Abstract
Macrophages play a major role in almost all stages of the complex process of wound healing. It has been previously shown that the incorporation of a hypo-osmotic shock step, in the process of monocyte-concentrate preparation from a blood unit, induces monocyte/macrophage activation. As the macrophages are produced using a unique, closed and sterile system, they are suitable for local application on ulcers in elderly and paraplegic patients. Enhanced phagocytosis by the activated cells, as well as increased secretion of cytokines such as IL-1, IL-6, were detected in a recent study which are in accord with the very encouraging clinical results. In the present study, we used DNA microarrays to analyse the differential gene expressions of the hypo-osmotic shock-activated monocytes/macrophages and compare them to non-treated cells. Of the genes that exhibited differences of expression in the activated cell population, 94% (68/72) displayed increased activity. The mRNA levels of 43/68 of these genes (63%) were found to be 1·5-fold or higher (1·5–7·98) in the activated macrophages cell population as compared to the non-treated cells. Only four genes were found to have lower mRNA levels in the activated cells, with ratios of expression of 0·62–0·8, which may suggest that the changes are insignificant. A significant number of the genes that showed increased levels of expression is known to be directly involved in macrophage function and wound healing. This may correlate with the increased secretion of different cytokines by the activated macrophages depicted previously. Other groups of genes expressed are known to be involved in important pathways such as neuronal growth and function, developmental defects and cancer. The hypo-osmotic shock induces a gene expression profile of cytokines and receptors in the activated cells. These may evoke potential abilities to produce a variety of protein products needed in the wound healing process and may bring to light possibilities for other therapeutic applications of these cells.
Keywords: cytokines, gene expression, hypo-osmotic shock, macrophages
INTRODUCTION
The monocytes, which constitute 1–6% of the nucleated peripheral blood cells, egress the blood stream into tissues and differentiate into a variety of macrophages, depending on the nature of the signals in the local microenvironment [1,2].
Their phagocytic abilities and diverse secretory potential place the monocytes/macrophages in the centre of the wound healing process [1,3–5]. This biologically complex sequence of events involves cellular and molecular processes such as cell migration, inflammation, angiogenesis, collagen synthesis and deposition and re-epithelialization [5,6]. The monocytes/macrophages have key functions in almost every stage of the process. Upon initiation of the inflammation stage, the macrophages secrete IL-1 that induces the rapid recruitment of inflammation cells from the circulation into the wound [7–9]. As phagocytes, they help in the digestion of bacteria and debridement [10] and, in the later stages of the wound-healing process, they secrete IL-6, which is believed to influence endothelial cell proliferation and the initiation of angiogenesis [11–13]. The macrophage serves as the coordinator of this proliferative process, by producing growth factors such as PDFG BB, TGF-α, TGF-β1, VEGF, FGF, EGF and IGF-1, necessary in the wound-healing process [5,8,14].
It seems that the age of the macrophages may play a role in the process. Application of rabbit-antimouse macrophage serum to wounds of young mice delayed wound healing, reducing it to the rate observed in old mice [15]. Moreover, wound repair was enhanced in old mice by local injection of monocytes/macrophages derived from young mice [16].
We have developed a method for preparation of human activated monocytes/macrophages from randomly collected blood units in a closed, sterile system, which are used successfully, within 24 h, in the treatment of human chronic decubital ulcers [17,18]. The macrophage preparation process includes a step of hypo-osmotic shock which induces monocyte/macrophage activation, without having to introduce additional substances (LPS, Inf-γ, etc.) to the cell culture [19–23]. The hypo-osmotic shock was found to be an effective trigger for monocyte/macrophage activation, as depicted using phagosytosis, and secretion of IL-1 and IL-6 as test parameters [24]. To further investigate the possibility that other products of the activated monocyte/macrophage may have been triggered but not yet secreted into the surrounding medium, the changes in mRNA levels of the cells submitted to hypo-osmotic shock were studied using DNA microarrays. The present report summarizes our observation of increased expression of mRNA of interleukins, growth factors and receptors in the hypo-osmotic shock-treated cells as compared to non-treated controls.
MATERIALS AND METHODS
Preparation of monocytes/macrophages
Monocytes/macrophages were prepared in a closed sterile system as previously described [17] (USA patent no. 6,146,890, D. Danon) with some modifications. Briefly, a whole blood unit collected into a triple blood bag system was separated into packed red blood cells, white blood cells (WBC) and plasma. The bags containing the plasma and WBC layer were connected to the macrophage preparation system using a sterile connecting device (Terumo Sterile Tubing Welder SC-201 A, Tokyo, Japan). Transfer of 60 ml CaCl2 (80 mm) to the plasma bag induced a coagulation that was completed after 2h of incubation at 37°C. The obtained autologous serum served as the growth medium for the monocytes/macrophages.
A control sample was prepared from each blood unit by attaching an additional culture bag to the preparation system. The WBC layer was divided equally into the two culture bags, of which only one was submitted to hypo-osmotic shock by the addition of 100 ml distilled water. After 45 s, 10 ml of a 10-fold concentrated saline solution was added to re-establish isotonicity. In parallel, 110 ml of isotonic saline was transferred into the control culture bag. The set was centrifuged at 600 g for 10 min and the supernatant of both bags was decanted. Equal volumes of autologous serum (30 ml) were then transferred into each bag and incubated for 1h at 37°C, after which the supernatant of the culture bags was decanted. The bags were rinsed with fresh serum and transferred into the ‘sink bag’, rinsing away all the non-adherent cells. At this stage, a layer of monocytes/macrophages adhered to the inner surface of both culture bags. A new volume (30 ml) of serum was placed into each bag, which was then incubated for 24 h. Following incubation, the cells were harvested by rubbing the inner surfaces of the plastic bags against each other. The cell suspension was transferred into 50 ml sterile polyethylene tubes and centrifuged at 600 g for 10 min The supernatant was discarded and the cells were resuspended in 1 ml of autologous serum. Cells were counted in a Newbauer haemocytometer after suspension in trypan blue (1:1 in 0·4% trypan blue solution) for evaluation of cell count and percentage of vitality. They were then centrifuged at 600 g for 6 min and the cell pellet was frozen at –70°C for further studies. Pools of monocytes/macrophages from 12 blood units was collected and frozen. Pools of the control saline-washed cells contained 5 × 107 cells and those from the hypo-osmotic shock-treated cells contained 7·5 × 107 cells.
RNA extraction
Total RNA was isolated using TRIZOL reagent (Gibco BRL, Life Technologies, NY, USA). Homogenization of the cells was performed by the addition of 1 ml of reagent per 1 × 107 cells. The cell suspension was divided into 1-ml tubes. RNA was extracted by adding 0·2 ml chloroform to each tube, mixing and storing the samples for 2–3 min at RT, followed by a 15-min centrifugation at 12 000 g in 4°C. RNA precipitation was achieved by transferring the aqueous phase into a new tube, adding 0·5 ml isopropanol, mixing and storing the samples for 5–10 min at RT. Centrifugation at 12 000 g in 4°C was performed for 10 min. RNA was washed by mixing the pellet with 1 ml 75% ethanol and centrifuging at 7500 g 4°C for 5 min. The RNA pellet was air-dried for 5–10 min and then dissolved in 50 μl by pipetting and incubating at 55–60°C for 10 min. After solubilization, all samples were frozen at –70°C.
DNA microarrays
DNA array hybridization. We used the cytokine/receptor (268 genes) array of human cDNAs spotted on a nylon membrane (Atlas Filter Arrays, Clontech, Palo Alto, CA, USA). The filters also included housekeeping control cDNAs and negative controls spotted in duplicate dots. A complete list of the cDNAs and controls as well as their accession numbers is available at http://www.clontech.com/atlas/genelists/index.html. Total RNAs were treated with DNaseI according to the manufacturer’s instructions and used for cDNA synthesis. A 3-μl mix containing 5 μg of total RNA and 1 μl of ×10CDS primer mix (specific for each filter array, provided by the manufacturer Clontech, USA) was incubated at 70°C for 2 min followed by additional incubation at 48°C for 2 min. To this mix, 8 μl of master mix (containing 2 μl × 5 reaction buffer, 1 μl × 10 dNTP mix, 3·5 μl [α-32P]dATP (3000 Ci/mmol, 10 mCi/ml, Amersham/Pharmacia Biotech, Amersham, UK), 0·5 μl 100 mm DTT and 1 μl MMLV Reverse Transcriptase (50 U/μl) was added, mixed and incubated for 25 min at 48°C. The reaction was terminated by adding 1 μl of × 10 termination mix at room temperature. The radioactive-labelled cDNA mix was fractionated on a Chroma Spin-200 column (Clontech Laboratories Inc., USA) and fractions that comprise the first peak of radioactivity were pooled for each cDNA synthesis reaction. In each set of hybridization, equal counts were taken for control and experimental labelled cDNA probes. The labelled cDNA probe was then mixed with 1/10th vol. of × 10 denaturing solution (1 m NaOH, 10 mm EDTA) and incubated at 68°C for 20 min. Following the incubation, 5 μl (1 μg/μl) of cot-1 DNA and equal volume of × 2 neutralizing solution (1 m NaH2PO4, pH 7·0) were added and incubated at 68°C for 10 min. Denatured, labelled cDNA was then added to 5 ml of ExpressHyb solution (Clontech) with 1 mg of sheared salmon sperm DNA (Sigma, St Louis, MO, USA) and mixed. This hybridization solution was added to the Atlas cDNA expression array membrane, which was prehybridized in 10 ml of expressHyb hybridization solution at 68°C for 1 h. Hybridization proceeded overnight at 68°C in a roller bottle. Membranes were washed once with prewarmed 2XSSC/1%SDS for 30 min and once or twice with 0·5XSSC/0·5%SDS for 30 min at 68°C with constant agitation. The membranes were exposed to Fuji X-ray films at –70°C with intensifying screens.
Analysis of hybridization signals. The cDNA microarray autoradiograms were scanned and the images were analysed using AtlasImage 1·01 software (Clontech, USA). The background was calculated using default external background calculation that takes into consideration the background signals at blank space between the different panels of the arrays. Signal threshold was set as background-based signal threshold. The signal intensities were normalized globally using the sum method (AtlasImage, Clontech, USA). A report of differentially expressed genes was generated based on ratio and intensity differences.
Interleukins
Samples of supernatants were collected and stored at –60°C for further interleukins titration by ELISA kits. In order to refer the interleukins production to the number of monocytes, they were marked with CD-14 fluorescent antibody (CD-14 is a common marker for both monocytes and macrophages, thus including all the relevant cell population).
Sample collection
The culture bags containing the adherent cells were incubated with 30 ml of autologous serum for 24 h. Aliquots of 1·5 ml supernatant were sampled from each bag after 24-h incubation. The level of interleukins in the serum before incubation served as baseline. All samples were centrifuged at 12 000 g for 5 min and the supernatant was transferred into clean Eppendorf tubes and stored at –60°C.
CD-14 marking
After 24h incubation, the cells were harvested by rubbing the inner surfaces of the plastic bags against each other; the cell suspension was transferred into 50-ml sterile polyethylene tubes and centrifuged at 600 g for 10 min. The supernatant was discarded and the cells were resuspended in 1 ml of serum. Cells were counted in a Newbauer haemocytometer after suspension in trypan blue (1:1 in 0·4% trypan blue solution) for evaluation of cell count and percentage of vital cells. The cells were concentrated to 10 × 106/ml by centrifugation (600 g for 10 min) and serum was added to the sediment, as required. The 50 μl suspension (containing 5 × 105 cells) was then transferred into a new conic tube. Ten microlitres of CD-14 was added (CD-14 FITC mouse antihuman, monoclonal, Ancell, Italy) and incubated at 4°C for 25 min in the dark. At the end of the incubation, the cells were washed twice with 1 ml cold PBS (600 g for 10 min) and resuspended in 1 ml cold PBS. The samples were measured for fluorescence by FACS for evaluation of macrophage percentage in the collected cell population.
Interleukins quantification
IL-1β and IL-6 were titrated using Edogen ELISA kits, Cambridge, MA, USA.
Samples were measured in duplicate and the mean result of the interleukin concentration in the supernatant was corrected to 106 monocytes. The level of the interleukins in pure serum of each blood unit was subtracted from each sample.
RESULTS
Table 1 summarizes 72 genes whose expression level ratios differed from the hypo-osmotic shock-induced cells (showed in Fig. 1) and the controls (Fig. 2), and a graphic comparison of the data is presented in Fig. 3.
Table 1.
Ratios of 72 genes whose expression levels were found to differ between the hypo-osmotic shock-activated cells and the controls.Bold characters indicate factors known to be related to wound healing
| Accession no. | Coordinate | Gene/protein name | Ratio |
|---|---|---|---|
| L38734 | 10 I | Ephrin–B2 precursor* | 0.62 |
| M10051; X02160 | 12 E | Insulin receptor | 0.72 |
| M26880 | 1 A | Ubiquitin1 | 0.76 |
| X97057 | 20 J | WNT-10B precursor; Wnt-12 | 0.8 |
| M65199 | 18 L | Endothelin ET2 | 1.25 |
| X01057; X01058; X01402 | 4 I | IL-2 receptor α subunit precursor (IL-2 receptor α subunit; IL2RA);TAC antigen; CD25 | 1.25 |
| M73980 | 21 G | Neurogenic locus notch protein homolog 1 precursor (NOTCH 1); translocation-associatednotch protein (TAN 1) | 1.26 |
| U04806; U03858 | 19 C | SL cytokine precursor, FLT3 ligand (FLT3LG) | 1.29 |
| M31159; M35878 | 11 O | Insulin-like growth factor-binding protein 3 precursor (IGFBP3) | 1.33 |
| U28811; U64791 | 17 G | Cysteine-rich fibrobl* growth factor receptor; Golgi membrane sialoglycoproteinMG160 (GLGI) | 1.34 |
| U14188 | 10 K | Ephrin A3 precursor (EFNA3); EPH-related receptor tyrosine kinase ligand 3; LERK-3;EHK1L; EFL2 | 1.34 |
| X74764 | 23 K | Neurotrophic tyrosine kinase receptor-related 3 TKT precursor | 1.35 |
| X53799 | 7 B | Macrophage inflamatory protein 2α-(MIP -2α); growth-regulated protein beta (GRO-beta) | 1.36 |
| L20861 | 20 H | Wnt –5a | 1.36 |
| M24545 | 7 D | Monocyte chemotactic protein 1 precursor (MCP1); monocyte chemotactic and activatingfactor – MCAF monocyte secretory protein JE; monocyte chemotactic protein 1; HC11;small inducible cytokine A2 (SCYA2) | 1.37 |
| Y00787 | 3 J | Monocyte-derived neutrophil chemotactic factor – (MDNCF); IL-8 precursor; T-cellchemotactic factor; neutrophil activating protein 1; lymphocyte-derived neutrophilactivating protein; protein3-10C | 1.38 |
| M74178 | 6 L | Macrophage stimulating protein ( MSP); hepatocyte growth factor-like protein | 1.38 |
| X78686 | 7 E | Granulocyte chemotactic protein 2 (GCP2); neutrophil activating peptide ENA-78 | 1.38 |
| L20471 | 22 F | Basigin precursor (BSG);leucocyte activation antigen M6; collagenase stimulatory factor;extracellular matrix metalloproteinase inducer (EMMPRIN); 5F7; CD147 antigen | 1.38 |
| X03438 | 6 J | Granulocyte colony –stimulating factor precursor (G-CSF); plutiprotein; CSF3 | 1.38 |
| J03634 | 16 F | Erythroid differentiation protein | 1.39 |
| X01394 | 14 I | Tumour necrosis factor | 1.4 |
| M21121 | 7 F | RANTES proinflammatory cytokine | 1.4 |
| U32659 | 4 E | IL-17 precursor; cytotoxic T-lymphocyte- associated antigen 8 (CTLA8) | 1.42 |
| X04602; M14584 | 3 HIL-6 precursor; B-cell stimulatory factor 2 (BSF2); interferon beta-2 (IFNB2);hybridoma growth factor | 1.43 | |
| X54936 | 18 N | Placenta growth factor precursor 1 and 2 | 1.46 |
| U06863 | 22 H | Follistsatin-related protein precursor | 1.46 |
| K00558 | 1 E | Brain-specific tubulin α subunit | 1.46 |
| X14454 | 6 E | Interferon regulatory factor 1 | 1.47 |
| X06233 | 6 I | MRP-14 (calcium binding protein in macrophages MIF-related) | 1.48 |
| U90875 | 12 L | Cytotoxic ligand TRAIL receptor | 1.49 |
| X72304 | 22 E | β thromboglobulin-like protein | 1.5 |
| M11220 | 6 K | Granulocyte-macrophage colony stimulating factor (GM-CSF) | 1.54 |
| D10924 | 22 M | Stromal cell derived factor 1 receptor (SDF1 receptor); fusin; CXCR4; leucocyte-derivedseven transmembrane domain receptor (LESTR); LCR1 | 1.58 |
| M95667 + M11730 | 18 G | ERBB2 receptor protein-tyrosine kinase; neu proteo-oncogene; c-erbB2 + HER2 receptor | 1.59 |
| M15530 | 6 G | B-cell growth factor (BCGF1) | 1.59 |
| D16431 | 22 K | Hepatoma-derived growth factor | 1.59 |
| M22489 | 15 J | Bone morphogenic protein 2a | 1.63 |
| L06801 | 4 B | IL-13 precursor; NC30 | 1.67 |
| X06234 | 6 H | Migration inhibitory facto-related protein 8 (MRP-8) calcium-binding protein in macrophagesMIF-related) calgranulina; leucococyte L1 complex light subunit; S100 calcium-bindingprotein A8; cystic fibrosis antigen (CFAG) | 1.68 |
| M32977; M27281 | 19 F | Vascular endothelial growth factor precursor (VEGF); vascular permeability factor (VPF) | 1.69 |
| D10923 | 22 L | G-protein-coupled receptor HM74 | 1.7 |
| L15344 | 4 C | IL-14 precursor; high molecular weight B-cell growth factor | 1.71 |
| X56932 | 1 H | 23-kD highly basic protein; 60S ribosomal protein LI3A | 1.76 |
| L08187 | 9 I | Cytokine receptor EB13 | 1.77 |
| X53655; M37763 | 9 D | Neurotrophin-3 precursor (NT-3); neurotrophic factor (HDNF); nerve growth factor (NGF-2) | 1.81 |
| M60316 | 16 C | Bone morphogenic protein 7 precursor (BMP7); osteogenic protein 1(OP1) | 1.82 |
| M62402 | 12 D | Insulin-like growth factor binding protein 6 precursor (IGF-binding protein 6; IGFBP6;IBP6) | 1.9 |
| M19154; M22045;M22046; Y00083 | 15 G | TGFβ-2 precursor; gliobl*oma-derived T-cell suppressor factor; bsc-1 cell growth inhibitor;polyergin; cetermin | 1.9 |
| M61176 | 8 L | Brain-derived neutrophic factor (BDNF) | 1.93 |
| M96955 + M96956 | 18 B | TDGF1 + TDGF3 + CRGF; CRG | 1.99 |
| U84401 | 20 F | Smoothened; GX | 2.18 |
| X04434; M24599 | 12 F | IGF I receptor | 2.21 |
| M57627 | 3 L | IL-10 precursor; CSIF | 2.25 |
| M25667 | 10 E | Neuromodulin; axonal membrane protein GAP-43; PP46; protein F1;calmodulin-binding protein p57 | 2.33 |
| M86528 + S41522 +S41540 + S41541 | 11 I | NT-4 NT-5 + NT6-α + NT6-β + NT6-γ | 2.36 |
| L00587 | 22 C | Calcitonin receptor | 2.44 |
| K02770 | 3 C | IL-1β precursor; catabolin | 2.44 |
| M22488 + U50330 | 15 I | Bone morphogenetic protein 1 BMP1 + procollagen C-proteinase (pCP-2) | 2.49 |
| M35410 | 11 N | IGF BP-2 | 2.5 |
| M32315 + M55994 | 14 K | TNF-receptor + TNF-receptor 2; TBP2 | 2.51 |
| X03541 | 9 J | High affinity nerve growth factor receptor precursor; trk-1 transforming tyrosine kinaseprotein; p140-TRKA; p68-trk-T3 oncoprotein | 2.61 |
| X02851 | 3 B | IL-1α precursor; haematopoietin-1 | 2.66 |
| U36223 | 17 B | FGF-8; AIGF; HBGF8 | 2.67 |
| M23452 | 6 N | MIP1-α; tonsillar lymphocyte LD78 α protein; GOS19-1 protein; PAT 464.2; SIS -β; smallinducible cytokine A3 (SCYA3) | 2.7 |
| D11086 | 4 K | IL-2 receptor γ; cytokine receptor common γ chain precursor; p64 | 2.78 |
| U95299 | 21 I | Notch 4 | 3.15 |
| X74979 | 23 J | Epithelial discoidin domain receptor 1 precursor (EDDR1; DDR1); cell adhesion kinase (CAK); TRKE; RTK6 | 3.25 |
| M84747 | 5 E | IL-9 receptor precursor | 3.36 |
| X02812; J05114 | 15 F | TGF β | 3.4 |
| X00351 | 1 G | Cytoplasmic β actin | 4.04 |
| U02687 | 19 G | Stem cell tyrosine kinase 1 (STK1); FL cytokine receptor precursor; tyrosine-protein kinasereceptor flt3; CD135 antigen | 4.51 |
| M13982 | 3 F | IL-4 precursor; B-cell stimulatory factor 1 (BSF-1); lymphocyte stimulatory factor 1 | 7.98 |
Fig. 1.
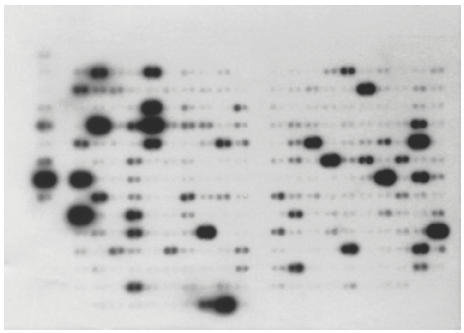
Differential hybridization of monocytes/macrophages activated by hypo-osmotic shock, to the cytokine/receptor filters.
Fig. 2.
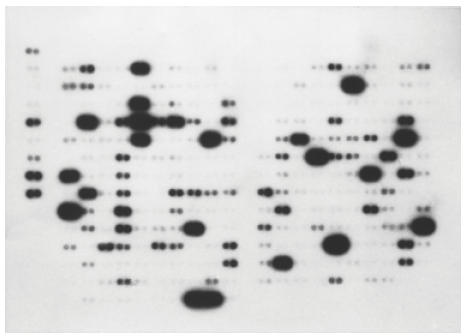
Differential hybridization of control monocytes/macrophages to cytokine/receptor filters.
Fig. 3.
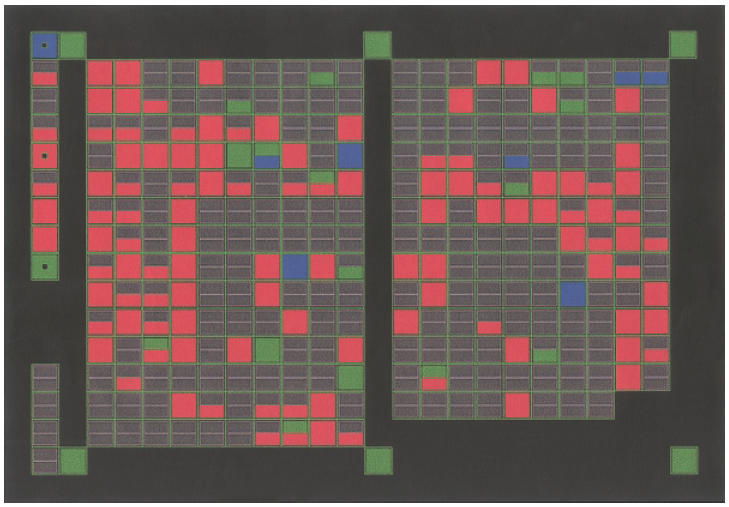
Comparative hybridization of control and hypo-osmotic shock activated monocytes/macrophages, using the AtlasImageTM software.The blue boxes indicate genes expressed at lower levels in the activated cell population, compared with the controls. The red boxes indicate genes expressed at higher levels in the activated cell population. The green boxes indicate genes with no significant difference in expression levels in both groups and the grey boxes indicate genes expressed at a background level.
Among the genes that exhibited differences of expression in the activated cell population, 94% (68/72) displayed increased activity and only four genes were found to have lower mRNA levels in the activated cells, with ratios of expression of 0·62–0·8, which may suggest that the changes are insignificant.
The mRNA levels of 43/68 (63%) of genes that expressed increased activity were found to be 1·5-fold or higher (1·5–7·98) in the activated macrophages cell population compared to the non-treated cells. To correlate between the gene profiles and the relevant cytokines production, the secretion of IL-1, IL-6, TGB-β and GM-CSF by both the hypo-osmotic shock-treated macrophages and the controls were measured. The levels of IL-1 measured in the supernatant of the hypo-osmotic shock-treated monocytes/macrophages after 24 h of incubation, ranged between 1·7 and 123 times higher than those of the controls (P = 0·004) in all blood samples. (Table 2). The augmentation in the levels of IL-6 was even more pronounced, ranging from 6·6 to 175-fold increase in the hypo-osmotic shock-treated macrophages, compared to their matched controls (P = 0·001) (Table 2).
Table 2.
Levels of IL-1 and IL-6 secreted by monocytes/macrophages activated by hypo-osmotic shock and their respective controls, measured in the supernatant of eight individual blood samples, after 24h of incubation time
| IL-1 (pg/ml) | IL-6 (pg/ml) | |||
|---|---|---|---|---|
| Blood sample no. | Control | Osmotic shock | Control | Osmotic shock |
| 1 | 1 | 20 | 35 | 348 |
| 2 | 1 | 123 | 33 | 1523 |
| 3 | 4 | 204 | 114 | 20000 |
| 4 | 7 | 393 | 383 | 20000 |
| 5 | 55 | 697 | 935 | 20000 |
| 6 | 30 | 53 | 5·9 | 500 |
| 7 | 39 | 72 | 26·8 | 289 |
| 8 | 12 | 29 | 134 | 883 |
A statistically significant difference was found in both interleukins between the stimulated cells and their controls, P = 0·004 and P = 0·001, respectively.
Interestingly enough, no increase was found in the levels of TGF-β, as measured after the 24-h incubation, in both cell preparations from seven different blood units tested (Table 3). However, the levels of TGF-β in the cell growth medium (serum) were relatively high. Levels of GM-CSF were not detected in the osmotic-shock or the control cell preparations after incubation of the cells for 24 h, nor were they found in the serum medium before incubation (data not shown).
Table 3.
Levels of TGF β levels in hypo-osmotic shock stimulated monocytes/macrophages and their controls, measured in the supernatantof the individual blood samples, after 24h of incubation time
| TGF-β (pg/ml) | |||
|---|---|---|---|
| Blood sample no. | Serum | Control | Osmotic shock |
| 1 | 250 | 279 | 294 |
| 2 | 240 | 260 | 264 |
| 3 | 433 | 481 | 567 |
| 4 | 260 | 250 | 221 |
| 5 | 683 | 733 | 650 |
| 6 | 375 | 423 | 481 |
| 7 | 433 | 365 | 375 |
There was no statistically significant effect of the hypo-osmotic shock stimulation on secretion of TGF-β to the supernatant.Note: high levels of TGF-β are detected in the serum that serves as growth medium to the cells (controls and hypo-osmotic shock stimulated).
DISCUSSION
Activated human monocytes/macrophages, produced by hypo-osmotic shock, have been used successfully for the treatment of decubital chronic ulcers for few years [17,18]. This procedure is of special interest since no toxic additives or chemical reagents are used, making the clinical application safe. In addition, increased secretion of cytokines, such as IL-1, IL-6 as well as enhanced phagocytosis, were detected by the activated cells, all bearing relevance to the wound-healing process [24].
In this study, we used DNA microarrays to detect differential gene expressions of activated monocytes/macrophages and compared it to that of non-treated cells. Of the genes that exhibited differences of expression in the activated cell population, 94% (68/72) displayed increased activity, and only four genes were found to have lower mRNA levels in the activated cells, with ratios of expression of 0·62–0·8, which may suggested that the changes are insignificant. The mRNA levels of 68 genes were found to be increased in the activated macrophage cell population when compared with the non-treated cells. The expression ratios of 43 of these genes (63%) were found to be 1·5-fold or higher (1·5–7·98).
The genes, whose level of expression was significantly increased, can be divided into four groups:
Genes known to be directly involved in macrophage function and wound healing. These include TGF-β, MIF, FGF-8, TNF receptors, IGF BP, BMP1, IGF-I receptor, monocyte IL-1β, bone morphogenic protein 7, VEGF, MRP-8 and GM-CSF. The methodology and interpretation used in this study, which enabled detection of differentially expressed genes that are known to be involved in monocyte/macrophage activation, metabolism and effective functions, supports and validate the observations of their importance in the wound-healing process.
Genes characterized in this study that were not associated previously with macrophage function and wound healing should be studied further in depth in order to elucidate their specific role and potential clinical applications in the process. Several attractive genes were shown to be expressed preferentially in the activated cell population. One example is the expression of the FLT3 growth factor receptor tyrosine kinase (FLT3R, STK-1), which was shown to be significantly induced (4·51-fold) after activation. Interestingly, FLT3 ligand was recently shown to induce macrophage activation in mice [25]. In bovine bone marrow culture, the combination of FLT 3 ligand with GM-CSF and IL-4 (both found to be induced in our study) resulted in the generation of dendritic cells [26]. Calcitonin receptor mRNA was also induced in the hypo-osmotic shock-treated cells. However, the calcitonin receptor protein is usually expressed on osteoclasts, rather than monocytes/macrophages [27]. Moreover, IL-4, as well as other cytokines such as IL-1α, which were also found to be induced in a very significant way in our study, were associated with osteoclast formation in a monoblastic cell line [28,29]. It will be interesting, therefore, to study whether osteoclast activation can also be induced by the hypo-osmotic shock protocol.
The third group includes genes associated with neuronal development and function. These include nerve growth factor receptor (TRK), high affinity nerve growth factor receptor precursor, neuronal growth protein GAP-43, NT-4, NT-6 and nerve growth factor (NGF-2). The increased expression of mRNAs encoding ‘neuronal’ genes can be explained by their parallel involvement in both the nervous system and the monocyte/ macrophage lineage of the haematopoietic system. Alternatively, a functional cross-talk between monocytes/macrophages and neuronal cells can be proposed. Indeed, a great body of evidence has accumulated in recent years concerning the involvement of monocytes/macrophages and macrophage-secreted factors in the nerve regeneration processes [30]. The identification of the various proteins up-regulated in monocyte/ macrophage activation by differential expression might lead to the characterization of molecules with therapeutic potential in neuroprotection and neuroregeneration.
The fourth group of genes found to be expressed preferentially in the activated monocytes/macrophages includes genes that encode components of developmental controlling pathways such as Notch, Wnt and smoothened. These key pathways are central to the development and function of various tissues and genetic aberrations in several genes encoding different components of these pathways were associated in developmental defects and cancer. Interestingly, glycogen synthase kinase-3β (GSK-3β), a key player in the Wnt/Wingless signalling pathway, was shown recently to be involved in the signalling pathway induced by the tumour-necrosis factor α (TNF-α) proinflammatory cytokine [31].
Several reservations should be borne in mind when the results of DNA microarray experiments are considered. The analysis of mRNA levels does not necessarily reflect the level and activity of the respective protein product, as translational and post-translational processes can have a major effect on the product. The comparison performed in this study between gene expression ratio and the measurement of cytokine levels illustrates the discrepancy between mRNA and protein levels in some examples. For some cytokines, such as IL-1 and IL-6, there was a good correlation between the increased expression at both the mRNA and the protein levels. For other cytokines such as GM-CSF and TGF-β no such correlation was found. The results obtained from mRNA studies should therefore be complemented by analyses of protein levels and function.
We restricted our conclusions to those genes where the expression ratio was higher than 1·5, having in mind the technical limitations inherent in the methodology and the potential sample to sample variability. However, many genes which demonstrated only borderline increased expression are known to be involved meaningfully in macrophage activities, thus one should not always expect to find unequivocal quantitative correlation. For instance, when the interleukins IL-6 and IL-1 were measured by both the DNA microarray and ELISA methods, no quantitative correlation was found. While there was a 6·6–175-fold increase in the levels of IL-6 secreted by the activated cells, at the protein level, the mRNA precursor was only 1·4-fold higher than that of the controls. Conversely, the IL-1 mRNA was expressed 2·4 times higher in the activated cells, with a 1·7–123-fold increase at the protein level. We decided to include those uncertain candidates in the data presented to enable other researchers access to this information. One should bear in mind that the cells activated by hypo-osmotic shock in our system are not composed of a pure macrophage population, but contain certain numbers of other cells such as lymphocytes and granulocytes. The gene profile reported in this study may therefore represent the expression profile of that mixture, enriched for activated monocytes/ macrophages. As the clinical utilization of these cell preparations has already been demonstrated, the characterization of the various genes contributing to the therapeutic effects is important, irrespective of the exact cellular origin. As an example, monocytes and macrophages were shown to express both IL-4 and IL-13 receptors [32]. While IL-4 mRNA was significantly increased in the hypo-osmotic treated cells (7·98-fold), IL-13 mRNA was elevated only moderately. Whether the source of IL-4 or IL-13 in the activated cells is the monocytes/macrophages or some other ‘contaminating’ cells remains to be tested. However, a more important issue, in our opinion, is to study the correlation and contribution of these cytokines to the therapeutic utility of the activated cell mixture.
The system described was designed to create activated monocytes/macrophages for clinical use from random blood units donated by healthy volunteers. Further studies are necessary where activation of the macrophage population is performed using agents such as LPS and interferon-γ and compared to the gene expression profiles described in the present results. This will strengthen our understanding and shed more light on the basic macrophage biology and their potential therapeutic applications in other fields.
Acknowledgments
We are grateful for the Arison Dorsman family’s donation to the Center of DNA Chips In Paediatric Oncology. G. Rechavi holds the Gregorio and Dora Shapiro Chair for Hematologic Malignancies, Sackler School of Medicine, Tel Aviv University.
REFERENCES
- 1.Stein M, Keshav S. The versatility of monocytes/macrophages. Clin Exp Allergy. 1992;22:19–27. doi: 10.1111/j.1365-2222.1992.tb00110.x. [DOI] [PubMed] [Google Scholar]
- 2.Gordon S, Lawson L, Rabinovitz S, et al. Current topics in microbiology and immunology. Vol. 181. Berlin: Springer-Verlag; 1992. Antigen markers of macrophage activation in murine tissues; pp. 1–34. [DOI] [PubMed] [Google Scholar]
- 3.Leibovitch SI, Ross R. The role of the macrophage in wound repair: a study with hydrocortisone and antimacrophage serum. Am J Pathol. 1975;78:71–91. [PMC free article] [PubMed] [Google Scholar]
- 4.Hunt TK, Knighton DR. Studies on inflammation and wound healing: angiogenesis and collagen synthesis stimulated in vivo by resident and activated wound monocytes/macrophages. Surgery. 1984;96:48–54. [PubMed] [Google Scholar]
- 5.Di Pietro LA. Wound healing: the role of macrophage and other immune cells. Shock. 1995;4:233–40. [PubMed] [Google Scholar]
- 6.Clark RA. Basics of cutaneous wound repair. J Dermatol Surg Oncol. 1993;19:693–706. doi: 10.1111/j.1524-4725.1993.tb00413.x. [DOI] [PubMed] [Google Scholar]
- 7.Di Padova F, Pozzi C, Tondre MY, et al. Selective and early increase of IL-1 inhibitors, IL-6 and cortisol after elective surgery. Clin Exp Immunol. 1991;85:137–42. doi: 10.1111/j.1365-2249.1991.tb05694.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Rappolee DA, Werb Z. Macrophage derived growth factors. In: Evans DA, Patel VL, editors. Current topics in microbiology and immunology. Vol. 181. Berlin: Springer-Verlag; 1992. pp. 87–140. [DOI] [PubMed] [Google Scholar]
- 9.Dinarello CA. Biology of interleukin 1. FASEB J. 1988;2:108–15. [PubMed] [Google Scholar]
- 10.Aderem A, Underhill DM. Mechanisms of phagocytosis in monocytes/macrophages. Annu Rev Immunol. 1999;17:593–623. doi: 10.1146/annurev.immunol.17.1.593. [DOI] [PubMed] [Google Scholar]
- 11.Nishida T, Nakamura M, Mishima H, et al. Interleukin-6, 11. facilitates corneal epithelial wound closure in vivo. Arch Ophthalmol. 1992;110:1292–93. doi: 10.1001/archopht.1992.01080210110036. [DOI] [PubMed] [Google Scholar]
- 12.Romeo MB, Reichner JS, Albina JE. Interleukin-6 activity in wounds. Am J Physiol. 1994;266:R1840–R1844. doi: 10.1152/ajpregu.1994.266.6.R1840. [DOI] [PubMed] [Google Scholar]
- 13.Wong GG, Clark SC. Multiple actions of interleukin 6 with a cytokine network. Immunol Today. 1988;9:137–9. doi: 10.1016/0167-5699(88)91200-5. [DOI] [PubMed] [Google Scholar]
- 14.Singer AJ, Clark RAF. Cutaneous wound healing. N Engl J Med. 1999;341:738–46. doi: 10.1056/NEJM199909023411006. [DOI] [PubMed] [Google Scholar]
- 15.Cohen BJ, Danon D, Roth GS. Wound repair in mice as influenced by age and antimacrophage serum. J Gerontol. 1987;42:295–301. doi: 10.1093/geronj/42.3.295. [DOI] [PubMed] [Google Scholar]
- 16.Danon D, Kowach MA, Roth GS. Promotion of wound repair in old mice by local injection of monocytes/macrophages. Proc Natl Acad Sci USA. 1989;86:2018–20. doi: 10.1073/pnas.86.6.2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Danon D, Madjar J, Edenov E, et al. Treatment of human ulcers by application of monocytes/macrophages prepared from a blood unit. Exp Gerontol. 1997;32:633–41. doi: 10.1016/s0531-5565(97)00094-6. [DOI] [PubMed] [Google Scholar]
- 18.Danon D, Frenkel O, Diamantshtein L, et al. A case report: macrophage treatment of pressure sores in paraplegia. J Wound Care. 1998;7:281–3. doi: 10.12968/jowc.1998.7.6.281. [DOI] [PubMed] [Google Scholar]
- 19.Gordon S, Unkeless JC, Cohn ZA. Induction of macrophage plasminogen activator by endotoxin stimulation and phagocytosis. Evidence for a two-stage process. J Exp Med. 1974;140:995–1010. doi: 10.1084/jem.140.4.995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Becker S. Influence of interferon on human monocyte to macrophage development. Cell Immunol. 1984;84:145–9. doi: 10.1016/0008-8749(84)90085-6. [DOI] [PubMed] [Google Scholar]
- 21.Wright SD, Ramos RA, Tobias PS, et al. CD-14, a receptor for complexes of lipopolysaccharide (LPS) and LPS binding protein. Science. 1990;249:1431–3. doi: 10.1126/science.1698311. [DOI] [PubMed] [Google Scholar]
- 22.Lake FR, Noble PW, Henson PM, et al. Functional switching of macrophage responses to tumor necrosis factor-α (TNF-α) by interferons. J Clin Invest. 1994;93:1661–9. doi: 10.1172/JCI117148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lazarov-Spigler O, Solomon AS, Ben-Zeev-Brann A, et al. Transplantation of activated monocytes/macrophages overcomes central nervous system regrowth failure. FASEB J. 1996;10:1–7. doi: 10.1096/fasebj.10.11.8836043. [DOI] [PubMed] [Google Scholar]
- 24.Frenkel O, Shani E, Ben-Bassat I, Brok-Simoni F, Shinar E, Danon D. Activation of human monocyte/macrophages by hypo-osmotic shock. Clin Exp Immunol. 2001;127:103–9. doi: 10.1046/j.1365-2249.2001.01496.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Dannaeus K, Johanson A, Nilsson K, et al. Flt3 ligand induces the outgrowth of Mac-1+B220+ mouse bone marrow progenitor cells restricted to macrophage activation that co-express early B cell-associated genes. Exp Hematol. 1999;27:1646–54. doi: 10.1016/s0301-472x(99)00106-x. [DOI] [PubMed] [Google Scholar]
- 26.Hope JC, Werling D, Collins RA, et al. Flt-3 ligand, in combination with bovine granulocyte-macrophage-colony-stimulating factor and interleukin-4, promotes the growth of bovine bone marrow derived dendritic cells. Scand J Immunol. 2000;51:60–6. doi: 10.1046/j.1365-3083.2000.00646.x. [DOI] [PubMed] [Google Scholar]
- 27.Quinn JM, Morfis M, Lam MH, et al. Calcitonin receptor antibodies in the identification of osteoclasts. Bone. 1999;25:1–8. doi: 10.1016/s8756-3282(99)00094-0. [DOI] [PubMed] [Google Scholar]
- 28.Kaji Y, Ikeda K, Ikeda T, et al. IL-4 but not vitamin D(3), induces monoblastic cell line U differentiate into multinucleated giant cells on osteoclast. J Cell Physiol. 2000;182:214–21. doi: 10.1002/(SICI)1097-4652(200002)182:2<214::AID-JCP10>3.0.CO;2-F. 10.1002/(sici)1097-4652(200002)182:2<214::aid-jcp10>3.3.co;2-6. [DOI] [PubMed] [Google Scholar]
- 29.Lader CS, Flanagan AM. Prostaglandin E2, interleukin 1 alpha, and tumor necrosis factor-alpha increase human osteoclast formation and bone resorption in vitro. Endocrinology. 1998;139:3157–64. doi: 10.1210/endo.139.7.6085. [DOI] [PubMed] [Google Scholar]
- 30.Brown MC, Perry VH, Lunn ER, et al. Macrophage dependence of peripheral sensory nerve regeneration possible involvement of nerve growth factor. Neuron. 1991;6:359–70. doi: 10.1016/0896-6273(91)90245-u. [DOI] [PubMed] [Google Scholar]
- 31.Hoeflich KP, Luo J, Rubie EA, et al. Requirement for glycogen synthase kinase-3 beta in cell survival and TNF-α activation. Nature. 2000;406:86–90. doi: 10.1038/35017574. 10.1038/35017574. [DOI] [PubMed] [Google Scholar]
- 32.Hart PH, Bonder CS, Balogh J, et al. Differential responses of human monocytes and monocytes/macrophages to IL-4 and IL-13. J Leukoc Biol. 1999;66:575–8. [PubMed] [Google Scholar]


