Abstract
A number of studies have found increased levels of antibodies to human endogenous retroviruses (HERVs) in autoimmune rheumatic diseases. It is not clear whether this immune response is driven by the HERV itself or by cross-reactions with an exogenous virus or an autoantigen. To address this question, we examined the antibody response to the Env protein of two closely related members of the HERV-K family, HERV-K10 and IDDMK1,222. By immunoblotting of recombinant proteins, antibodies were found in 32–47% of 84 sera from patients with autoimmune rheumatic disease, and 29% of 35 normal controls. Epitope mapping with overlapping 15mers identified multiple reactive peptides on both antigens, with one (GKTCPKEIPKGSKNT) containing immunodominant epitope(s). By ELISA, the median titre of antibody to this peptide was significantly increased in 39 patients with SLE compared to 39 healthy controls and 86 patients with other rheumatic diseases (P < 0·003).
We have shown that there is a high frequency of IgG antibodies to HERV-K env sequences in human sera, both in health and autoimmune rheumatic disease, and that the response is to multiple epitopes. This supports the hypothesis that the autoimmune response to HERV-K is antigen-driven and may be an early stage in the chain of events that leads to tolerance breakdown to other autoantigens.
Keywords: HERV-K, autoimmunity, lupus, Sjogren’s syndrome, rheumatoid arthritis
INTRODUCTION
Retroviral elements homologous to infectious retroviruses can be found integrated as endogenous proviruses in the genome of most vertebrates. In primates, such integration is thought to have happened several times over the past 30 million years [1,2]. Endogenous retroviral related sequences represent up to 5% of the human genome [3]. They are transmitted genetically in a Mendelian fashion and for this reason are found in the genome of all cells of the host. In many cases, human endogenous retroviruses (HERVs) contain deletions, stop codons or frame shifts such that no protein is expressed. In other instances both transcripts and proteins have been described. These include ERV-3 [4–8], HERV-W [9,10] and a number of members of the HERV-K family [11–14].
Amongst the HERVs, HERV-K10 is one of the best characterized. It was completely sequenced in 1986 as a full-length 9·4 kb provirus flanked by LTRs at both ends [11]. This element is present in multiple copies (about 30) in the human genome [12], with most of them having open reading frames for Gag proteins and for a truncated envelope protein [15]. At least one member of this family has an open reading frame for a full length envelope protein [16]. Recently, what was claimed to be a new human endogenous retrovirus termed IDDMK1,222 [17] was detected in culture supernatants of leucocyte-infiltrated islet cells from two patients with type 1 diabetes, though its proposed link with the disease has been challenged by several studies [18–23]. Nevertheless, this sequence remains of interest because antibodies to HERV-K Env protein have been detected in up to 30% of healthy individuals [24].
Although it is still controversial that functional, replicating endogenous retrovirus particles are expressed in human tissue, there is little doubt that HERV-encoded proteins are present in some tumours, cell lines and inflamed tissues from patients with autoimmune disease [25,26]. Several studies have shown that antibodies reactive with Gag or Env sequences from both HERVs and infectious retroviruses such as HIV and HTLV-I can be detected in human sera. In general, these antibodies are found with a higher frequency and titre in patients with autoimmune disease [27–30]. It has been suggested that those reactive with HIV-1 or HTLV-I Gag are driven by cross-reactions with endogenous retroviruses, autoantigens or, conceivably, infection with an exogenous human retrovirus [31,32]. One obvious candidate would be human retrovirus-5 (HRV-5) [33,34].
In this report, we investigate antibodies to truncated envelope proteins of two members of the HERV-K family, one 100% identical to the original published sequence of HERV-K10 and the second 100% identical to IDDMK1,222. We mapped their epitopes to establish whether the antibodies are driven by these HERV-K retrovirus sequences themselves or a cross-reaction with another similar sequence.
MATERIALS AND METHODS
Serum samples
Blood for serum and for DNA extraction was collected from patients attending Charing Cross Hospital and diagnosed according to standard criteria [31] with informed consent and with approval from the local Ethics Committee. Eighteen serum samples from patients with newly diagnosed type 1 diabetes were a kind gift from Professor Marco Londei, Kennedy Institute. Normal sera were from healthy volunteers which were coded to prevent identification of the donors.
Polymerase chain reaction amplification (PCR)
Genomic DNA was extracted using a Qiagen QIAamp Blood Midi Kit (Qiagen, Crawley, West Sussex, UK) according to the manufacturer’s specification. One μl of DNA template was used in a 50-μl reaction volume containing 50 mm KCl, 10 mm TrisHCl (pH 8·3), 1·5 mm MgCl2, 10 pmoles of each oligonucleotide primer U3 (forward primer 5′-ATGGTAACACCAGTCA CATGGATGG-3′, reverse primer 5′-ACAGCTGGACT CACTTGTGCAC-3′), 10 mm of each dNTP and 2·5 units of Taq polymerase (Bio/Gene Ltd, Kimbolton Cambridgeshire, UK). Cycle conditions were: 1min at 94°C, 35 cycles of 94°C for 30 s, 55°C for 30 s, and 72°C for 1min and a final extension at 72°C for 5 min.
Cloning and sequencing
PCR products were cloned into pCR 2.1 vector (Invitrogen, Groningen, The Netherlands) according to the manufacturer’s recommendations and sequenced using an Applied Biosystems 377 automated DNA sequencer.
Construction of recombinant prokaryotic expression vector
Two of the sequences, one identical to HERV-K10 and one to IDDMK1,222 [17] were subcloned from pCR 2.1 into pTrcHis2B (Invitrogen, Groningen, The Netherlands). To generate two different restriction enzyme sites (NCoI, XbaI) at both ends, the fragments were amplified by PCR using primers containing these sites. For HERV-K10, forward primer, 5′-GAGAACCATG GTAACACCAGTCACATGGATGG-3′ and reverse primer; 5′-TTTGTTCTAGACCTAATATCACCGCACTATTGGC-3′ were used. For IDDMK1,222 the same forward primer as above was used but with a different reverse primer, 5′-TTTGTTCTA GATCTATAATAGTTCCGAATTCATT-3′, to encompass the longer open reading frame. The reaction was performed as above using the same PCR cycles.
The PCR products and the vector pTrcHis2B were digested with NCoI and XbaI and, after purification using the Geneclean Spin Kit (Anachem, Luton Beds LU2 0EB, UK), the fragments were subcloned into the dephosphorylated vector. Epicurian Coli BL21-CodonPlus (DE3)-RIL (Stratagene, Amsterdam Zuidoost, The Netherlands) was transformed with the plasmid. The fusion protein expression was induced with IPTG 1 mm, and purified using Ni2+-NTA beads (Qiagen, Crawley, West Sussex, UK), according to the manufacturer’s recommendations.
Western blot
Proteins were separated on 12% SDS-polyacrylamide gels (SDS polyacrylamide gel electrophoresis [PAGE]) and electrophoretically transferred to Immobilon PVDF membranes (Millipore, Hertfordshire, UK). Blots were incubated with human sera at a dilution of 1:100 and stained indirectly by using peroxidase-conjugated goat antihuman antibodies (Bio-Rad, Hertfordshire, UK). Reactive bands were visualized by chemiluminescence using ECL (Amersham Pharmacia Biotech UK Ltd, Little Chalfont, Bucks, UK) and Hyper film ECL (Amersham Pharmacia Biotech UK Ltd, Little Chalfont, Bucks, UK).
Epitope mapping
The complete HERV-K10 env protein was synthesized as a series of 15mers overlapping by eight amino acids on a derivatized cellulose membrane (SPOTs Kit, Sigma-Genosys, Pampisford, Cambridge, UK) using Fmoc Chemistry according to the manufacturer’s recommendations. Table 1 shows the 27 peptide sequences synthesized. On the same membrane, eight additional peptides corresponding to parts of IDDMK1,222 sequence which differed from that of HERV-K10 sequence (marked with a prime; Table 1). The membrane was incubated with serum at a dilution of 1:100.
Table 1.
Sequences of peptides synthesized for epitope mapping spanning the open reading frame of HERV-K10
| Spot | Mol. wt. | Peptide sequence | ||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | 1794 | M | V | T | P | V | T | W | M | D | N | P | I | E | V | Y |
| 2 | 1793 | M | D | N | P | I | E | V | Y | V | N | D | S | I | W | V |
| 2′ | 1779 | M | D | N | P | I | E | V | Y | V | N | D | S | V | W | V |
| 3 | 1745 | Y | V | N | D | S | I | W | V | P | G | P | I | D | D | R |
| 4 | 1622 | V | P | G | P | I | D | D | R | C | P | A | K | P | E | E |
| 4′ | 1610 | V | P | G | P | T | D | D | R | C | P | A | K | P | E | E |
| 5 | 1717 | R | C | P | A | K | P | E | E | E | G | M | M | I | N | I |
| 6 | 1772 | E | E | G | M | M | I | N | I | S | I | G | Y | R | Y | P |
| 7 | 1678 | I | S | I | G | Y | R | Y | P | P | I | C | L | G | R | A |
| 7′ | 1659 | I | S | I | G | Y | H | Y | P | P | I | C | L | G | R | A |
| 8 | 1495 | P | P | I | C | L | G | R | A | P | G | C | L | M | P | A |
| 9 | 1627 | A | P | G | C | L | M | P | A | V | Q | N | W | L | V | E |
| 10 | 1651 | A | V | Q | N | W | L | V | E | V | P | T | V | S | P | I |
| 11 | 1763 | E | V | P | T | V | S | P | I | S | R | F | T | Y | H | M |
| 11′ | 1764 | E | V | P | T | V | S | P | N | S | R | F | T | Y | H | M |
| 12 | 1784 | I | S | R | F | T | Y | H | M | V | S | G | M | S | L | R |
| 13 | 1750 | M | V | S | G | M | S | L | R | P | R | V | N | Y | L | Q |
| 14 | 1928 | R | P | R | V | N | Y | L | Q | D | F | S | Y | Q | R | S |
| 15 | 1856 | Q | D | F | S | Y | Q | R | S | L | K | F | R | P | K | G |
| 16 | 1727 | S | L | K | F | R | P | K | G | K | P | C | P | K | E | I |
| 17 | 1655 | G | K | P | C | P | K | E | I | P | K | E | S | K | N | T |
| 17′ | 1587 | G | K | T | C | P | K | E | I | P | K | G | S | K | N | T |
| 18 | 1800 | I | P | K | E | S | K | N | T | E | V | L | V | W | E | E |
| 18′ | 1728 | I | P | K | G | S | K | N | T | E | V | L | V | W | E | E |
| 19 | 1648 | T | E | V | L | V | W | E | E | C | V | A | N | S | A | V |
| 19′ | 1676 | T | E | V | L | V | W | E | E | C | V | A | N | S | V | V |
| 20 | 1678 | E | C | V | A | N | S | V | V | I | L | Q | N | N | E | F |
Additional peptides (marked with a prime) were also synthesized so that the open reading frame (minus the last 5 amino acids) of IDDMK1,222 was also covered. Amino acid substitutions which differentiate IDDMK1,222 are marked in bold.
Elisa
Two synthetic 15mer polypeptides each with an additional cysteine at the C terminus were prepared and purified by Advanced Biotechnology Centre (Imperial College, Charing Cross Hospital, London, UK). The peptide identified as containing immunodominant epitope(s) of HERV-K10 protein was peptide 17′ (GKTCPKEIPKGSKNT-C). The peptide was diluted in PBS at 10 μg/ml and coated on to microtiter wells (Brand Products, Munc-Immuno Plate MaxiSorp Surface) overnight at room temperature, every two peptide-coated wells alternating with two uncoated (PBS only) as controls for nonspecific binding. The wells were blocked with PBS containing 2% Casein and 0·05% Tween-20 (Blocking Solution). Serum samples were diluted in the blocking solution (1/50) and added to the wells in quadruplicate (two coated wells and two uncoated wells) for 2 h at room temperature. A titration curve was done on each plate with a strongly reactive serum to ensure reproducibility between plates. The plates were washed with PBS plus 0·05% Tween-20 and, after blocking again with the blocking solution for 10 min, antibody binding was detected using peroxidase-conjugated goat antihuman antibodies (Bio-Rad) diluted in Blocking Buffer (1:3500), with an incubation for 1 h at room temperature. After seven washes with 0·05% Tween-20 PBS, substrate solution (KPL TMB) was added for a few minutes and stopped with H2SO4 (1:16). The absorbance at 450 nm was measured with a Victor 1420 Multilabel Counter (Wallac and Berthold, Crownhill, Milton Keynes MK8 0AB, UK). The absorbance on the uncoated wells has been taken as negative and the mean was subtracted from the mean absorbance of the coated wells.
RESULTS
Cloning of the HERV-K10 and IDDMK1,222 envelope ORFs and their relationship with other family members
To obtain sequences identical to those published for HERV-K10 and IDDMK1,222, a total of 21 clones were amplified from peripheral blood DNA from two individuals. An alignment of the predicted amino acid sequence of the HERV-K10 clone, the IDDMK1,222 clone and the remaining 19 are shown in Fig. 1. Most of them show an open reading frame (ORF). For HERV-K10 a stop codon is at position 144 and for IDDMK1,222 at position 154, which predicts theoretical molecular weights of 16kDa and 17kDa for the two proteins, respectively. Four other sequences have stop codons within the region cloned. Interestingly, a total of 15 sequences have open reading frames extending beyond the first 156 amino acids.
Fig. 1.
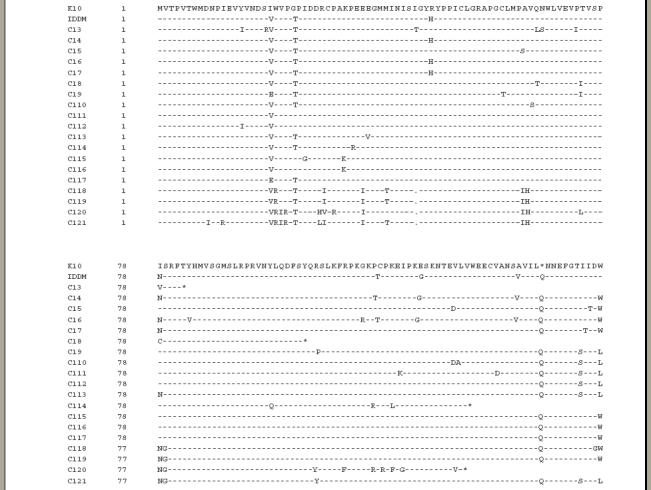
Alignment of the env amino sequence of HERV-K endogenous retroviruses. Dashes indicate identical amino acids compared with HERV-K10. Gaps are represented by dots and the stop codons by an asterisk.
All the sequences have a similarity of 82–98% with HERV-K10, with IDDMK1,222 being 95% identical. The stop codon on the 154th position is the only apparently distinct feature of IDDMK1,222. The amino acid histidine position 48 is shared by three other elements. The tyrosine at position 115, the glycine at 123 and the valine at 140 are each shared by two other elements.
Antibodies to HERV-K10 and IDDMK1,222 Env by Western blot
IDDMK1,222 and HERV-K10 were expressed in bacteria and the proteins purified on nickel agarose. The two proteins were mixed in approximately equal quantities, judged by the density of staining on a Coomassie stained gel and then dissolved in loading buffer. Because the proteins were of different sizes, it was possible to compare reactivity with IDDMK1,222 Env (mol. wt. 19·4 kDa including the tags) with that of HERV-K10 Env (mol. wt. 18·3 kDa). On the gel, the fragments appeared larger than the predicted size (around 25 kDa), presumably because of the conditions of the migration. An example of a Western blot obtained is shown in Fig. 2. The identity of the cloned proteins was confirmed by their reactivity with the monoclonal antibody to c-myc which reacted with the c-myc tags. Of 161 sera tested, antibodies to one or both proteins are seen in 29% of 35 sera from healthy individuals and 22% of 18 sera from patients with diabetes (Table 2). All of the groups with autoimmune rheumatic disease have antibodies detected more frequently, ranging from 32% in SLE to 47% in Sjogren’s syndrome, but no disease group has a statistically significant increase in the prevalence of antibodies compared to sera from healthy controls.
Fig. 2.
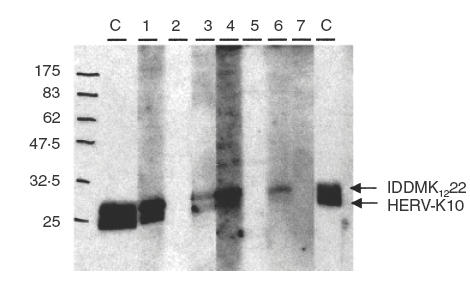
Detection of antibodies to HERV-K10 and IDDMK1,222 Env protein in human sera. The membrane is cut into nine strips and reacted (1:100 dilution) with two SLE sera (1, 2), with three RA sera (3–5), with two normal control sera (6, 7) and with monoclonal anticMyc antibodies as a positive control (C).
Table 2.
Prevalence of IDDMK1,222 or/and HERV-K10 antibodies (Ab) in patients with auto-immune rheumatic diseases, diabetes and normal controls
| Anti-IDDMK1,222 | Anti-HERV-K10 | Anti-IDDMK1,222 | Anti-IDDMK1,222 | |
|---|---|---|---|---|
| Ab | Ab alone | Ab alone | and anti-HERV-K10 Ab | or/and anti-HERV-K10 |
| Systemic lupus erythematosus (44) | 0 | 1(2·3%) | 13(29·5%) | 14(31·8%) |
| Rheumatoid arthritis (26) | 2(7·7%) | 0 | 8(30·8%) | 10(38·5%) |
| Sjogren’s syndrome (17) | 4(23·5%) | 0 | 4(23·5%) | 8 (47%) |
| Polymyositis (21) | 2 (9·5%) | 0 | 7(33·3%) | 9(42·8%) |
| Diabetes (18) | 1(5·5%) | 1(5·5%) | 2(11·1%) | 4(22·1%) |
| Normal (35) | 3 (8·6%) | 0 | 7(20%) | 10(28·6%) |
Epitope mapping
Epitope mapping was performed on peptides? from both the HERV-K10 and IDDMK1,222 Env proteins. Figure 3a shows an example of a serum strongly positive by Western blot reacting with the membrane. Several peptides have reacted with the antibodies. The membrane was also stained with a serum with a negative reaction on Western blot. In this case none of the spots has reacted (Fig. 3b). Figure 4 shows a summary of the positively reacting spots after staining with four different sera. Two peptides reacted with the four sera, number 15 (QDFSYQRSLKFRPKG) and 17′ (GKTCPKEIPKGSKNT). Interestingly peptide 17′, derived from the IDDMK1,222 sequence, gave a much stronger reaction with all four sera than did peptide 17 (GKPCPKEIP KESKNT) derived from the HERV-K10 sequence though it differed by only two amino acids, a threonine as opposed to a proline at position 3 and a glycine as opposed to a glutamic acid at position 11.
Fig. 3.
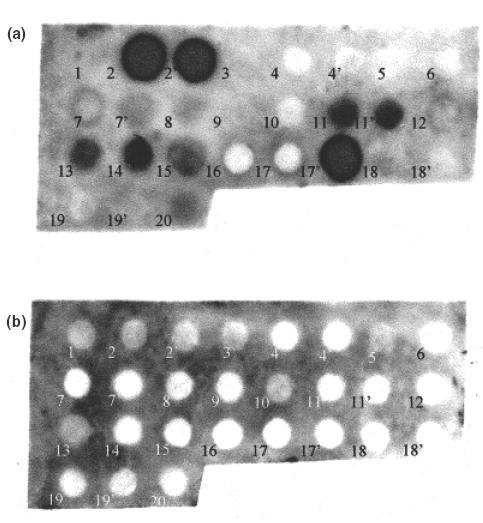
Epitope mapping, staining of the membrane with a serum, dilution 1/100, (a) from a patient with antibodies to HERV-K10 and IDDMK1,222 on Western blot, and (b) a serum from a patient, who had a negative reaction on Western blot. Nineteen peptides were synthesized for the epitope mapping (Table 1), corresponding to the complete HERV-K10 Env sequence with an overlap of eight amino acids. On the same membrane, eight additional peptides corresponding to part of IDDMK1,222 which differed from that of HERV-K10 sequence, are marked with a prime (2′, 4′, 7′, 11′, 17′, 18′ and 19′).
Fig. 4.
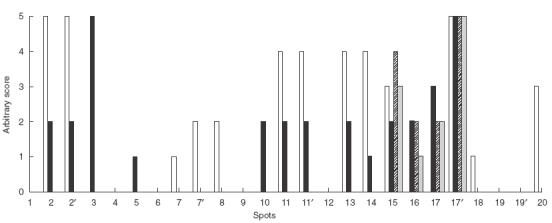
Epitope mapping base on spot staining by polyclonal antibodies from four different sera (four patients: □ A27, ■ A7, A2,
A2, A49).The strength of each reaction was given as an arbitary score based on visualization of the density of staining of the spot above background. For an example see serum A27 in Fig. 3a.
A49).The strength of each reaction was given as an arbitary score based on visualization of the density of staining of the spot above background. For an example see serum A27 in Fig. 3a.
Quantification of antibodies to the immunodominant epitopes of HERV-K10 and IDDMK1,222 by ELISA
The ELISA peptide 17′ correlated strongly with the results of Western blot (Fig. 5) suggesting that ELISA with this peptide was a quantitative and reproducible means of measuring anti-HERV-K antibodies. In SLE, antibodies to peptide 17′ were significantly elevated (P < 0·003), but levels were similar to controls in diabetes, polymyositis, RA and SS (Fig. 6).
Fig. 5.
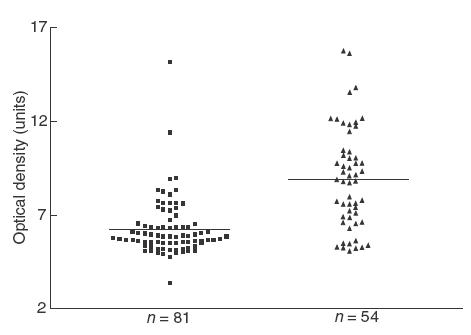
Comparison between the results obtained by Western blot (WB) and the optical density detected by ELISA, plates coated with the peptide 17′. ■ Sample negative by WB; ▴ sample positive by WB.
Fig. 6.
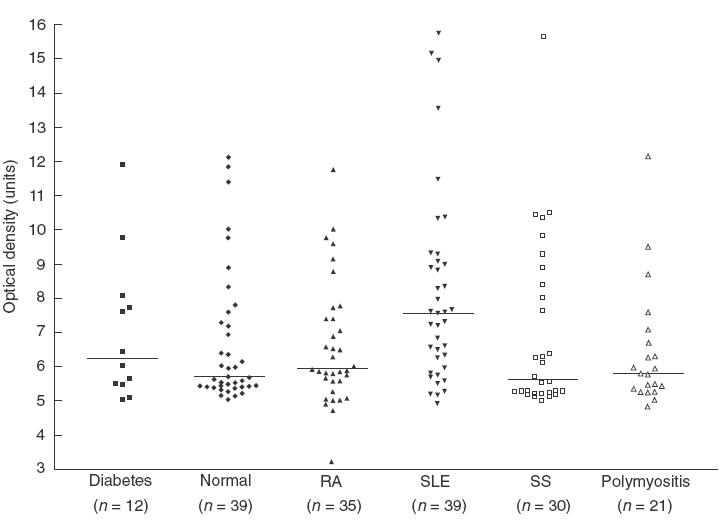
The level of anti-HERV-K10/anti-IDDMK1,222 peptide antibodies in patients with diabetes, normal healthy controls, rheumatoid arthritis (RA), systemic lupus erythematosus (SLE), Sjogren’s syndrome (SS) and polymyositis determined by ELISA. The plates were coated with peptide 17′.
DISCUSSION
Autoantibodies to endogenous retroviruses remain an unexplained phenomenon in healthy subjects and in patients with autoimmune rheumatic disease. To explore the mechanisms involved we have cloned and expressed open reading frames from the envelope of two members of the HERV-K family: HERV-K10 and IDDMK1,222. The 21 different sequences obtained confirm previous reports that there are multiple members of this family. It was particularly striking that 14 of the 21 had no stop codons within 154 amino acids analysed. Although sequencing the rest of the envelope gene of these clones was beyond the scope of this study, this finding suggests that very long open reading frames, perhaps even encompassing an entire functional retroviral envelope, might be more common in the HERV-K10 family than was previously thought. Analysis of the sequences amplified also challenges the original report [17] that IDDMK1,222 is distinct from other members of the HERV-K family. As has been argued by others [18–20], IDDMK1,222 is 95% identical to HERV-K10 over the env open reading frame; a higher degree of homology than most of the other HERV-K10 related sequences identified in our study.
Having identified two clones that were each identical to the published sequences of HERV-K10 and IDDMK1,222, we were then in a position to analyse the immune response to each of their respective open reading frames by immunoblotting. As expected, given the high degree of aminoacid sequence homology between the two proteins, there was a close correlation between the sera containing antibodies to HERV-K10 and those containing antibodies to IDDMK1,222, with the latter being a slightly more common target for the immune response. As with previous studies [19,24] we found no increase in the prevalence of antibodies to the envelope sequences in patients with diabetes. Patients with SLE and Sjogren’s syndrome did have a higher frequency of anti-Env antibodies by immunoblotting but this did not reach statistical significance. By ELISA, using a peptide containing immunodominant epitope(s), the increased frequency and titre reached statistical significance in SLE.
The finding of antibodies to retroviral antigens in SLE and/or Sjogren’s syndrome has been reported in previous studies. This has been shown for exogenous sequences such as the p24 Gag protein of HIV-1 [28,29,35] and for peptides representing the immunodominant epitopes of P19 of HTLV-I [31]. In these studies all of the patients had no evidence of infection with either of these viruses. It was speculated that these antiretroviral antibodies were cross-reactive with endogenous retroviruses, a previously undescribed exogenous retrovirus or possibly autoantigens such as the cellular ribonucleoproteins which are reported to have sequence similarity to Gag proteins [25]. Antibodies to endogenous retroviruses have also been described, again with SLE and Sjogren’s syndrome being the most frequent diagnosis. The antigens include the Gag of HRES-1 [31], the Env of ERV-3 [8] and consensus sequences from C-type retroviruses [36]. A common theme of many of these studies is that the epitopes are on arginine [30] or proline [8,31,36] rich sequences. It was even shown in one study [36] that nonviral synthetic polyproline sequences themselves preferentially react with antibodies from SLE serum samples. Although these studies are often cited as supporting a role for retroviruses in autoimmune rheumatic disease there are no data available to implicate which antigen drives this immune response or indeed whether it is retroviral in origin at all.
To examine the origin of antibodies to human endogenous retroviruses we chose to work with the HERV-K10 and IDDMK1,222 Env because serum samples contained antibodies in sufficiently high concentration for further analysis. By epitope mapping we were able to address the following questions. Are antibodies to these envelope sequences simply those that are preferentially reactive with proline or arginine-rich peptides? Are these antibodies cross-reactive and driven by an autoantigen or by an exogenous retrovirus such as HRV-5 [33,34]? Is there evidence that these antibodies are driven by expression of HERV-K10 proteins themselves?
The epitope mapping showed multiple reactive peptides suggesting that the antibody reaction was driven by the envelopes of HERV-K family members. Two sequences reacted with all four positive sera tested; number 15 (QDFSYQRSLKFRPKG) and 17′ (GKTCPKEIPKGSKNT). Neither is proline or arginine rich, though 17′ is lysine rich (four lysines in 15 amino acids). There is no common motif and neither has a sequence similar to that of HRV-5 Pol protein or Gag, which have now been cloned (Griffiths et al. in preparation). It is conceivable that there could be similarities to a putative HRV-5 Env, but this part of the virus remains to be identified. A database search has not revealed any similarity with any of the known autoantigens. There is therefore no evidence that these antibodies are driven by a cross-reactive antigen.
The finding of antibodies reactive with multiple unrelated epitopes of the HERV-K family strongly supports the hypothesis that anti-HERV antibodies, like autoantibodies to other cellular proteins, are antigen driven (reviewed in [37)]. Such antigen drive depends on loss of tolerance to these sequences. How tolerance is broken remains unexplained but the finding of IgG antibodies detectable by Western blot in up to 30% of healthy individuals suggests that this occurs quite readily. There is circumstantial evidence that HERV expression is up-regulated by pro-inflammatory cytokines [38,39] or ultraviolet light [40]. Therefore HERV-K proteins may well be expressed frequently during the lifetime of a healthy individual and preferentially in patients with inflammatory disease. Potential molecular mimicry of HERV-K Gag with cellular RNPs for example [41] could implicate HERV-K related sequences in the long chain of events that leads to autoimmunity in rheumatic disease.
Acknowledgments
This work was supported by the Arthritis Research Campaign.
We would like to thank Dr Roswitha Lower (Paul-Ehrlich-Institut, Paul-Ehrlich-Strasse 51–59, D-63225 Langen, Frankfurt, Germany) for positive control samples and for her helpful advice and Dr Ingrid Lundberg (Department of Medicine, Karolinska Institutet, Rheumatology Clinic, Karolinska Hospital, Stockholm, Sweden) for the serum samples from patients with polymyositis.
REFERENCES
- 1.Shih A, Coutavas EE, Rush MG. Evolutionary implications of primate endogenous retroviruses. Virology. 1991;182:495–502. doi: 10.1016/0042-6822(91)90590-8. [DOI] [PubMed] [Google Scholar]
- 2.Medstrand P, Mager DL. Human-specific integrations of the HERV-K endogenous retrovirus family. J Virol. 1998;72:9782–7. doi: 10.1128/jvi.72.12.9782-9787.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Venter JC, Adams MD, Myers EW, et al. The sequence of the human genome. Science. 2001;291:1304–51. doi: 10.1126/science.1058040. [DOI] [PubMed] [Google Scholar]
- 4.O’Connell C, O’Brien S, Nash WG, Cohen M. ERV3, a full-length human endogenous provirus: chromosomal localization and evolutionary relationships. Virology. 1984;138:225–35. doi: 10.1016/0042-6822(84)90347-7. [DOI] [PubMed] [Google Scholar]
- 5.Cohen M, Powers M, O’Connell C, Kato N. The nucleotide sequence of the env gene from the human provirus ERV3 and isolation and characterization of an ERV3-specific cDNA. Virology. 1985;147:449–58. doi: 10.1016/0042-6822(85)90147-3. [DOI] [PubMed] [Google Scholar]
- 6.Kato N, Pfeifer-Ohlsson S, Kato M, et al. Tissue-specific expression of human provirus ERV3 mRNA in human placenta: two of the three ERV3 mRNAs contain human cellular sequences. J Virol. 1987;61:2182–91. doi: 10.1128/jvi.61.7.2182-2191.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Venables PJ, Brookes SM, Griffiths D, Weiss RA, Boyd MT. Abundance of an endogenous retroviral envelope protein in placental trophoblasts suggests a biological function. Virology. 1995;211:589–92. doi: 10.1006/viro.1995.1442. 10.1006/viro.1995.1442. [DOI] [PubMed] [Google Scholar]
- 8.Li JM, Fan WS, Horsfall AC, et al. The expression of human endogenous retrovirus-3 in fetal cardiac tissue and antibodies in congenital heart block. Clin Exp Immunol. 1996;104:388–93. [PubMed] [Google Scholar]
- 9.Blond JL, Beseme F, Duret L, et al. Molecular characterization and placental expression of HERV-W, a new human endogenous retrovirus family. J Virol. 1999;73:1175–85. doi: 10.1128/jvi.73.2.1175-1185.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Blond JL, Lavillette D, Cheynet V, et al. An envelope glycoprotein of the human endogenous retrovirus HERV-W is expressed in the human placenta and fuses cells expressing the type D mammalian retrovirus receptor. J Virol. 2000;74:3321–9. doi: 10.1128/jvi.74.7.3321-3329.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ono M, Yasunaga T, Miyata T, Ushikubo H. Nucleotide sequence of human endogenous retrovirus genome related to the mouse mammary tumor virus genome. J Virol. 1986;60:589–98. doi: 10.1128/jvi.60.2.589-598.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ono M. Molecular cloning and long terminal repeat sequences of human endogenous retrovirus genes related to types A and B retrovirus genes. J Virol. 1986;58:937–44. doi: 10.1128/jvi.58.3.937-944.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Tonjes RR, Lower R, Boller K, et al. HERV-K: the biologically most active human endogenous retrovirus family. J Acquir Immune Defic Syndr Hum Retrovirol. 1996;13(Suppl):S261–7. doi: 10.1097/00042560-199600001-00039. [DOI] [PubMed] [Google Scholar]
- 14.Willer A, Saussele S, Gimbel W, et al. Two groups of endogenous MMTV related retroviral env transcripts expressed in human tissues. Virus Genes. 1997;15:123–33. doi: 10.1023/a:1007910924177. 10.1023/a:1007910924177. [DOI] [PubMed] [Google Scholar]
- 15.Mueller-Lantzsch N, Sauter M, Weiskircher A, et al. Human endogenous retroviral element K10 (HERV-K10) encodes a full-length gag homologous 73-kDa protein and a functional protease. AIDS Res Hum Retroviruses. 1993;9:343–50. doi: 10.1089/aid.1993.9.343. [DOI] [PubMed] [Google Scholar]
- 16.Lower R, Boller K, Hasenmaier B, et al. Identification of human endogenous retroviruses with complex mRNA expression and particle formation. Proc Natl Acad Sci USA. 1993;90:4480–4. doi: 10.1073/pnas.90.10.4480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Conrad B, Weissmahr RN, Boni J, Arcari R, Schupbach J, Mach B. A human endogenous retroviral superantigen as candidate autoimmune gene in type I diabetes. Cell. 1997;90:303–13. doi: 10.1016/s0092-8674(00)80338-4. [DOI] [PubMed] [Google Scholar]
- 18.Lan MS, Mason A, Coutant R, et al. HERV-K10s immune-mediated (Type 1) diabetes. [discussion 16]. Cell. 1998;95:14–16. doi: 10.1016/s0092-8674(00)81777-8. [DOI] [PubMed] [Google Scholar]
- 19.Lower R, Tonjes RR, Boller K, et al. Development of insulin-ependent diabetes mellitus does not depend on specific expression of the human endogenous retrovirus HERV-K. [discussion 16]. Cell. 1998;95:11–14. doi: 10.1016/s0092-8674(00)81776-6. [DOI] [PubMed] [Google Scholar]
- 20.Murphy VJ, Harrison LC, Rudert WA, et al. Retroviral superantigens and type 1 diabetes mellitus. [discussion 16]. Cell. 1998;95:9–11. doi: 10.1016/s0092-8674(00)81775-4. [DOI] [PubMed] [Google Scholar]
- 21.Kim A, June HS, Wong L, et al. Human endogenous retrovirus with a high genomic sequence homology with IDDMK(1,2)22 is not specific for Type I (insulin–dependent) diabetic patients but ubiquitous. Diabetologia. 1999;42:413–18. doi: 10.1007/s001250051173. 10.1007/s001250051173. [DOI] [PubMed] [Google Scholar]
- 22.Muir A, Ruan QG, Marron MP, She JX. The IDDMK(1,2)22 retrovirus is not detectable in either mRNA or genomic DNA from patients with type 1 diabetes. Diabetes. 1999;48:219–22. doi: 10.2337/diabetes.48.1.219. [DOI] [PubMed] [Google Scholar]
- 23.Jaeckel E, Heringlake S, Berger D, Brabant G, Hunsmann G, Manns MP. No evidence for association between IDDMK(1,2)22, a novel isolated retrovirus, and IDDM. Diabetes. 1999;48:209–14. doi: 10.2337/diabetes.48.1.209. [DOI] [PubMed] [Google Scholar]
- 24.Badenhoop K, Donner H, Neumann J, et al. IDDM patients neither show humoral reactivities against endogenous retroviral envelope protein nor do they differ in retroviral mRNA expression from healthy relatives or normal individuals. Diabetes. 1999;48:215–18. doi: 10.2337/diabetes.48.1.215. [DOI] [PubMed] [Google Scholar]
- 25.Garry RF. New evidence for involvement of retroviruses in Sjogren’s syndrome and other autoimmune diseases [editorial] Arthritis Rheum. 1994;37:465–9. doi: 10.1002/art.1780370405. [DOI] [PubMed] [Google Scholar]
- 26.Blomberg J, Nived O, Pipkorn R, Bengtsson A, Erlinge D, Sturfelt G. Increased antiretroviral antibody reactivity in sera from a defined population of patients with systemic lupus erythematosus. Correlation with autoantibodies and clinical manifestations. Arthritis Rheum. 1994;37:57–66. doi: 10.1002/art.1780370109. [DOI] [PubMed] [Google Scholar]
- 27.Phillips PE, Johnston SL, Runge LA, Moore JL, Poiesz BJ. High IgM antibody to human T-lymphotropic virus type I in systemic lupus erythematosus. J Clin Immunol. 1986;6:234–41. doi: 10.1007/BF00918703. [DOI] [PubMed] [Google Scholar]
- 28.Talal N, Dauphinee MJ, Dang H, Alexander SS, Hart DJ, Garry RF. Detection of serum antibodies to retroviral proteins in patients with primary Sjogren’s syndrome (autoimmune exocrinopathy) Arthritis Rheum. 1990;33:774–81. doi: 10.1002/art.1780330603. [DOI] [PubMed] [Google Scholar]
- 29.Talal N, Garry RF, Schur PH, et al. A conserved idiotype and antibodies to retroviral proteins in systemic lupus erythematosus. J Clin Invest. 1990;85:1866–71. doi: 10.1172/JCI114647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Banki K, Maceda J, Hurley E, et al. Human T-cell lymphotropic virus (HTLV)-related endogenous sequence, HRES-1, encodes a 28-kDa protein: a possible autoantigen for HTLV-I gag-reactive autoantibodies. Proc Natl Acad Sci USA. 1992;89:1939–43. doi: 10.1073/pnas.89.5.1939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Brookes SM, Pandolfino YA, Mitchell TJ, et al. The immune response to and expression of cross-reactive retroviral gag sequences in autoimmune disease. Br J Rheumatol. 1992;31:735–42. doi: 10.1093/rheumatology/31.11.735. [DOI] [PubMed] [Google Scholar]
- 32.Talal N, Flescher E, Dang H. Are endogenous retroviruses involved in human autoimmune disease? J Autoimmun. 1992;5(Suppl):61–6. doi: 10.1016/0896-8411(92)90020-q. [DOI] [PubMed] [Google Scholar]
- 33.Griffiths DJ, Venables PJ, Weiss RA, Boyd MT. A novel exogenous retrovirus sequence identified in humans. J Virol. 1997;71:2866–72. doi: 10.1128/jvi.71.4.2866-2872.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Griffiths DJ, Cooke SP, Herve C, et al. Detection of human retrovirus 5 in patients with arthritis and systemic lupus erythematosus [see comments] Arthritis Rheum. 1999;42:448–54. doi: 10.1002/1529-0131(199904)42:3<448::AID-ANR9>3.0.CO;2-8. [DOI] [PubMed] [Google Scholar]
- 35.Dang H, Takei M, Isenberg D, et al. Expression of an interspecies idiotype in sera of SLE patients and their first-degree relatives. Clin Exp Immunol. 1988;71:445–50. [PMC free article] [PubMed] [Google Scholar]
- 36.Bengtsson A, Blomberg J, Nived O, Pipkorn R, Toth L, Sturfelt G. Selective antibody reactivity with peptides from human endogenous retroviruses and nonviral poly (amino acids) in patients with systemic lupus erythematosus. Arthritis Rheum. 1996;39:1654–63. doi: 10.1002/art.1780391007. [DOI] [PubMed] [Google Scholar]
- 37.Venables PJ. Epitope mapping of ribonucleoprotein antigens: answers without questions? [Editorial] Clin Exp Immunol. 1993;94:225–6. doi: 10.1111/j.1365-2249.1993.tb03435.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Larsson E, Venables P, Andersson AC, et al. Tissue and differentiation specific expression on the endogenous retrovirus ERV3 (HERV-R) in normal human tissues and during induced monocytic differentiation in the U-937 cell line. Leukemia. 1997;11(Suppl):142–4. [PubMed] [Google Scholar]
- 39.Sibata M, Ikeda H, Katumata K, Takeuchi K, Wakisaka A, Yoshoki T. Human endogenous retroviruses: expression in various organs in vivo and its regulation in vitro. Leukemia. 1997;11(Suppl):145–6. [PubMed] [Google Scholar]
- 40.Hohenadl C, Germaier H, Walchner M, et al. Transcriptional activation of endogenous retroviral sequences in human epidermal keratinocytes by UVB irradiation. J Invest Dermatol. 1999;113:587–94. doi: 10.1046/j.1523-1747.1999.00728.x. 10.1046/j.1523-1747.1999.00728.x. [DOI] [PubMed] [Google Scholar]
- 41.Tenenbaum SA, Voss TG, Garry RF, Gallaher WR. Sequence similarities between retroviral proteins and components of the spliceosome. AIDS Res Hum Retroviruses. 1994;10:521–2. doi: 10.1089/aid.1994.10.521. [DOI] [PubMed] [Google Scholar]


