Abstract
This study aims to determine the influence of the polymorphism within the intron 2 of the interleukin-1 receptor antagonist gene (IL-1RN*) on the outcome of severe sepsis, and to assess its functional significance by correlating this polymorphism with the total production of interleukin-1 receptor antagonist (IL-1Ra) protein determined in stimulated peripheral blood mononuclear cells (PBMC). A group of 78 patients with severe sepsis (51 survivors and 27 nonsurvivors) was compared with a healthy control group of 130 blood donors, and 56 patients with uncomplicated pneumonia. We found a significant association between IL-1RN* polymorphism and survival. Thus, after adjusting for age and APACHE II score, multiple logistic regression analysis showed that patients homozygotes for the allele *2 had a 6·47-fold increased risk of death (95% CI 1·01–41·47, P = 0·04). Besides, compared with patients homozygous or heterozygous for the allele *1, IL-1RN*2 homozygotes produced significantly lower levels of IL-1Ra from their PBMC. Our results suggest that insufficient production of this cytokine might contribute, among other factors, to the higher mortality rate found in severe sepsis patients with the IL-1RN*2 homozygous genotype.
Keywords: interleukin-1Ra, gene polymorphism, cytokine production, peripheral blood, mononuclear cells, sepsis, outcome
INTRODUCTION
Polymorphic gene sequences of certain cytokines could be potential markers of susceptibility and clinical outcome in different human infectious diseases [1–3]. Genetic variations of TNF-α and TNF-β genes have been associated with fatal outcome in patients with severe sepsis [4] and septic shock [5]. Polymorphisms of the genes of the IL-1 complex have been scarcely evaluated in sepsis, and limited conclusions are available so far [6]. In one single study [7], association between the polymorphic region within intron 2 of the IL-1Ra (IL-1RN*) gene and susceptibility to suffer severe sepsis, but not disease outcome, was found; nevertheless, authors did not evaluate the functional significance of such association in terms of cytokine production by blood cells. This is a very interesting issue according to recent findings of our group revealing that, apart from nuclear factor κB (NF-κB) activation, the IL-1Ra production was the only cytokine whose plasma levels were of value in predicting mortality in patients with sepsis [8].
The polymorphic region within intron 2 of the IL-1RN* gene contains a variable numbers of tandem repeats (VNTR) of 86 bp (see Fig. 1a); five alleles of the IL-1RN* have been reported (*1 to *5), corresponding to 2, 3, 4, 5 and 6 copies of the 86-bp sequence, respectively [9]; the most frequent allele was designated as *1. Since three potential protein-binding sites are located in the 86-bp sequence, it is likely that the number of repeats may influence gene transcription and protein synthesis. It was reported that IL-1Ra plasma levels are coordinately regulated by both IL-1RN* and IL-1β genes [10], and that in vitro activated peripheral blood mononuclear cells (PBMCs) from healthy IL-1RN*2 allele carriers produce higher levels of IL-1Ra than the noncarriers [11], but these observations should be confirmed in different ethnic populations.
Fig. 1.
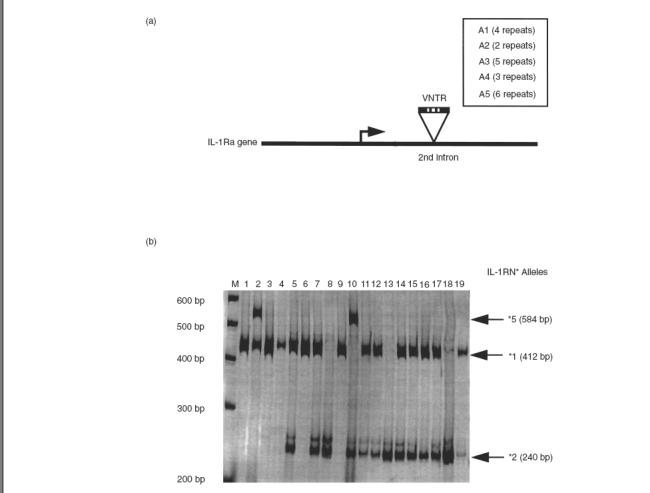
Interleukin-1 receptor antagonist genotypes. (a) Scheme of the IL-1RN* polymorphism; variable number of tandem repeats (VNTR) for each allele are indicated in box. (b) Agarose gel (2%) displaying IL-1RN* genotypes in 19 healthy controls: homozygotes *1/1 (lanes 1, 3, 4, 6 and 9); homozygotes *2/2 (lanes 8, 13 and 18); heterozygotes *1/5 (lane 2); heterozygotes *1/2 (lanes 5, 7, 11, 12, 14, 15, 16 and 17 and 19); heterozygotes *2/5 (lane 10). M = base pair marker.
To our knowledge, no study has evaluated the genetic interindividual differences in the production of IL-1Ra after various stimuli in patients with sepsis. We performed a case-control study in patients with severe sepsis and ethnically matched healthy subjects to determine the influence of IL-1RN* polymorphism on mortality, and to assess the functional significance of this polymorphism by correlating ex vivo IL-1Ra production with IL-1RN* genotypes.
MATERIALS AND METHODS
Patients
From an original cohort of 200 critically ill patients diagnosed with sepsis, consecutively admitted to the Intensive Care Unit (ICU) over a 15-month period, 78 patients fulfilled the consensus criteria for the definition of severe sepsis (ACCP/SCCM Consensus Conference) [12], and they form the basis of the study. The time between admission and inclusion in the study varied from 1 to 12 days (median, 5 days). Written informed consent was obtained from patients or their relatives, and the study was approved by the local ethics committee. Severity of illness was assessed on enrolment using the Acute Physiologic and Chronic Health Evaluation (APACHE II) [13], and the individual predicted mortality by converting the score into probability of death using logistic regression [14]. Primary sites of infection were complicated pneumonia (32 patients), abdominal abscesses (17 patients), and infections of either cutaneous (n = 11), biliary tract (n = 7), urinary tract (n = 8), or neurological origin (n = 2). In all patients, empirical antibiotic treatment was judged adequate. The exclusion criteria were: undrained source of surgical sepsis, malignancy and chronic diseases, treatment with steroids or immunosuppressive drugs, liver dysfunction, renal insufficiency, AIDS and gestation. Patients were followed for 30 days after inclusion. To compare the genotype distribution and allele frequencies in patients with severe sepsis, a group of 56 patients with a community-acquired pneumonia admitted to the Internal Medicine Service who did not met sepsis criteria served as reference. Also 130 healthy unrelated blood donor subjects (control group) were included. All patients and control subjects were of Spanish Caucasian origin and resident in the Madrid region; none of them had known Jewish ancestry. For all patients, blood samples were taken on entry into the study, that is at the time of systolic hypotension, and every 6 h thereafter for 24 h. A single blood sample was obtained from each control subject and patient with pneumonia.
Genotyping of subjects for the IL-1RN* polymorphism
Genomic DNA was extracted by standard methods using the Genomic Blood DNA Purification Kit (Amershan Pharmacia Biotech). Genotyping was performed by a polymerase chain reaction (PCR)-based method with our own modifications [15]. Briefly, 2 ng of DNA was used as template in 25 μl of PCR reaction; the reaction sample contains 2·5 units of Biotools DNA Polymerase (Biotools B & M Laboratories, S.A., Spain), dNTPs and oligonucleotides specific for framework sequences flanking the polymorphic region of exon 2. Primers were synthesized by Isogen (Isogen Bioscience BV, Maarseen, The Netherlands): sense 5′-CTCAGCAACACTCCTAT-3′, and antisense 5′-TCCT GGTCTG-CAGGTAA-3′. The PCR conditions consist in the initial denaturation at 94°C for 3 min, followed by 30 cycles of 94°C for 30 s, 56°C for 30 s and 72°C for 30 s, and final extension at 72°C for 5 min. Resulting PCR products of 410 bp (IL-1RN*1, four repeats), 240 bp (IL-1RN*2, two repeats), 500 bp (IL-1RN*3, five repeats), 325 bp (IL-1RN*4, three repeats) and 595 bp (IL-1RN*5, six repeats) were run on 2% agarose gels stained with ethidium bromide (0·1%) to reveal the alleles present. A molecular ladder of 100 bp (Gibco/BRL, Life Technologies SA, Spain) was used to estimate the size of the PCR fragments (Fig. 1b).
Ex vivocytokine production from stimulated PBMC
Cells from 30 healthy controls and 78 patients with severe sepsis were used for evaluating cytokine production, according to our own protocol [16]. Briefly, cells were cultured at a concentration of 2·0 × 106 cells/ml, and incubated during 18 h in RPMI 1640 medium supplemented with 2% penicillin-streptomycin, 5% l-glutamine and 2% AB serum (Gibco, Grand Island, NY), at 37°C under 5% CO2 humidified air, in the presence or absence of either lipopolysaccharide (LPS from E. coli serotype 055:B5, Sigma Chem. Co., St. Louis, MO) or a combination of LPS plus IL-4 (both at a concentration of 25 μg/ml). After incubation, with or without stimulation, cells were counted in a Neubauer chamber, being the cell viability higher than 95%. The supernatants were collected and frozen at –70°C.
Cytokine assay
ELISA-kits were used for measurements of IL-1β (Medgenix Diagnostics™, Fleurus, Belgium) and IL-1Ra (Quantikine™, R & D systems, Minneapolis, Minn, USA) in plasma and in the supernatants of stimulated or nonstimulated PBMC. The detection limit of the assays was 20 pg/ml (IL-1β) and 250 pg/ml (IL-1Ra). Both assays presented an interassay and intra-assay CV lower than 9·2% and 5·1%, respectively.
Statistical analysis
Allele and genotype frequencies were compared by Χ2 test. Variables were tested for their association with mortality using a Χ2 test for categorical data, and the Mann–Whitney U-test for numerical data. Genotype frequencies in patients and controls were not significantly different from those predicted under Hardy–Weinberg equilibrium (Linkage Utility programs provided by Dr Jurg Ott, Laboratory of Statistical Genetics, Rockefeller University, NY). The strength of association between genotypes and outcome was expressed by the Odds ratios (OR). Comparison between expected and observed mortality was made by the Poisson distribution. A multiple logistic regression model including age, IL-1RN* genotypes, and the APACHE II-derived probability of dying was used to determine the risk of mortality of patients homozygous for IL-1RN*2.
RESULTS
The allele frequencies and genotype distributions in the study population are shown in Table 1. No significant difference in the IL-1RN* allele frequency or genotype distribution was seen between patients with severe sepsis, patients with uncomplicated pneumonia or healthy control volunteers (Pearson x2 = 2·1 and 2·4, respectively, P = NS).
Table 1.
Demographic data, IL-1RN* genotypes and allele frequencies in the study population
| Healthy donors (n = 130) | Pneumonia patients (n = 56) | Severe sepsis patients (n = 78) | P | |
|---|---|---|---|---|
| Gender (M/F) | 65/65 | 28/27 | 40/38 | NS |
| Age (years) | 48 ± 19 (29–65) | 58 ± 16 (32–70) | 54 ± 13 (36–74) | NS |
| IL-1RN* genotypes | ||||
| 1/1 | 60 (46) | 24 (43) | 32 (41) | NS |
| 1/2 | 54 (42) | 26 (46) | 33 (42) | |
| 2/2 | 12 (9) | 5 (9) | 13 (17) | |
| 1/5 | 3 (2) | 1 (2) | 0 | |
| 2/5 | 1 (1) | 0 | 0 | |
| IL-1RN* alleles | ||||
| Allele 1 | 177 (68) | 75 (67) | 97 (62) | NS |
| Allele 2 | 79 (30) | 36 (32) | 59 (38) | |
| Allele 5 | 4 (2) | 1 (1) | 0 |
Age is mean ± s.d. (extreme values). Genotype and allele frequencies are expressed as absolute numbers, with relative values, expressed as percentage, in parentheses. Statistical analysis of the genotype and allele frequencies were done by chi-square test. NS = not significant.
The observed mortality rate of 34·6% (27/78 patients did not survive) was in agreement with the predicted risk of death (37·2%) based on the APACHE II score [14]. The clinical characteristics of the survivors and nonsurvivors patients, genotype, allele frequencies and average serial of cytokine concentrations in plasma over the first 24 h are compared in Table 2. The mean age and APACHE II scores were significantly higher, and plasma IL-1Ra concentrations were lower in patients who died. The apparent severity of illness at the time of admission, expressed as mean APACHE II score, was comparable in the three genotypic groups (20·7 ± 2·1 for allele 2 homozygotes, 20·2 ± 2·4 for heterozygotes, and 20·5 ± 1·9 for allele 1 homozygotes, P > 0·05). In contrast, a significant association between the genotype *2/2 with poor prognosis was found (Table 2). Patients with the homozygous genotype IL-1RN*2 had significantly higher mortality rate than those carrying other genotypes (77% versus 27% for *1/2 heterozygotes and 25% for *1/1 homozygotes, respectively; P = 0·001, x2 = 12·4). When compared with patients carrying at least one IL-1RN*1 allele (*1/1 homozygous and *1/2 heterozygous genotype), the IL-1RN*2 homozygous genotype was associated with mortality with an OR of 9·41 (P = 0·002; 1 d.f., Χ2 = 12·37; 95% CI 2·31–38·31). To find out whether homozygous genotype IL-1RN*2 might be an independent predictor of mortality, a multiple logistic regression model was performed. After adjusting for age and APACHE II score, patients homozygotes for IL-1RN*2 had a 6·47-fold increased risk of death (OR 6·47, 95% CI 1·01–41·47, 1 d.f., P = 0·04).
Table 2.
Characteristics of the severe sepsis group
| Severe sepsis patients (n = 78) | Non-survivors (n = 27) | Survivors (n = 51) | P | |
|---|---|---|---|---|
| Gender (M/F) | 40/38 | 13/14 | 27/24 | NS |
| Age, mean ± s.d. (range) | 53·6 ± 9·6 (27–74) | 59·3 ± 6·9 (45–74) | 50·6 ± 9·4 (27–73) | < 0·001a |
| APACHE II score, mean ± s.d. (range) | 20·4 ± 2·2 (16–25) | 21·9 ± 2·1 (16–25) | 18·5 ± 1·7 (16–23) | < 0·001a |
| Positive blood culture/local culture, no. | 32/46 | 12/15 | 20/31 | NS |
| Gram-negative/Gram-positive bacteria, no. | 29/38 | 10/14 | 19/24 | NS |
| Mixed flora, no. (%), | 11 | 3 | 8 | |
| Plasma IL-1β (pg/ml) | 41 (0–110) | 52 (0–110) | 36 (0–88) | NS |
| Plasma IL-1Ra (ng/ml) | 1·6 (0·7–3·6) | 1·3 (0·7–1·9) | 1·8 (1·0–3·6) | < 0·001a |
| IL-1RN* genotypes | ||||
| *1/1 | 32 (41) | 8 (30) | 24 (47) | < 0·002b |
| *1/2 | 33 (42) | 9 (33) | 24 (47) | |
| *2/2 | 13 (17) | 10 (37)c | 3 (6) | |
| IL-1RN* Alleles | ||||
| Allele 1 | 97 (62) | 25 (46) | 59 (71) | NS |
| Allele 2 | 59 (38) | 29 (54) | 30 (29) |
Plasma cytokine values are geometric mean (and range) of the 4 blood samples drawn during the first day. Genotype and allele frequencies are shown as absolute values, with relative values, expressed as percentage, in parentheses.
Comparison by Mann–Whitney U-test;
comparison by Χ2 test;
relative risk of mortality for IL-1RN
2/2 carriers is 9·4, P ≤ 0·002, Χ2 = 12·4; 95% CI: 2·31–38·1. NS = not significant.
Table 3 summarizes plasma levels of IL-1Ra, as well as the PBMC IL-1Ra and IL-1β production, in 30 healthy subjects and in all patients with sepsis, according to their genotypes and clinical outcome. No significant association between IL-1RN* polymorphism and plasma IL-1Ra levels was found in healthy individuals and patients with sepsis. In contrast, the ex vivo production of IL-1Ra from LPS-stimulated PBMC was closely related to the IL-1RN* alleles. Thus, healthy subjects carrying one or two copies of IL-1RN*1 (genotypes 1/1 and 1/2) had significantly higher production of IL-1Ra, in both unstimulated and stimulated culture conditions, compared with those homozygous for IL-1RN*2. Similarly, patients with the allele *1 (genotypes 1/1 and 1/2), produced 1·8-fold higher levels of IL-1Ra in unstimulated PBMC, and 3·1-fold higher after stimulation with LPS, compared with patients homozygous for IL-1RN*2. Regarding the patient’s outcome, production of IL-1Ra in survivors were 1·5-fold higher in unstimulated cells, and 2·7-fold higher after stimulation with LPS (P < 0·01 versus nonsurvivors). In contrast, no significant association of allele or genotype distribution of IL-1RN* with production of IL-1β in PBMC was detected either in healthy subjects or in patients with severe sepsis.
Table 3.
Plasma levels and ex vivo cytokine production from PBMC related to IL-1RN* genotypes in healthy subjects and patients with severe sepsis.Cytokine production according to desease outcome in severe sepsis was also shown
| Healthy blood donors Genotype distribution | Severe sepsis patients Genotype distribution | Severe sepsis patients Clinical outcome | ||||
|---|---|---|---|---|---|---|
| (*1/1, *1/2) (n = 19) | *2/2 (n = 11) | (*1/1, *1/2) (n = 65) | *2/2 (n = 13) | Survivors (n = 51) | Non-survivors (n = 27) | |
| Plasma IL-1Ra (ng/ml) | 0·3 ± 0·1 | 0·4 ± 0·2 | 1·7 ± 0·5 | 1·4 ± 0·4 | 1·8 ± 0·5 | 1·3 ± 0·2§ |
| (0·1 – 0·5) | (0·1 – 0·6) | (0·8 – 2·6) | (0·7 – 2·8) | (1·0 – 2·6) | (0·7 – 1·9) | |
| IL-1Ra production (ng/106 cells) | ||||||
| Non-stimulated | 2·1 ± 0·5 | 1·7 ± 0·4* | 4·3 ± 0·5 | 2·4 ± 0·4* | 4·1 ± 0·5 | 2·8 ± 0·5‡ |
| (0·8 – 3·4) | (0·8 – 3·1) | (3·4 – 5·5) | (1·8 – 3·0) | (3·1 – 5·3) | (1·8 – 3·7) | |
| LPS induced | 7·2 ± 0·6 | 3·1 ± 0·5** | 12·8 ± 1·0 | 4·1 ± 0·6† | 12·2 ± 1·1 | 4·5 ± 0·7§ |
| (5·7 – 8·9) | (1.8 – 4·4) | (10·5 – 14·7) | (2·9 – 5·5) | (9·8 – 14·6) | (3·0 – 6·2) | |
| LPS + IL-4 induced | 8·1 ± 0·5 | 3·8 ± 0·6** | 16·9 ± 1·2 | 5·0 ± 0·7† | 16·2 ± 1·4 | 5·2 ± 0·6§ |
| (6·8 – 10·3) | (2·3 – 5·6) | (14·5 – 19·8) | (3·4 – 7·0) | (13·1 – 19·2) | (3·8 – 6·9) | |
| IL-1β production (ng/106 cells) | ||||||
| Non-stimulated | 0·04 ± 0·01 | 0·05 ± 0·02 | 0·22 ± 0·16 | 0·36 ± 0·20 | 0·38 ± 0·20 | 0·45 ± 0·25 |
| LPS induced | 0·05 ± 0·02 | 0·07 ± 0·03 | 0·45 ± 0·22 | 0·58 ± 0·35 | 0·55 ± 0·30 | 0·40 ± 0·25 |
| LPS + IL-4 induced | 0·05 ± 0·02 | 0·06 ± 0·04 | 0·55 ± 0·30 | 0·61 ± 0·26 | 0·65 ± 0·35 | 0·60 ± 0·30 |
Values of cytokines are mean ± s.d.; in brackets the 95% confidence intervals of IL-1Ra values.
P ≤ 0·01, and
P ≤ 0·001 between healthy blood donors in each group;
P ≤ 0·001 between severe sepsis patients in each genotypic group;
P ≤ 0·01 and
P ≤ 0·001 between survivors and nonsurvivors.
DISCUSSION
This study reveals, for the first time, to our knowledge, that homozygosity for allele 2 of the IL-1RN* is strongly associated with a decreased production of IL-1Ra from culture of PBMC in patients with severe sepsis, and appears to be an independent risk factor for mortality. For identical age and rank of APACHE II score, patients with genotype *2/2 had a 6·47-fold increased risk of death. However, the interesting findings reported here should be interpreted cautiously since they are observational and provide no direct cause-and-effect relationship between the gene polymorphism studied and the poor clinical outcome observed.
A predisposition to develop severe sepsis, but not a worse prognosis, was linked to the IL-1RN*2 allele in a cohort of patients living in Germany [7], though the possible functional implication of such an association was not explored. In contrast, the current study shows no difference, either in allele frequency or genotype distribution for the IL-1RN* polymorphism between patients with severe sepsis and the two comparative groups, that is patients with uncomplicated pneumonia and healthy volunteers. Therefore, the possibility that the IL-1RN* polymorphism may cause a predisposition to sepsis in our ethnic group (Spanish-based white-Caucasian population) can be ruled out. This apparent discrepancy could be explained by differences in clinical conditions between patients in both series, and by ethnic differences in linkage of this particular cytokine gene polymorphism to disease severity, which has been reported in other diseases such as systemic lupus erythematosus [17,18] and ulcerative colitis [19,20].
We chose to look at the functional significance of the IL-1RN* polymorphism by correlating plasma IL-1Ra levels, and the ability of PBMC to produce IL-1β and IL-1Ra with IL-1RN* genotype. Whereas plasma IL-1Ra concentrations did not relate to any genotype, carriage of the IL-1RN* 2/2 genotype was associated with a decreased production of IL-1Ra from PBMC in patients with severe sepsis and ethnically matched healthy controls. Since the initial severity of illness, measured as APACHE II score at the time of admission, was similar in the three genotyped groups, it seems unlikely that the extent of systemic inflammation and monocyte deactivation might lead to a depression of the PBMC function in patients with severe sepsis, independent from the carriage of IL-1RN*2. Therefore, despite only 13 patients with the genotype 2/2 being enroled in this study, our data suggest that a genetic polymorphism in the IL-1RN* may contribute to the difference observed in IL-1Ra production capacity. In fact, our results are in line with a previous study reporting that production of IL-1Ra by the colonic mucosa was reduced in patients with ulcerative colitis with at least one copy of the allele 2 of IL-1RN* [21], but differ from those in other study [11] showing that carriage of the allele 2 was related to a high in vivo secretion of IL-1Ra in healthy white subjects from Australia. One possible explanation for that is, not only the different ethnic background of these populations, but also the fact that different cell preparation techniques and stimuli to elicit IL-1Ra release from PBMC were used in the two studies and might have resulted in different responses.
These results also indicate that IL-1Ra and IL-1β production have different genetic regulation since there was no association of IL-1RN* alleles with the PBMC release of IL-1β. The mechanism by which the IL-1RN* polymorphism, which lies in the noncodified nucleotide sequence of the cytokine gene, influences IL-1Ra production is unknown. There is evidence from in vitro studies that transcription or messenger RNA stability of IL-1Ra does not appear to be affected by carriage of allele 2 of IL-1RN* [22]. Nevertheless, it cannot be ruled out that the genotype 2/2 is not responsible for IL-1Ra secretion but is in linkage disequilibrium with another DNA variation, yet undiscovered, within the structural part of the IL-1RN* that regulates the transcription and directly influences IL-1Ra protein synthesis by PBMC.
In conclusion, the present work provides evidence that homozygosity of the IL-1RN*2 is associated with a decreased production of IL-1Ra in PBMC and higher mortality risk during severe sepsis. These findings are consistent with the hypothesis that individuals producing lower amounts of IL-1Ra are afforded a lower level of protection against fatal outcome than subjects producing higher levels. Hence, although reproducibility of our findings in other cohorts of patients of different ethnicity will be necessary, these results suggest that genotyping data offer a better risk assessment of patients with severe sepsis when they are assigned to study protocols for anti-inflammatory treatments.
Acknowledgments
This work was supported by grants from FIS (no. 0168/99, Ministerio Sanidad, and Universidad Autónoma de Madrid, Spain) to FA; and from DGES (PB98-0090, Ministerio Educación y Cultura, Spain) to CM. We thank the patient care staff and our patients for their willing involvement in the study.
References
- 1.Hill AV. The immunogenetics of human infectious diseases. Annu Rev Immunol. 1998;16:593–617. doi: 10.1146/annurev.immunol.16.1.593. [DOI] [PubMed] [Google Scholar]
- 2.Hurme M, Lahdenpohja N, Santtila S. Gene polymorphisms of interleukin 1 and 10 in infectious and autoimmune diseases. Ann Med. 1998;30:469–73. doi: 10.3109/07853899809002488. [DOI] [PubMed] [Google Scholar]
- 3.Dinarello CA. Proinflammatory and anti-inflammatory cytokines as mediators in the pathogenesis of septic shock. Chest. 1997;112(Suppl.):321S–329S. doi: 10.1378/chest.112.6_supplement.321s. [DOI] [PubMed] [Google Scholar]
- 4.Stüber F, Peterson M, Bokelmann F, Schade U. A genomic polymorphism within the tumour necrosis factor locus influences plasma tumour necrosis factor-α concentrations and outcome of patients with severe sepsis. Crit Care Med. 1996;24:381–4. doi: 10.1097/00003246-199603000-00004. [DOI] [PubMed] [Google Scholar]
- 5.Mira JP, Cariou A, Grall F, et al. Association of TNF2, a TNF promoter polymorphism, with septic shock susceptibility and mortality: a multicenter study. JAMA. 1999;282:561–8. doi: 10.1001/jama.282.6.561. [DOI] [PubMed] [Google Scholar]
- 6.Van Deventer SJH. Cytokine and cytokine receptor polymorphisms in infectious diseases. Intensive Care Med. 2000;26(Suppl.):S98–S102. doi: 10.1007/s001340051125. [DOI] [PubMed] [Google Scholar]
- 7.Fang MX, Schröder A, Hoeft A, Stüber F. Comparison of two polymorphisms of the interleukin-1 gene family: Interleukin-1 receptor antagonist polymorphism contributes to susceptibility to severe sepsis. Crit Care Med. 1999;27:1330–4. doi: 10.1097/00003246-199907000-00024. [DOI] [PubMed] [Google Scholar]
- 8.Arnalich F, García-Palomero E, López J, Jiménez M, Madero R, Renart J, Vázquez JJ, Montiel C. Predictive value of nuclear factor κ-B activity and plasma cytokine levels in patients with sepsis. Infect Immun. 2000;68:1942–5. doi: 10.1128/iai.68.4.1942-1945.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Tarlow JK, Blakemore AIF, Lennard A, Solari R, Highes HN, Steinkasserer A, Duff GW. Polymorphism in the human IL-1 receptor antagonist gene intron 2 is caused by variable numbers of an 86-bp tandem repeat. Human Genet. 1993;91:403–4. doi: 10.1007/BF00217368. [DOI] [PubMed] [Google Scholar]
- 10.Hurme M, Santtila S. IL-1 receptor antagonist (IL-1Ra) plasma levels are co-ordinately regulated by both IL-1Ra and IL-1β genes. Eur J Immunol. 1998;28:2598–602. doi: 10.1002/(SICI)1521-4141(199808)28:08<2598::AID-IMMU2598>3.0.CO;2-K. [DOI] [PubMed] [Google Scholar]
- 11.Danis VA, Millington M, Hyland VJ, Grennan D. Cytokine production by normal human monocytes: inter-subject variation and relationship to an IL-1 receptor antagonist (IL-1Ra) gene polymorphism. Clin Exp Immunol. 1995;99:303–10. doi: 10.1111/j.1365-2249.1995.tb05549.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bone RC, Balk RA, Cerra FB, et al. The Members of the ACCP/SCCM Consensus Conference Committee. Definitions for sepsis and organ failure and guidelines for the use of innovative therapy in sepsis. Chest. 1992;101:1644–55. doi: 10.1378/chest.101.6.1644. [DOI] [PubMed] [Google Scholar]
- 13.Knaus WA, Draper EA, Wagner DP, Zimmermann JR. APACHE II: a severity of disease classification system. Crit Care Med. 1985;13:389–93. [PubMed] [Google Scholar]
- 14.Beck DH, Taylor BL, Millar B, Smith GB. Prediction of outcome from intensive care: a prospective cohort study comparing Acute Physiologic and Chronic Health Evaluation II and III prognostic systems in a United Kingdom intensive care unit. Crit Care Med. 1997;25:9–15. doi: 10.1097/00003246-199701000-00006. [DOI] [PubMed] [Google Scholar]
- 15.García-Palomero E, Cuchillo-Ibañez I, García AG, Renart J, Albillos A, Montiel C. Greater diversity than previously thought of chromaffin cell Ca2+ channels, derived from mRNA identification studies. FEBS Lett. 2000;481:235–9. doi: 10.1016/s0014-5793(00)01984-0. [DOI] [PubMed] [Google Scholar]
- 16.Arnalich F, Hernánz A, Vázquez JJ, Amores A. Cell-mediated immune response and cytokine production in idiopathic senile anorexia. Mech Ageing Dev. 1994;77:67–74. doi: 10.1016/0047-6374(94)90047-7. [DOI] [PubMed] [Google Scholar]
- 17.Blakemore AIF, Tarlow JK, Cork MJ, Gordon C, Emery P, Duff GW. Interleukin-1 receptor antagonist gene polymorphism as a disease severity factor in systemic lupus erythematosus. Arthritis Rheum. 1994;37:1380–5. doi: 10.1002/art.1780370917. [DOI] [PubMed] [Google Scholar]
- 18.Danis VA, Millington M, Huang Q, Grennan D. Lack of association between an interleukin-1 receptor antagonist gene polymorphism and systemic lupus erythematosus. Dis Markers. 1994;12:135–9. doi: 10.1155/1994/464787. [DOI] [PubMed] [Google Scholar]
- 19.Carter MJ, Di Giovine FS, Jones S, Mee J, Camp NJ, Lobo AJ, Duff GW. Association of the interleukin-1 receptor antagonist gene with ulcerative colitis in Northern European Caucasians. Gut. 2001;48:461–7. doi: 10.1136/gut.48.4.461. 10.1136/gut.48.4.461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hacker CT, Gomolka M, Keller E, et al. Lack of association between an interleukin-1 receptor antagonist gene polymorphism and ulcerative colitis. Gut. 1997;40:623–7. doi: 10.1136/gut.40.5.623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Andus T, Daig R, Vogl D, et al. Imbalance of the interleukin-1 system in the colonic mucosa – association with intestinal inflammation and interleukin-1 receptor antagonist genotype 2. Gut. 1997;41:651–7. doi: 10.1136/gut.41.5.651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Clay FE, Tarlow JK, Cork MJ, Cox A, Nicklin MJH, Duff GW. Novel interleukin-1 receptor antagonist exon polymorphisms and their use in allele-specific mRNA assessment. Human Genet. 1996;97:723–6. doi: 10.1007/BF02346180. [DOI] [PubMed] [Google Scholar]


