d-galactopyranoside was added to initiate the reactions. After incubation at 37°C for 1–2 h, reactions were stopped by adding 500 μl of 1 M sodium carbonate. The reaction product was measured by determination of the absorbance at 420 nm. Luciferase activity was determined by using the Luciferase assay kit from Promega. Radioluminescence was measured with a Packard TRI-CARB scintillation counter in the single photon mode.
Sequencing.
The 1.7-kb XbaI–XbaI fragment of phPAH9-CAT was subcloned as 0.4-kb XbaI–BglII, 0.8-kb BglII–BamHI, and 0.5-kb BamHI–XbaI fragments. The inserts in these three subclones were sequenced from both ends by using pUC sequence primers 5′-GTAAAACGACGGCCAGT-3′ and 5′-CAGGAAACAGCTATGAC-3′ (New England Biolabs) with an automated laser fluorescence DNA sequencer (Pharmacia). Sequence data were analyzed by using the Wisconsin Package Version 9.0, Genetics Computer Group (GCG).
Gel Mobility Shift Assay.
The complementary unphosphorated oligonucleotides (synthesized by Lofstrand Laboratories, Gaithersburg, MD) were annealed by heating at 90°C for 2 min and cooling at room temperature for 10 min. The annealed oligonucleotides were labeled with [γ-32P]ATP (3,000 Ci/mmol, 10 mCi/ml, DuPont/NEN) by using T4 polynucleotide kinase (New England Biolabs). Labeled oligonucleotides were purified by using a Qiaquick nucleotide removal kit (Qiagene, Chatsworth, CA). Labeled oligonucleotides (3 × 105 cpm) and 50 ng of purified HNF1 were mixed in the binding buffer containing 25 mM Tris⋅HCl (pH 8.0), 50 mM KCl, 6.25 mM MgCl2, 0.5 mM EDTA, 10% glycerol, 1 mM DTT, 2 μg of BSA, and 1.5 μg of poly(dI⋅dC) (Pharmacia Biotech). The solution was incubated at room temperature for 10 min. Subsequently, the sample was loaded onto a 6% DNA retardation gel (NOVEX, San Diego) and electrophoresed in 0.25× TBE buffer (22.5 mM Tris/22.5 mM boric acid/0.5 mM EDTA, pH 8.3). The gel was fixed, dried, and subjected to autoradiography overnight.
RESULTS
Inability of DCoH to Transactivate the 9-kb Human PAH 5′ Flanking Fragment.
To investigate whether DCoH can modulate the expression of the human PAH gene, we carried out cotransfection assays in CHO cells. This cell line was chosen because it does not express the proteins DCoH or HNF1 (14). A mammalian expression plasmid, pBJ5-DCoH that contains an insert encoding DCoH, was used to express DCoH in CHO cells. In a typical experiment, 5 μg of this plasmid was transfected into 7.3 × 106 CHO cells; the specific enzyme activity of the expressed DCoH was comparable to that of a rat liver extract. Expression of DCoH in CHO cells transfected with pBJ5-DCoH was further confirmed by Western blot assay. A reporter plasmid, phPAH9-CAT or phPAH1.5-CAT, was used in which the reporter gene CAT was driven by a 9-kb or a 1.5-kb human PAH 5′ flanking DNA fragment (simply referred as 9-kb or 1.5-kb fragment, respectively, in this article). The 9-kb fragment has previously been shown to be able to confer tissue- and developmental-specific expression of the reporter gene in transgenic mice (20), an indication that it contains sufficient cis information to direct liver-specific expression of the human PAH gene. When 5 μg of phPAH9-CAT or phPAH1.5-CAT was transfected into CHO cells, respectively, a basal CAT activity was detected, which made the cotransfection assays possible. Cotransfecting 5 μg of phPAH9-CAT or phPAH1.5-CAT with 2–20 μg of pBJ5-DCoH into CHO cells did not enhance the expression of the reporter gene, suggesting that DCoH on its own cannot transactivate either the 9-kb or the 1.5-kb fragment.
Transactivation of the Human PAH 9-kb 5′ Flanking Fragment by HNF1.
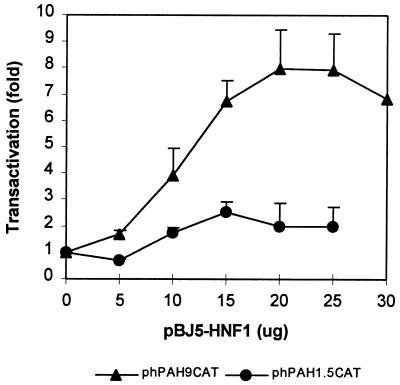
Because DCoH itself could not transactivate either the 9-kb or 1.5-kb fragments, we examined the possibility that DCoH can enhance the human PAH gene expression through potentiation of the transcriptional activity of HNF1. The prerequisite for this possibility is the existence of functional HNF1 response elements in the human PAH gene regulatory region. Although a HNF1 binding site has not been found in the human PAH gene, a liver-specific hormone-inducible enhancer at −3.5 kb that contains two functional HNF1 binding sites and regulates the PAH gene promoter activity has been identified in mice (23). In addition, in HNF1 knockout mice, the hepatic PAH gene was silent, and the mice suffered from pronounced phenylketonuria (24), demonstrating the involvement of HNF1 in the expression of the mouse PAH gene. Because the human PAH gene and the mouse PAH gene have a similar tissue-specific expression pattern, it seemed probable that HNF1 could also be involved in the liver-specific expression of the human PAH gene. Therefore, we carried out transactivation studies with the reporter plasmid and a HNF1 expressing plasmid, pBJ5-HNF1. As shown in Fig. 1, HNF1 transactivated the 9-kb fragment in a dose-dependent manner with a maximum of nearly 8-fold activation. Other HNF1-unrelated plasmids such as pBJ5-DCoH and pBJ5 had no transactivation activity on the 9-kb fragment, indicating that the observed effects were HNF1-specific. In contrast, HNF1 had only marginal effects on the 1.5-kb fragment. Sequence analysis of the 1.5-kb fragment did not detect any segments with significant homology to the HNF1 binding sites consensus sequence GTTAATNATTAAC (unpublished data). These results suggest that HNF1 is involved in the liver-specific expression of the human PAH gene and that the HNF1 binding sites are located upstream of the 1.5-kb fragment.
Figure 1.
Dose-dependent transactivation of HNF1 on the 9-kb fragment. CHO cells were cotransfected with 5 μg of phPAH9CAT or phPAH1.5CAT and the indicated amount of pBJ5-HNF1. pSV-β-Galactosidase (2 μg) was included in all transfections as an internal control. The total amount of DNA included was kept constant by adjusting the amount of pBJ5 lacking an insert. The data shown are means ± SD (n = 2–6).
Potentiation of the Transactivation Activity of HNF1 by DCoH.
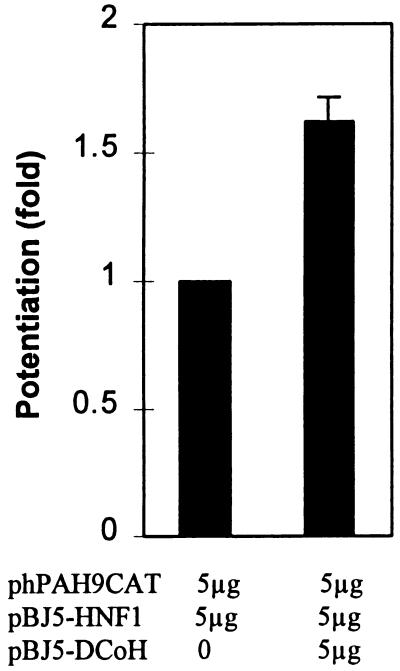
We further examined whether the observed HNF1 effects would be enhanced by DCoH. As shown in Fig. 2, DCoH did potentiate by 1.6 ± 0.1-fold the transactivation effects of HNF1 on the 9-kb fragment. Although this potentiation effect is modest, it was reproducible in the cotransfection experiments. As a control, the parent plasmid pBJ5 did not show the potentiation effect. A similar level of potentiation by DCoH on HNF1’s transactivation on a luciferase reporter plasmid with three copies of the HNF1 binding site of the β-fibrinogen gene upstream of a promoter has also been observed (25).
Figure 2.
Potentiation by DCoH of the HNF1-mediated transactivation. Cotransfection was as described in Materials and Methods. The data shown are mean ± SD (n = 6).
Location of the HNF1 Binding Sites.
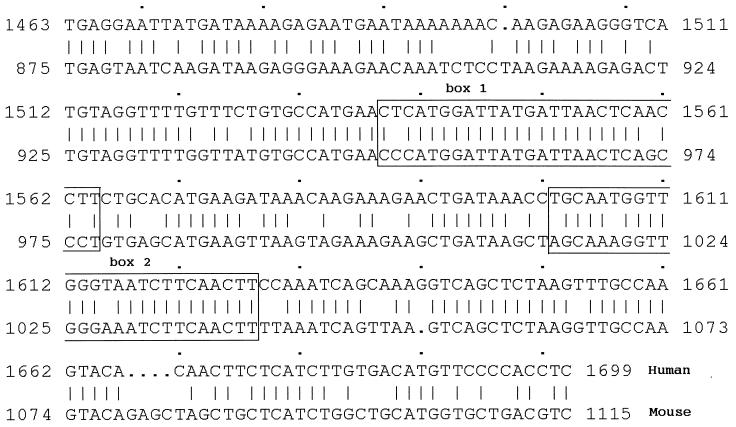
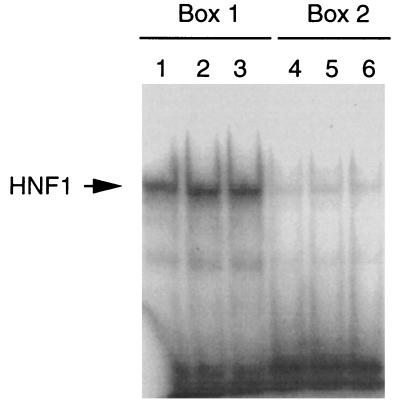
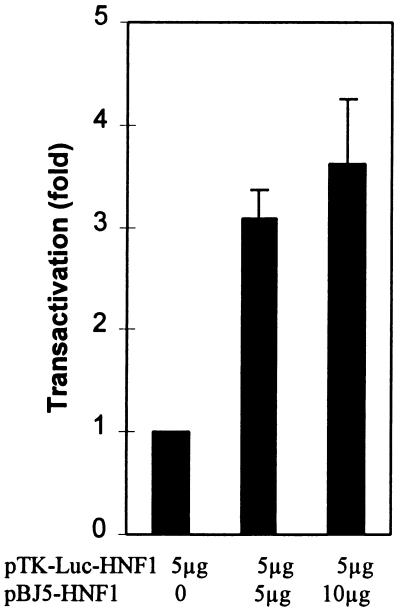
To locate the HNF1 binding sites, we cotransfected CHO cells with various deletion constructs of the 9-kb fragment with or without the HNF1-expressing plasmid. To construct the deletion plasmids, we first analyzed the physical map of the 9-kb fragment. The restriction endonucleases used to construct the deletion plasmids and to determine the orientation of the subclones are depicted in Fig. 3. As can be seen, removal of the distal 5′ region such as the 1.5-kb XbaI–XbaI region did not affect the transactivation activity by HNF1. However, a deletion plasmid lacking the 1.7-kb XbaI–XbaI region lost more than 70% of its ability to be transactivated by HNF1. Deletion of the 3.8-kb BglII–BglII region, which includes the 1.7-kb XbaI–XbaI segment, also caused the same degree of loss of activity. These results suggest that the 1.7-kb fragment at about 3.5 kb upstream of the human PAH gene harbors the HNF1 binding response elements. Early observations from transfection assays of the 9-kb fragment of the human PAH gene carried out in human hepatoma cells also suggested that the region of −4.0 to −1.5 kb contains important cis elements (21). Deletion of the region upstream of −4 kb had negligible effects on the human PAH promoter activity. However, deletion close to −1.5 kb resulted in 80% loss of activity. To define the HNF1 binding sites, we sequenced the complete 1.7-kb fragment. Interestingly, when the 1.7-kb fragment was compared with the published tissue-specific hormone-inducible enhancer of the mouse PAH gene (23), we found a 77.5% similarity (Fig. 4). In contrast, the sequences flanking this highly homologous segment have a similarity of 40–43%. Moreover, this similarity is even higher than that between the human and mouse PAH proximal promoter (unpublished data). This highly homologous segment is located within the 0.5-kb BamHI–XbaI fragment at about −3.5 kb, which has the same position as the mouse PAH enhancer. Two functional HNF1 binding sites were previously identified in the mouse PAH enhancer (23). At the corresponding positions of the human PAH 9-kb fragment, two very similar segments are present (Fig. 4, boxes 1 and 2). In box 1, only the ends of the boxed sequences differ by three bases. The sequences indicated in box 2 are less homologous. There are two base differences in the center of the HNF1 binding site in addition to another one at one end. To test whether sequences of box 1 and box 2 from the human PAH gene bind with HNF1, we carried out gel mobility shift assays using purified HNF1. As shown in Fig. 5, both the segments in box 1 (lane 1–3) and box 2 (lane 4–6) of the human PAH gene fragment were shifted by HNF1. Apparently, the binding site in box 1 has a relatively higher affinity for HNF1 than that in box 2. We also tested whether the HNF1 binding sites in the human PAH gene can confer HNF1 response to a heterologous promoter. The 0.5-kb BamHI–XbaI fragment that is very similar to the mouse PAH enhancer was inserted upstream of the thymidine kinase promoter of the reporter vector pTK-Luc to construct pTK-Luc-HNF1. Cotransfection assays showed (Fig. 6) that this fragment activated the thymidine kinase promoter activity 3- to 4-fold in the presence of HNF1. The enhanced expression of luciferase protein was confirmed by Western blot assays (data not shown).
Figure 4.
Sequence comparison of the 1.7-kb segment from the human 9-kb fragment with the mouse PAH gene enhancer. Similarity between the 1.7-kb segment from the human 9-kb fragment and the mouse PAH gene enhancer was analyzed by the program bestfit, Wisconsin Package Version 9.0, Genetics Computer Group (GCG). The first base of the second XbaI site (see Fig. 3) in the 9-kb fragment was numbered 1. The HNF1 binding sites defined in the mouse PAH gene enhancer and the human homologous sequences are boxed. The mouse sequences in the boxes correspond to the HNF1 footprint regions (23).
Figure 5.
Gel mobility shift assay of the HNF1 binding sites. Gel mobility shift assays were carried out as described in Materials and Methods. The same amount of purified HNF1 (50 ng) and 32P-labeled oligonucleotide (3 × 10 5cpm) corresponding to the sequence of the 9-kb fragment in boxes 1 and 2 was used in the assay. For each oligonucleotide, assays were performed in triplicate.
Figure 6.
Activation of the heterologous thymidine kinase promoter by human PAH HNF1 binding sites. The 0.5-kb BamHI–XbaI fragment containing human HNF1 binding sites were inserted upstream of thymidine kinase protomer of the luciferase vector pTK-Luc to construct pTK-Luc-HNF1. HNF1 response was examined by cotransfecting CHO cells with pTK-Luc-HNF1 in the presence or absence of pBJ5-HNF1. The data shown are means ± SD (n = 3).
DISCUSSION
We have shown that the 9-kb human PAH 5′ flanking fragment can be transactivated by HNF1 in a dose-dependent manner and that DCoH potentiated the transactivation effects of HNF1 by 1.6-fold. This observation suggests that with respect to the phenylalanine hydroxylation system, DCoH/PHS in vivo may have a dual function. On one hand, through its carbinolamine dehydratase activity, it is involved in regenerating the cofactor tetrahydrobiopterin, whose oxidation is coupled to the hydroxylation of phenylalanine; on the other hand, as the dimerization cofactor of HNF1, it up-regulates the expression of the PAH gene by enhancing the transactivation activity of HNF1 on the PAH gene. Accordingly, deficiencies in DCoH/PHS may possibly have two effects on the phenylalanine hydroxylation system: a deficiency in the regeneration of tetrahydrobiopterin and a lower PAH activity caused by a decreased rate of synthesis of PAH. It should be noted that because our recent findings indicate that the enzymatic and transcriptional activities of PHS are not strictly coupled (25), these two effects may not always occur simultaneously with all PHS deficiencies.
Previous studies on the human (21) and mouse (23) PAH genes revealed a lack of similarity between their potential regulatory regions. In brief, the mouse PAH gene promoter and upstream region, up to −7 kb, showed extremely weak activity in transitory expression assays, irrespective of the origin of the host cell. In contrast, the human PAH gene promoter and upstream regions, up to −9 kb, were active in cotransfection assays, and the −121-bp proximal promoter still retained a significant level of activity only in hepatic cells. Our finding that there is a high degree of identity between the 5′ flanking sequence of the human PAH gene and the mouse PAH enhancer at the same position suggests that these two genes also share some similarities, at least with respect to the involvement of HNF1 in their regulation. But even with this respect, these two genes show differences. The activity of the mouse PAH gene enhancer required hormones such as dexamethasone or cAMP, whereas the human PAH HNF1 response elements were functional in the absence of added hormones.
The mutations causing phenylketonuria or hyperphenylalaninemia that have already been characterized are only found in the coding sequence or the exon–intron splice junctions of the human PAH gene (26). However, mutations in the regulatory region could also lead to disease. For example, natural mutations in the C/EBP binding site in the human factor IX promoter are known to cause hemophilia B (27), and natural mutations in the Sp1 and ATF sites in the human retinoblastoma gene result in hereditary retinoblastoma (28). The search for mutations in the regulatory region of the human PAH gene has been hampered by the lack of information about cis elements. The structural characterization of the human proximal PAH promoter allowed the detection of three mutations in this region. Although these mutations did not correlate with phenylketonuria or hyperphenylalaninemia (29), the possibility could not be excluded that disease-causing mutations might occur in other regulatory regions. The identification of the HNF1 binding site-containing regulatory cis element in the human PAH 9-kb 5′ flanking fragment in the present work makes it possible to detect potential mutations in this region. In regard to such a possibility, special attention should be paid to classical phenylketonuria or hyperphenylalaninemic patients with no mutations in the PAH coding sequence or the exon–intron splice junctions.
Acknowledgments
We are grateful to Dr. S. Woo for providing the plasmids phPAH1.5CAT and phPAH9CAT. We thank Dr. G. Johnen for providing purified HNF1 and critical reading of the manuscript. We also thank C. Falke and A.-M. Vasquez for technical assistance.
ABBREVIATIONS
- HNF1
hepatocyte nuclear factor 1
- PAH
phenylalanine hydroxylase
- DCoH/PHS
dimerization cofactor of HNF1/phenylalanine hydroxylase stimulator
- CHO
Chinese hamster ovary
- CAT
chloramphenicol acetyltransferase
Footnotes
Data deposition: The sequence reported in this paper has been deposited in the GenBank database (accession no. AF033857).
References
- 1.Kaufman S. Adv Enzymol Relat Areas Mol Biol. 1993;67:77–264. doi: 10.1002/9780470123133.ch2. [DOI] [PubMed] [Google Scholar]
- 2.Kaufman S. J Biol Chem. 1970;245:4751–4759. [PubMed] [Google Scholar]
- 3.Huang C Y, Kaufman S. J Biol Chem. 1973;248:4242–4251. [PubMed] [Google Scholar]
- 4.Lazarus R A, Benkovic S J, Kaufman S. J Biol Chem. 1983;258:10960–10962. [Google Scholar]
- 5.Kaufman S. Proc Natl Acad Sci USA. 1963;50:1085–1093. doi: 10.1073/pnas.50.6.1085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Adler C, Ghisla S, Rebrin I, Haavik J, Heizmann C W, Blau N, Kuster T, Curtius H C. Eur J Biochem. 1992;208:139–144. doi: 10.1111/j.1432-1033.1992.tb17167.x. [DOI] [PubMed] [Google Scholar]
- 7.Citron B A, Kaufman S, Milstien S, Naylor E W, Greene C L, Davis M D. Am J Hum Genet. 1993;53:768–774. [PMC free article] [PubMed] [Google Scholar]
- 8.Davis M D, Kaufman S. FEBS Lett. 1991;285:17–20. doi: 10.1016/0014-5793(91)80714-e. [DOI] [PubMed] [Google Scholar]
- 9.Davis M D, Ribeiro P, Tipper J, Kaufman S. Proc Natl Acad Sci USA. 1992;89:10109–10113. doi: 10.1073/pnas.89.21.10109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Curtius H C, Adler C, Rebrin I, Heizmann C, Ghisla S. Biochem Biophys Res Commun. 1990;172:1060–1066. doi: 10.1016/0006-291x(90)91554-6. [DOI] [PubMed] [Google Scholar]
- 11.Davis M D, Kaufman S, Milstien S. Proc Natl Acad Sci USA. 1991;88:385–389. doi: 10.1073/pnas.88.2.385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Citron B A, Davis M D, Milstien S, Gutierrez J, Mendel D B, Crabtree G R, Kaufman S. Proc Natl Acad Sci USA. 1992;89:11891–11894. doi: 10.1073/pnas.89.24.11891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hauer C R, Rebrin I, Thony B, Neuheiser F, Curtius H C, Hunziker P, Blau N, Ghisla S, Heizmann C W. J Biol Chem. 1993;268:4828–4831. [PubMed] [Google Scholar]
- 14.Mendel D B, Khavari P A, Conley P B, Graves M K, Hansen L P, Admon A, Crabtree G R. Science. 1991;254:1762–1767. doi: 10.1126/science.1763325. [DOI] [PubMed] [Google Scholar]
- 15.Mendel D B, Crabtree G R. J Biol Chem. 1991;266:677–680. [PubMed] [Google Scholar]
- 16.Hansen L P, Crabtree G R. Curr Opin Genet Dev. 1993;3:246–253. doi: 10.1016/0959-437x(93)90030-s. [DOI] [PubMed] [Google Scholar]
- 17.Zhao G, Xia T, Song J, Jensen R A. Proc Natl Acad Sci USA. 1994;91:1366–1370. doi: 10.1073/pnas.91.4.1366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Endrizzi J A, Cronk J D, Wang W, Crabtree G R, Alber T. Science. 1995;268:556–559. doi: 10.1126/science.7725101. [DOI] [PubMed] [Google Scholar]
- 19.Ficner R, Sauer U H, Stier G, Suck D. EMBO J. 1995;14:2034–2042. doi: 10.1002/j.1460-2075.1995.tb07195.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wang Y, DeMayo J L, Hahn T M, Finegold M J, Konecki D S, Lichter-Konecki U, Woo S L. J Biol Chem. 1992;267:15105–15110. [PubMed] [Google Scholar]
- 21.Wang Y, Hahn T M, Tsai S Y, Woo S L. J Biol Chem. 1994;269:9137–9146. [PubMed] [Google Scholar]
- 22.Seed B, Sheen J Y. Gene. 1988;67:271–277. doi: 10.1016/0378-1119(88)90403-9. [DOI] [PubMed] [Google Scholar]
- 23.Faust D M, Catherin A M, Barbaux S, Belkadi L, Imaizumi-Scherrer T, Weiss M C. Mol Cell Biol. 1996;16:3125–3137. doi: 10.1128/mcb.16.6.3125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Pontoglio M, Barra J, Hadchouel M, Doyen A, Kress C, Bach J P, Babinet C, Yaniv M. Cell. 1996;84:575–585. doi: 10.1016/s0092-8674(00)81033-8. [DOI] [PubMed] [Google Scholar]
- 25.Johnen G, Kaufman S. Proc Natl Acad Sci USA. 1997;94:13469–13474. doi: 10.1073/pnas.94.25.13469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Nowacki P, Byck S, Prevost L, Scriver C R. Nucleic Acids Res. 1997;25:139–142. doi: 10.1093/nar/25.1.139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Crossley M, Brownlee G G. Nature (London) 1990;345:444–446. doi: 10.1038/345444a0. [DOI] [PubMed] [Google Scholar]
- 28.Sakai T, Ohtani N, McGee T L, Robbins P D, Dryja T P. Nature (London) 1991;353:83–86. doi: 10.1038/353083a0. [DOI] [PubMed] [Google Scholar]
- 29.Svensson E, Wang Y, Eisensmith R C, Hagenfeldt L, Woo S L. Eur J Hum Genet. 1993;1:306–313. doi: 10.1159/000472429. [DOI] [PubMed] [Google Scholar]