Abstract
CD14 is a pattern recognition receptor on the membranes of monocytes and macrophages for several microbial products, of which lipopolysaccharide (LPS) is the best known. A shed form of CD14 is present in serum. As the CD14 gene promoter polymorphism –159C/T and some bacterial infections may affect the sCD14 levels, we compared the impact of both the CD14 promoter polymorphism and Helicobacter pylori infection on serum sCD14 levels in 201 dyspeptic patients (group 1) who had undergone gastroscopy, and 127 staff members (group 2) with no endoscopy. sCD14 was measured from the sera by a commercial enzyme immunoassay (EIA), and CD14 genotyping was carried out with PCR. Helicobacter pylori infection was detected by serology and/or culture or PCR. sCD14 levels were elevated in the subjects carrying the T allele (CT or TT genotype) in both groups when compared with subjects with the CC genotype. Overall, H. pylori-positive subjects tended to have higher sCD14 levels compared with H. pylori-negative subjects. In group 1 consisting of dyspeptic patients, those with gastric ulcer, gastric erosion or duodenal ulcer had significantly elevated levels of sCD14 compared with the patients with normal endoscopic findings or macroscopic gastritis. The recent use of NSAIDs was also associated with enhanced sCD14. Thus, we were able to show several factors, one genetic and the other environmental (H. pylori infection and mucosal lesion), to have an impact on sCD14.
Keywords: CD14, polymorphism, Helicobacter Pylori
INTRODUCTION
CD14 is a pattern recognition receptor for several microbial products and apoptotic cells [1,2]. It is mainly expressed on monocytes, neutrophils and hepatocytes. Additionally, a soluble form of the CD14 molecule (sCD14) may be found in serum due to shedding from cell membranes. It has also been suggested that hepatocytes, in particular, could be a source of sCD14 production [3,4]. The CD14 gene is localized on chromosome 5q31.1, and a C-to-T transition polymorphism at position –159 in the promoter region of the CD14 gene may influence the sCD14 level [5].
The activation of monocytes/macrophages by LPS mainly takes place through a CD14-dependent pathway [6]. sCD14 mediates the effects of lipopolysaccharide (LPS) and also of other microbial products to the cells lacking membrane-bound CD14 [7–9]. Moreover, sCD14 may be an important molecule in modulating LPS-induced apoptosis in endothelial cells [10,11]. sCD14 is elevated in many chronic infectious and inflammatory conditions, including borreliosis, tuberculosis, periodontal disease and Kawasaki disease [12–15]. sCD14 levels in plasma rise rapidly by 45–75% during endotoxaemia, which is compatible with the criteria for an acute-phase protein [16,17].
Chronic Helicobacter pylori infection is one of the most widespread bacterial infections in humans. There are controversial reports of the association of H. pylori infection with other systemic acute phase reactants, such as C-reactive protein [18]. Helicobacter pylori LPS is able to bind to CD14 and activate monocytes [19]. However, there are no reports on the effect of H. pylori infection on sCD14 levels. We decided to study whether a promoter gene polymorphism and chronic H. pylori infection are able to modulate sCD14 levels systemically, as this would give information about the factors influencing the sCD14 level. If the sCD14 level is enhanced in H. pylori infection, this may also evaluate the significance of H. pylori LPS, as sCD14 may reflect its potency.
MATERIALS AND METHODS
This study was approved by the Ethical Committees of the University Hospital of Oulu, Oulu City Hospital and the Deaconess Institute of Oulu. Informed consent to participate in the study was obtained from all study subjects.
Two separate groups of adult subjects of Finnish origin were recruited. Group 1 consisted of 201 patients (aged 53·6 ± 14·1 years; 118 females, 83 males) with gastrointestinal symptomatology scheduled for upper gastrointestinal endoscopy. They consisted of unselected consecutive patients. However, we also aimed to include an adequate number of peptic ulcer patients. Individual histories were taken concerning smoking habits and recent use (within 1 week) of non-steroidal anti-inflammatory analgetics (NSAIDs). Only the patients with no previous H. pylori eradication therapy were included in the study. Group 2 consisted of 127 control subjects (university and laboratory staff, aged 46·7 ± 9·2 years; 80 females, 47 males), from whom no data were collected concerning possible dyspeptic symptoms or visits to gastroenterologists. Serum samples were obtained from each subject, and H. pylori-specific IgG antibodies were measured with EIA (Pyloriset EIA, Orion Diagnostica, Espoo, Finland). Soluble CD14 was measured with a Human sCD14 EIA kit (R & D Systems, Minneapolis, MN, USA) according to the manufacturer’s instructions. In group 1, multiple biopsies were obtained during gastroscopy, and H. pylori was cultured from one antral biopsy specimen. One antral biopsy of H. pylori-IgG-positive subjects was additionally subjected to H. pylori PCR if the cultures remained negative. Briefly, total DNA from the specimen was extracted with a QIAamp DNA Mini Kit (Qiagen, Valencia, CA, USA). The extracted DNA was subsequently amplified with primers specific for the H. pylori 16S rRNA gene (5′-GCT AAG AGA TCA GCC TAT GTC C-3′ and 5′-TGG CAA TCA GCG TCA GGT AAT G-3′) as described earlier [20]. cagA+ strains were detected by PCR methods using two sets of primers: (i) CAGAF (5′-GAT AAC AGG CAA GCT TTT GAG G-3′) and CAGAR (5′-CTG CAA AAG ATT GTT TGG CAG A-3′) as described earlier [21]; and (ii) D008 (5′-ATA ATG CTA AAT TAG ACA ACT TGA GCG A-3′) and R008 (5′-TTA GAA TAA TC ACA AAC ATC ACG CCA T-3′) according to Miehlke et al.[22]. The strain was considered cagA-positive if either of the primer sets gave a positive amplification product.
In group 2, the subjects were considered to be H. pylori- positive if H. pylori-specific antibodies were detected in their serum. In group 1, H. pylori positivity was determined based on either the presence of antibodies, or a positive culture, or PCR. CD14 genotyping was carried out on DNA extracted from peripheral blood leucocytes.
A new, simple, bi-directional, allele-specific, single-tube PCR method was developed to determine −159C/T alleles in the promoter region of the CD14 gene of the study subjects. Target sequences for partly overlapping allele-specific primers (cfors for the C allele and trevs for the T allele) were obtained from GenBank (Accession number X74984). An additional mismatch was inserted at the penultimate 3′ nucleotide (underlined) of the allele-specific primers to increase the specificity of the PCR reaction. Additionally, two previously described outer primers were used [23]. The primer sequences were as follows:
cfors: 5′-CTC CAG AAT CCT TCC TGT TAC GAC-3′
trevs: 5′-TGT AGG ATG TTT CAG GGA GGG GTA-3′
cdp1: 5′-TTG GTG CCA ACA GAT GAG GTT CAC-3′ [23]
cdp2: 5′-TTC TTT CCT ACA CAG CGG CAC CC-3′ [23]
PCR was performed in a total volume of 10μl (50–100 ng genomic DNA, 1 × PCR buffer with 1·5 mm MgCl, 0·2 mm nucleotides, 0·5 U Taq polymerase and 0·5 μm CD14 primers). The following reaction conditions were used: an initial denaturation at 95°C for 5 min followed by 30 cycles at 95°C for 30 s, at 60°C for 30 s, and at 72°C for 1 min. The final extension step was at 72°C for 5 min. The PCR products were visualized by electrophoresis in 2% (w/v) agarose gel stained with ethidium bromide. The PCR method was validated by determining the CD14 alleles of 50 samples with the PCR-RFLP method described earlier [5].
Statistical analysis
The differences in the frequencies of the CD14 genotypes and alleles were analysed by the two-tailed χ2-test. The relationships between the serum CD14 levels, the CD14 genotypes and the disease outcome were tested with the Mann–Whitney U-test.
RESULTS
Establishment of a new bi-directional allele-specific PCR for CD14 (–159C/T) genotype determination
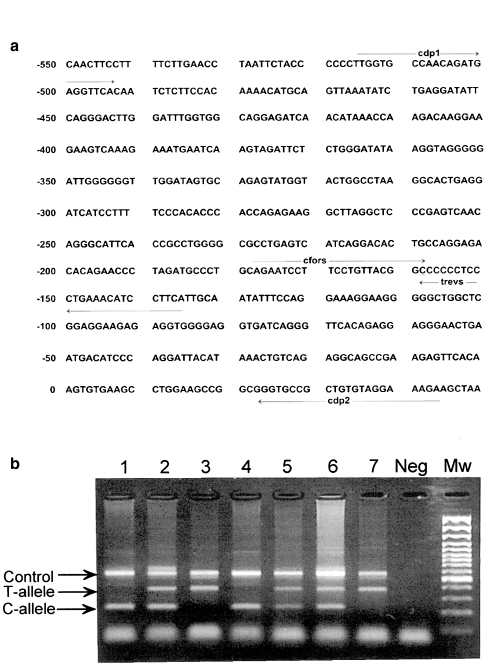
CD14 promoter region polymorphism was studied with a new bi-directional, allele-specific PCR method, which allows determination of both alleles in one PCR reaction tube, as the PCR amplimers representing the different alleles are of different sizes. This was made possible by designing the allele-specific primers to run in opposite directions, and to anneal with opposite strands of DNA. Non-specific amplification of the other allele was effectively prevented by designing an additional mismatch at the penultimate 3′ nucleotide of the allele-specific primer (Fig. 1a).
Fig. 1.
Primer locations in the promoter region of the CD14 gene (a) and a typical gel electrophoresis of CD14 (–159C/T) genotyping (b). The CC homozygous samples are shown in lanes 1 and 4, the CT heterozygous samples in lanes 2, 5 and 6, and the TT homozygous samples in lanes 3 and 7. Mw = 100 bp ladder.
The PCR assay developed here yields a 381 bp band for the T allele, a 227 bp band for the C allele and an additional 561 bp control product, as shown in Fig. 1(b). Identical results were obtained when the results of this new PCR, applied to 50 samples, were compared with the PCR-RFLP results of a previously described method [5].
Effect of CD14 genotypes and H. pylori infection on sCD14 levels
The median serum levels of sCD14 were significantly increased in group 1, which consisted of dyspeptic patients, compared with the control subjects of group 2 (1400 ng/ml versus 1270 ng/ml, P=0·001), as shown in Table 1. However, the median sCD14 level in the dyspeptic patients in group 1 with macroscopically-normal gastric mucosa, or with macroscopic gastritis alone (n = 76), was similar to that of the control subjects in group 2 (1265 versus 1270 ng/ml, P=ns). No significant differences in sCD14 levels were evident between patients with normal findings and those with macroscopic gastritis as the only finding in endoscopy in group 1.
Table 1.
Median (range) sCD14 serum levels (ng/ml) in Helicobacter pylori+ and H. pylori− subjects and in different –159C/T genotypes
| n | All genotypes median (range) | n | CC-genotype median (range) | n | CT and TT genotypes median (range) | CC versus CT/TT † | |
|---|---|---|---|---|---|---|---|
| Combined group (groups 1 and 2) | |||||||
| All subjects | 328 | 1315 (714–2620) | 146 | 1240 (714–2620) | 182 | 1400 (814–2600) | P < 0·001 |
| H. pylori+ | 161 | 1400 (855–2620) | 72 | 1300 (855–2620) | 89 | 1420 (890–2600) | P=0·016 |
| H. pylori− | 167 | 1280 (714–2000) | 74 | 1190 (714–2000) | 93 | 1350 (814–1970) | P < 0·001 |
| Hp−versus Hp+* | P < 0·001 | P=0·002 | P=0·025 | ||||
| Group 1 (dyspeptic patients) | |||||||
| All subjects | 201 | 1400 (875–2620) | 91 | 1260 (875–2620) | 110 | 1430 (890–2600) | P < 0·001 |
| H. pylori+ | 117 | 1410 (875–2620) | 52 | 1310 (875–2620) | 65 | 1440 (890–2600) | P = 0·015 |
| H. pylori− | 84 | 1300 (909–2000) | 39 | 1200 (909–2000) | 45 | 1400 (960–1970) | P = 0·001 |
| Hp−versus Hp+* | P = 0·013 | P = 0·019 | P = ns | ||||
| Group 2 (control subjects) | |||||||
| All subjects | 127 | 1270 (714–2010) | 55 | 1210 (714–1890) | 72 | 1300 (814–2010) | P = 0·018 |
| H. pylori+ | 44 | 1300 (855–2010) | 20 | 1290 (855–1890) | 24 | 1300 (112–0 2010) | P = ns |
| H. pylori− | 83 | 1240 (714–1850) | 35 | 1140 (714–1700) | 48 | 1300 (814–1850) | P = 0·015 |
| Hp−versus Hp+* | P = 0·05 | P = 0·08 | P = ns | ||||
Comparison between H. pylori+ subjects and H. pylori− subjects within each genotype (Mann–Whitney U-test).
Comparison between subjects with CC genotype versus CT or TT genotypes (Mann–Whitney U-test).
Median sCD14 concentrations were higher in the sera of subjects carrying the T allele (CT or TT genotypes) than in the subjects with no T allele (CC genotype) in the combined group (P < 0·001), and also when groups 1 and 2 were analysed separately (P=0·001 and P=0·018).
In H. pylori− subjects, the CC genotype was clearly associated with lower sCD14 levels than in the CT or TT genotypes (the combined group, P < 0·001; group 1, P=0·001; group 2, P=0·01), as shown in Table 1. The analysis of subjects positive for H. pylori separately in the three groups also revealed statistically significant associations between genotypes and sCD14 levels in the combined group (P=0·016) and in group 1 (P=0·015).
The H. pylori+ subjects had higher sCD14 levels than the H.pylori− subjects in the combined group (P < 0·001), and also when groups 1 and 2 were analysed separately with all the genotypes included (P=0·013 and P=0·05). The effect of H. pylori infection on sCD14 levels was also analysed separately according to the genotype (T-carrying and non-T genotype). The increase in sCD14 levels in H. pylori+versus H. pylori− subjects was most marked in the non-T genotype (=CC) in all three groups (combined group, P=0·002; group 1, P=0·019; and group 2, P=0·08). The increase was also significant in the T-carrying genotype in the combined group (P=0·025).
Use of NSAIDs affected sCD14, as the median levels were higher in patients who had used NSAIDs within 0–7 days compared with those who had not (1410 ng/ml versus 1320 ng/ml, P=0·015, group 1). The frequency of those who had used NSAIDs recently (within 1 week) was similar in both H. pylori+ and H. pylori− subjects (50/117 (42·7%) versus 36/84 (42·9%), P=ns). Smoking or bacterial strain virulence (cag± strain) did not affect the level of sCD14.
Effect of endoscopic findings on sCD14 levels
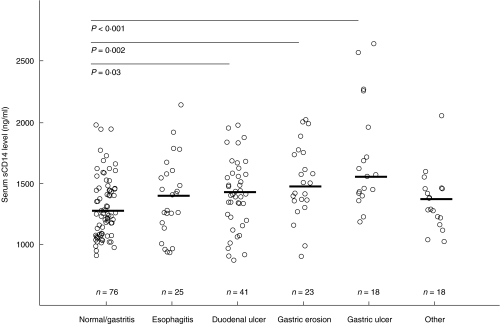
We evaluated sCD14 levels in the dyspeptic patients according to their major endoscopy findings (Fig. 2). The patients with a current gastric ulcer (17/18 H. pylori+) or gastric erosion (9/23 H. pylori+) had a higher median sCD14 level than the patients with normal mucosa or macroscopic gastritis (1540 versus 1265 ng/ml, P < 0·001; and 1460 versus 1265 ng/ml, P=0·002). The patients with a current duodenal ulcer (41/41 H. pylori+) also had moderately increased sCD14 levels (1420 ng/ml) compared with the patients with no findings or macroscopic gastritis only (P=0·03). In the group with erosion, the median sCD14 concentrations were significantly higher in the H. pylori+ patients compared with H. pylori− patients (1800 versus 1425 ng/ml, P=0·047). Moreover, the patients with oesophagitis had a slightly elevated median sCD14 level compared with the patients with normal findings or gastritis, but this difference was not significant.
Fig. 2.
Median soluble CD14 serum levels (ng/ml) in dyspeptic patients (group 1) with various endoscopic findings.
The effect of recent NSAID use in the groups with different endoscopy findings is shown in Table 2. No statistically significant differences were found in any of the subgroups, although sCD14 levels of recent NSAID users tended to be slightly higher in all other groups except that with duodenal ulcer.
Table 2.
The effect of recent use of NSAIDs on median soluble CD14 levels (ng/ml) in groups with different upper gastrointestinal endoscopy findings
| Recent use of NSAIDs | No recent use of NSAIDs | ||||
|---|---|---|---|---|---|
| Endoscopy finding | n | sCD14 Median (range) | P-value | n | sCD14 Median (range) |
| Normal or gastritis | 29 | 1380 (929–1970) | NS | 47 | 1210 (961–2030) |
| Esophagitis | 11 | 1430 (1170–2080) | NS | 14 | 1270 (938–1730) |
| Duodenal ulcer | 15 | 1400 (1020–1970) | NS | 25 | 1425 (890–2000) |
| Gastric erosion | 11 | 1510 (909–2000) | NS | 13 | 1450 (977–1950) |
| Gastric ulcer | 10 | 1690 (1230–2620) | NS | 8 | 1480 (1410–2290) |
| Other | 7 | 1400 (1060–2000) | NS | 11 | 1300 (960–1640) |
Frequencies of CD14 genotypes in the groups classified based on H. pylori status or disease outcome
Genotype frequencies were similar in the H. pylori+ and H. pylori− subjects in all three groups (Table 3). The effect of the CD14 genotype on susceptibility to peptic ulcer disease was also evaluated in group 1, which was divided into H. pylori− and H. pylori+ non-ulcer subjects and subjects with current or previous endoscopically-defined duodenal ulcer (DU) or gastric ulcer (GU). The frequency of the TT genotype tended to be increased in subjects with DU (21·7%) and GU (18·5%) compared with the H. pylori+ (15·7%) and H. pylori− (11·7%) subjects without peptic ulcers, but the differences were not statistically significant.
Table 3.
CD14 genotype frequencies in groups with different Helicobac- ter pylori-status and disease outcome n (%)
| CD14 genotypes | ||||
|---|---|---|---|---|
| n | CC | CT | TT | |
| Combined group | 328 | 146 (44·5) | 132 (40·2) | 50 (15·2) |
| H. pylori– | 167 | 74 (44·3) | 72 (43·1) | 21 (12·6) |
| H. pylori+ | 161 | 72 (44·7) | 60 (37·3) | 29 (18·0) |
| Group 1 (dyspeptic patients) | 201 | 91 (45·3) | 78 (38·8) | 32 (15·9) |
| Non-ulcer, Hp− | 77 | 35 (45·5) | 33 (42·9) | 9 (11·7) |
| Non-ulcer, Hp+ | 51 | 23 (45·1) | 20 (39·2) | 8 (15·7) |
| Duodenal ulcer | 46 | 18 (39·1) | 18 (39·1) | 10 (21·7) |
| Gastric ulcer | 27 | 15 (55·6) | 7 (25·9) | 5 (18·5) |
| Group 2 (control subjects) | 127 | 55 (43·3) | 54 (42·5) | 18 (14·2) |
| H. pylori− | 83 | 35 (42·2) | 38 (45·8) | 10 (12·0) |
| H. pylori+ | 44 | 20 (45·5) | 16 (36·4) | 8 (18·2) |
(P=ns).
DISCUSSION
sCD14 is a multi-functional molecule. In addition to being a receptor to LPS and other bacterial structures, it may modulate LPS-triggered apoptosis [10,11]. Products of H. pylori, including LPS, also enhance apoptosis of mucosal epithelial cells [24,25]. sCD14 may also regulate T- and B-lymphocyte activation and function [26,27].
Our results from a fairly large study population show that the subjects of Finnish origin with genotypes carrying the T allele at the –159 site of the promoter region of the CD14 gene have higher sCD14 levels than subjects with the non-T genotype. Thus, our findings confirm an earlier report suggesting that the CD14 level, at least the soluble form, is under genetic control [5]. In a recently published study, however, no association between CD14 genotypes and serum sCD14 levels were observed in a Japanese population, although the trend was similar to that seen in studies describing other ethnic groups [28].
The results presented here suggest that an H. pylori infection elevates sCD14 levels. In our study, this effect was most marked in subjects with the CC genotype. The factors that could contribute to serum sCD14 levels in H. pylori infection are currently unknown. As monocytes and neutrophils are locally accumulated and activated in H. pylori- infected gastric mucosa, they could be the source of a shed protein [29]. Elevated systemic sCD14 levels could thus reflect an enhanced local reaction to H. pylori LPS and other pathogen-related molecules. Lipopolysaccharides of many Gram-negative bacteria are powerful stimulators of monocytes and macrophages [30]. During Gram-negative sepsis, CD14 levels are greatly enhanced [16,17]. This increase may also be related to hepatocyte action, as endotoxaemia may also increase the shedding of CD14 from (rat) hepatocytes [3]. Despite the fact that H. pylori LPS is only a weak stimulator of monocytes, it has been suggested to play an important role in H. pylori infection [24]. Helicobacter pylori LPS is able to bind to mCD14 on monocytes, and to stimulate monocytes to secrete cytokines [19]. We should not exclude the possibility that small amounts of H. pylori LPS may leak into the circulation and, accordingly, stimulate hepatocytes and monocytes to shed soluble CD14. As the recent use of NSAIDs, as well as the presence of mucosal lesions (DU, GU or erosion), also enhanced sCD14 in our study, these results may suggest that a damaged mucosal epithelium may be involved in the systemic change found in sCD14. Alternatively, recent use of NSAIDs may reflect an ongoing inflammatory condition/infection outside the gastrointestinal tract, and the elevated sCD14 levels might be due to that process. Unfortunately, no appropriate data were collected from patients in group 1.
Though the net increase of sCD14 in H. pylori infection was moderate, our results may have interesting implications for the suggested extragastric pathology of chronic H. pylori infection. There are controversial reports of the linkage between H. pylori infection and atherosclerosis [18]. sCD14 contributes to lipid metabolism, as sCD14 transports lipids and LPS to high-density lipoprotein (HDL) [31,32]. Dysfunctional lipid metabolism is a characteristic feature of several infections [33]. Some studies have found an association between H. pylori infection and proatherogenic lipid levels, including a low HDL level [34,35].
LPS is probably one factor responsible for the lipid profile changes, but sCD14 and, especially, the distribution of CD14- positive monocytes may also be involved in the regulation of serum lipid levels [36,37]. In vitro experiments have shown that the availability of sCD14 may determine the efficacy of lipid transfer from cells to extracellular space [38]. Whether moderately elevated sCD14 in H. pylori infection contributes to lipid metabolism remains to be settled. Interestingly, an association between the TT genotype of the CD14 gene and myocardial infarction has been recently reported in three different studies [23,39,40].
In our study, the most markedly elevated sCD14 levels were found in patients with current gastric ulcer, which has been suggested to coincide with coronary heart disease more frequently than expected [41]. Elevated sCD14 levels have also been reported in chronic periodontal disease [12], which is associated with an increased susceptibility to atherosclerosis [42].
CD14 is one of the candidate genes for atopy. Elevated total serum IgE levels were found in skin test-positive adults with the CC genotype [43]. Accordingly, skin test-positive children with the TT genotype had lowered total serum IgE [5]. sCD14 may regulate human B-cell function by enhancing IgG1, and suppressing IgE production in activated tonsillar B cells and antigen-stimulated PBMC through CD40-dependent interaction [26]. There is currently an ongoing debate as to whether childhood exposure to environmental microbes (including H. pylori) is able to prevent atopic sensitization. LPS and other bacterial products from the gastrointestinal tract may have a role in the maturation of the Th1-type immune response [44]. Our finding of elevated sCD14 in adults with H. pylori infection speaks for the possibility that also during childhood infection, products of H. pylori might contribute to Th1/Th2 balance through CD14 action.
In conclusion, a CD14 promoter gene polymorphism, namely, the presence of the T allele, was associated with an increase in sCD14 levels. Helicobacter pylori infection was also associated with an elevated CD14 level, and this was especially true of the CC genotype. We were thus able to identify several factors, both genetic and environmental (mucosal damage caused by H. pylori or use of NSAIDs), that have an impact on sCD14.
Acknowledgments
The authors thank Eila Matkaselkä and Eeva-Liisa Heikkinen for excellent technical assistance. This work was supported by grants from the University of Oulu Foundation, the Finnish Cancer Foundation and the Cancer Society of Northern Finland (JK).
REFERENCES
- 1.Gregory CD. CD14-dependent clearance of apoptotic cells: relevance to the immune system. Curr Opin Immunol. 2000;12:27–34. doi: 10.1016/s0952-7915(99)00047-3. [DOI] [PubMed] [Google Scholar]
- 2.Kaisho T, Akira S. Dendritic-cell function in Toll-like receptor- and MyD88-knockout mice. Trends Immunol. 2001;22:78–83. doi: 10.1016/s1471-4906(00)01811-1. [DOI] [PubMed] [Google Scholar]
- 3.Liu S, Khemlani L, Shapiro R, et al. Expression of CD14 by hepatocytes: Upregulation by cytokines during endotoxemia. Infect Immun. 1998;66:5089–98. doi: 10.1128/iai.66.11.5089-5098.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Pan Z, Zhou L, Hetherington C, Zhang D-E. Hepatocytes contribute to soluble CD14 production, and CD14 expression is differentially regulated in hepatocytes and monocytes. J Biol Chem. 2000;275:36430–5. doi: 10.1074/jbc.M003192200. [DOI] [PubMed] [Google Scholar]
- 5.Baldini M, Lohman C, Halonen M, Erickson R, Holt P, Martinez F. A polymorphism in the 5′ flanking region of the CD14 gene is associated with circulating soluble CD14 levels and with total serum immunoglobulin E. Am J Respir Cell Mol Biol. 1999;20:976–83. doi: 10.1165/ajrcmb.20.5.3494. [DOI] [PubMed] [Google Scholar]
- 6.Wright S, Ramos R, Tobias P, Ulevitch R, Mathison J. CD14, a receptor for complexes of lipopolysaccharide (LPS) and LPS binding protein. Science. 1990;249:1431–3. doi: 10.1126/science.1698311. [DOI] [PubMed] [Google Scholar]
- 7.Haziot A, Rong G, Silver J, Goyert S. Recombinant soluble CD14 mediates the activation of endothelial cells by lipopolysaccharide. J Immunol. 1993;151:1500–7. [PubMed] [Google Scholar]
- 8.Loppnow H, Stelter F, Schönbeck U, et al. Endotocin activates human vascular smooth muscle cells despite lack of expression of CD14 mRNA or endogenous membrane CD14. Infect Immun. 1995;63:1020–6. doi: 10.1128/iai.63.3.1020-1026.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Pugin J, Schurer-Maly C-C, Leturcq D, Moriarty A, Ulevitch R, Tobias P. Lipopolysaccharide activation of human endothelial and epithelial cells is mediated by lipolysaccharide-binding protein and soluble CD14. Proc Natl Acad Sci USA. 1993;90:2744–8. doi: 10.1073/pnas.90.7.2744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Frey E, Finlay B. Lipopolysaccharide induces apoptosis in a bovine endothelial cell line via a soluble CD14 dependent pathway. Microb Pathogenesis. 1998;24:101–9. doi: 10.1006/mpat.1997.0178. [DOI] [PubMed] [Google Scholar]
- 11.Hu Y, Yee E, Harlan E, Wong F, Karsan A. Lipopolysaccharide induces the antiapoptotic molecules, A1 and A20, in microvascular endothelial cells. Blood. 1998;92:2759–65. [PubMed] [Google Scholar]
- 12.Hayashi J, Masaka T, Ishikawa I. Increased levels of soluble CD14 in sera of periodontitis patients. Infect Immun. 1999;67:417–20. doi: 10.1128/iai.67.1.417-420.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Juffermans N, Verbon A, van Deventer S, et al. Serum concentrations of lipopolysaccharide activity-modulating proteins during tuberculosis. J Infect Dis. 1998;178:1839–42. doi: 10.1086/314492. [DOI] [PubMed] [Google Scholar]
- 14.Lin B, Noring R, Steere A, Klempner M, Hu L. Soluble CD14 levels in the serum, synovial fluid, and cerebrospinal fluid of patients with various stages of Lyme disease. J Infect Dis. 2000;181:1185–8. doi: 10.1086/315357. [DOI] [PubMed] [Google Scholar]
- 15.Takeshita S, Nakatani K, Tsujimoto H, Kawamura Y, Kawase H, Sekine I. Increased levels of circulating soluble CD14 in Kawasaki disease. Clin Exp Immunol. 2000;119:376–81. doi: 10.1046/j.1365-2249.2000.01120.x. 10.1046/j.1365-2249.2000.01120.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Landmann R, Zimmerli W, Sansano S, et al. Increased circulating soluble CD14 is associated with high mortality in gram-negative septic shock. J Infect Dis. 1995;171:639–44. doi: 10.1093/infdis/171.3.639. [DOI] [PubMed] [Google Scholar]
- 17.Landmann R, Reber A, Sansano S, Zimmerli W. Function of soluble CD14 in serum from patients with septic shock. J Infect Dis. 1996;173:661–8. doi: 10.1093/infdis/173.3.661. [DOI] [PubMed] [Google Scholar]
- 18.Danesh J, Peto R. Risk factors for coronary heart disease and infection with Helicobacter pylori: meta-analysis of 18 studies. Br Med J. 1998;316:1130–2. doi: 10.1136/bmj.316.7138.1130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bliss CM, Jr, Golenbock D, Keates S, Linevsky J, Kelly C. Helicobacter pylori lipopolysaccharide binds to CD14 and stimulates release of interleukin-8, epithelial neutrophil-activating peptide 78, and monocyte chemotactic protein 1 by human monocytes. Infect Immun. 1998;66:5357–63. doi: 10.1128/iai.66.11.5357-5363.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Peek RM, Jr, Miller G, Tham K, et al. Detection of Helicobacter pylori gene expression in human gastric mucosa. J Clin Microbiol. 1995;33:28–32. doi: 10.1128/jcm.33.1.28-32.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Yamaoka Y, Kodama T, Gutierrez O, Kim J, Kashima K, Graham D. Relationship between Helicobacter pylori iceA, cagA, and vacA status and clinical outcome: studies in four different countries. J Clin Microbiol. 1999;37:2274–9. doi: 10.1128/jcm.37.7.2274-2279.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Miehlke S, Kibler K, Kim J, et al. Allelic variation in the cagA gene of Helicobacter pylori obtained from Korea compared to the United States. Am J Gastroenterol. 1996;91:1322–5. [PubMed] [Google Scholar]
- 23.Hubacek J, Rothe G, Pit’ha J, et al. C(-260)→T polymorphism in the promoter of the CD14 monocyte receptor gene as a risk factor for myocardial infarction. Circulation. 1999;99:3218–20. doi: 10.1161/01.cir.99.25.3218. [DOI] [PubMed] [Google Scholar]
- 24.Moran A. Helicobacter pylori lipopolysaccharide-mediated gastric and extragastric pathology. J Physiol Pharmacol. 1999;50:787–805. [PubMed] [Google Scholar]
- 25.Piotrowski J, Piotrowski E, Skrodzka D, Slomiany A, Slomiany B. Induction of acute gastritis and epithelial apoptosis by Helicobacter pylori lipopolysaccharide. Scand J Gastroenterol. 1997;32:203–11. doi: 10.3109/00365529709000195. [DOI] [PubMed] [Google Scholar]
- 26.Arias M, Rey Nores J, Vita N, et al. Human B cell function is regulated by interaction with soluble CD14: Opposite effects on IgG1 and IgE production. J Immunol. 2000;164:3480–5. doi: 10.4049/jimmunol.164.7.3480. [DOI] [PubMed] [Google Scholar]
- 27.Rey Nores J, Bensussan A, Vita N, et al. Soluble CD14 acts as a negative regulator of human T cell activation and function. Eur J Immunol. 1999;29:265–76. doi: 10.1002/(SICI)1521-4141(199901)29:01<265::AID-IMMU265>3.0.CO;2-G. [DOI] [PubMed] [Google Scholar]
- 28.Ito D, Murata M, Tanahashi N, et al. Polymorphism in the promoter of lipopolysaccharide receptor CD14 and ischemic cerebrovascular disease. Stroke. 2000;31:2661–4. doi: 10.1161/01.str.31.11.2661. [DOI] [PubMed] [Google Scholar]
- 29.Dixon MF. Helicobacter pylori gastritis: pathology and progression. In: Moran A, O’Morain C, editors. Pathogenesis and host response in Helicobacter pylori infections. Englewood, NJ: Normed-Verlag, Inc; pp. 110–8. [Google Scholar]
- 30.Landmann R, Knopf H-P, Link S, Sansano S, Schumann R, Zimmerli W. Human monocyte CD14 is upregulated by lipopolysaccharide. Infect Immun. 1996;64:1762–9. doi: 10.1128/iai.64.5.1762-1769.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wurfel M, Hailman E, Wright S. Soluble CD14 acts as a shuttle in the neutralization of lipopolysaccharide (LPS) by LPS-binding protein and reconstituted high density lipoprotein. J Exp Med. 1995;181:1743–54. doi: 10.1084/jem.181.5.1743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Yu B, Hailman E, Wright S. Lipopolysaccharide binding protein and soluble CD14 catalyze exchange of phospholipids. J Clin Invest. 1997;99:315–24. doi: 10.1172/JCI119160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Khovidhunkit W, Memon R, Feingold K, Grunfeld C. Infection and inflammation-induced proatherogenic changes of lipoproteins. J Infect Dis. 2000;181(Suppl):S462–72. doi: 10.1086/315611. [DOI] [PubMed] [Google Scholar]
- 34.Laurila A, Bloigu A, Näyhä S, Hassi J, Leinonen M, Saikku P. Association of Helicobacter pylori infection with elevated serum lipids. Atherosclerosis. 1999;142:207–10. doi: 10.1016/s0021-9150(98)00194-4. 10.1016/s0021-9150(98)00194-4. [DOI] [PubMed] [Google Scholar]
- 35.Niemelä S, Karttunen T, Korhonen T, et al. Could Helicobacter pylori infection increase the risk of coronary heart disease by modifying serum lipid concentrations? Heart. 1996;75:573–5. doi: 10.1136/hrt.75.6.573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Patino R, Ibarra J, Rodriguez R, et al. Circulating monocytes in patients with diabetes mellitus, arterial disease, and increased CD14 expression. Am J Cardiol. 2000;85:1288–91. doi: 10.1016/s0002-9149(00)00757-8. [DOI] [PubMed] [Google Scholar]
- 37.Rothe G, Herr A, Stöhr J, Abletshauser C, Weidinger G, Schmitz G. A more mature phenotype of blood mononuclear phagocytes is induced by fluvastatin treatment in hypercholesterolemic patients with coronary heart disease. Atherosclerosis. 1999;144:251–61. doi: 10.1016/s0021-9150(99)00061-1. [DOI] [PubMed] [Google Scholar]
- 38.Sugiyama T, Wright S. Soluble CD14 mediates efflux of phospholipids from cells. J Immunol. 2001;166:826–31. doi: 10.4049/jimmunol.166.2.826. [DOI] [PubMed] [Google Scholar]
- 39.Shimada K, Watanabe Y, Mokuno H, Iwama Y, Daida H, Yamaguchi H. Common polymorphism in the promoter of the CD14 monocyte receptor gene is associated with acute myocardial infarction in Japanese men. Am J Cardiol. 2000;86:682–4. doi: 10.1016/s0002-9149(00)01054-7. [DOI] [PubMed] [Google Scholar]
- 40.Unkelbach K, Gardemann A, Kostrzewa M, Philipp M, Tillmanns H, Haberbosch W. A new promoter polymorphism in the gene of lipopolysaccharide receptor CD14 is associated with low expired myocardial infarction in patients with low atherosclerotic risk profile. Arterioscler Thromb Vasc Biol. 1999;19:932–8. doi: 10.1161/01.atv.19.4.932. [DOI] [PubMed] [Google Scholar]
- 41.Sonnenberg A. Concordant occurrence of gastric and hypertensive diseases. Gastroenterology. 1988;95:42–8. doi: 10.1016/0016-5085(88)90288-0. [DOI] [PubMed] [Google Scholar]
- 42.Valtonen V. Role of infections in atherosclerosis. Am Heart J. 1999;138:S431–3. doi: 10.1016/s0002-8703(99)70269-3. [DOI] [PubMed] [Google Scholar]
- 43.Koppelman G, Reijmerink N, Stine O, et al. Association of a promoter polymorphism of the CD14 gene and atopy. Am J Respir Crit Care Med. 2001;163:965–9. doi: 10.1164/ajrccm.163.4.2004164. [DOI] [PubMed] [Google Scholar]
- 44.Matricardi P, Rosmini F, Riondino S, et al. Exposure of foodborne and orofecal microbes versus airborne viruses in relation to atopy and allergic asthma: epidemiological study. Br Med J. 2000;320:412–7. doi: 10.1136/bmj.320.7232.412. [DOI] [PMC free article] [PubMed] [Google Scholar]