Abstract
Idiopathic pulmonary fibrosis (IPF) is an inflammatory lung disease characterized by the accumulation of inflammatory cells and deposition of collagen, resulting in lung remodelling. High numbers of T cells are present in bronchoalveolar lavage fluid (BALF) of IPF patients, although the characteristics of these cells are yet to be determined. To elucidate the pathogenic mechanisms of IPF, we analysed the T cell receptor (TCR) of BALF lymphocytes in three patients with IPF and three healthy subjects as control. TCR repertoire of BALF lymphocytes and T cell clonality were examined by family PCR and Southern blot analysis, and single-strand conformation polymorphism (SSCP), respectively. We observed that the TCR repertoire in the lung was heterogeneous, both in the control subjects and three patients with IPF. SSCP analysis demonstrated an increase in the number of accumulated T cell clones in BALF of two of the three patients, but not in the healthy subject. Furthermore, junctional sequence analysis showed the presence of conserved amino acid motifs (ETGRSG, LAxG, QGQ, GxQP, GRxG, VAR, PGT, GTI, GGT, TGR, LxLxQ, SGQ) in the TCR-CDR 3 region of BAL lymphocytes in patients with IPF, whereas only two amino acid motifs (VTTG, GGE) were found in the control. Our findings suggest that T cells in BALF of patients with IPF expand oligoclonally in the lung, suggesting antigen stimulation of these cells.
Keywords: clonality, IPF, pathogenesis, T cell receptor, T lymphocytes
INTRODUCTION
Idiopathic pulmonary fibrosis (IPF) is a chronic and progressive pulmonary disorder, associated with both inflammation and fibrosis of the lung parenchyma [1,2]. Recent advances in biotechnology in the field of bronchoalveolar lavage (BAL) have provided considerable information on the cellular components of alveoli in this disease [3–5]. Pulmonary damage and fibrosis represent the consequences of immune response and inflammatory process. Previous studies have shown that lymphocytes (especially T cells) and alveolar macrophages play a central role in the pathogenesis of IPF, although the mechanism that triggers these cells has not been elucidated [6–8].
T cells recognize antigens in the context of MHC on antigen-presenting cells (APC) through an antigen receptor, the T cell receptor (TCR). Several groups investigating TCR genes in autoimmune diseases such as rheumatoid arthritis [9], Sjögren's syndrome [10,11] and multiple sclerosis [12], among other diseases, have demonstrated that T cells accumulate oligoclonally in the inflamed lesion. Furthermore, conserved amino acid motifs have been observed in the CDR3 region of TCR gene, whereas there was no skewed usage of TCR genes [10–13].
Several studies have examined TCR genes of BAL fluid (BALF) T cells in patients with various lung diseases such as sarcoidosis and bronchial asthma. Moller and colleagues [14] demonstrated an increased number of TCR BV8 T cells in BALF of patients with sarcoidosis. Zissel et al.[15] showed the predominant usage of TCR BV5, BV8, BV12, BV13S3 and BV19 genes in BALF. Bellocq and coworkers [16] found a number of TCR BV19-positive T cells in BALF of patients with sarcoidosis. In asthmatic patients, Hodges et al.[17] reported expansion of TCR BV5S2/3-positive T cells in BALF. In contrast, there was no dominant usage of TCR BV genes in BALF T cells in patients with non-atopic asthma [18]. These findings support the hypothesis that BALF T cells of patients with sarcoidosis and atopic bronchial asthma may be induced by antigen on antigen-presenting cells. To date, there are no reports on the TCR gene of BALF T cells, or possible triggering factors in patients with IPF.
The present study was conducted in order to investigate the pathogenesis of IPF. Our experiments included an analysis of the TCR BV repertoire and clonality of T cells infiltrating the lung. Our results showed that T cells in BALF of patients with IPF expand oligoclonally, suggesting antigen-driven stimulation. Furthermore, highly conserved amino acid sequence motifs were identified in the TCR BV CDR3 region of accumulated BALF T cells. These findings indicate that BALF T cells in patients with IPF recognize a limited epitope on antigens. Based on these findings, we discuss possible pathogenic mechanisms of IPF.
MATERIALS AND METHODS
Patients and histological examination
Three patients with IPF were referred to Tokyo Medical University Hospital. Each individual met the criteria for IPF diagnosis, including clinical features, laboratory findings on chest X-ray and chest CT and endoscopic biopsies. We also recruited three healthy subjects who had no respiratory-related complaints and a negative chest X-ray. The clinical characteristics of the three patients with IPF and the healthy subjects are summarized in Table 1. Written informed consent was obtained from all patients. A transbronchial lung biopsy was performed, and the tissue was stained with haematoxylin and eosin. No open lung biopsy was performed. Typing of HLA-DR alleles was performed using PCR combined with dot-blot hybridization with sequence-specific oligonucleotide probes (PCR-SSOP) following the protocol of the Eleventh Histocompatibility Workshop [10].
Table 1.
Characteristics of study population
| IPF-I | IPF-2 | IPF-3 | HS-1 | HS-2 | HS-3 | |
|---|---|---|---|---|---|---|
| Age (years) | 70 | 65 | 80 | 52 | 32 | 28 |
| Sex | M | F | M | M | M | M |
| Smoking | (+) | (−) | (+) | (+) | (+) | (−) |
| %VC (%predicted value) | 97·7 | 80·5 | 106·9 | ND | 118·0 | 133·4 |
| FEV1% | 79·5 | 88 | 77·3 | ND | 80·5 | 97·4 |
| %DLco | 48·2 | 57·9 | 74·6 | ND | 91·7 | 90·9 |
| PaO2 (mmHg) | 89·7 | 85·9 | 81·3 | ND | ND | ND |
| Chest radiographdiffuse reticular infiltrate pattern | (+) | (+) | (+) | (−) | (−) | (−) |
| BAL analyses | ||||||
| Total cell count (×105/ml) | 5 | 1·2 | 7 | 1 | 2·6 | 0·8 |
| Differential count (%) | ||||||
| Macrophages | 87·2 | 81·7 | 92·1 | 90·5 | 93·0 | 90·0 |
| Lymphocytes | 3·7 | 15·2 | 4·5 | 3·2 | 3·5 | 8·0 |
| Neutrophils | 6·8 | 2·9 | 1·8 | 4·9 | 3·0 | 2·0 |
| Eosinophils | 2·3 | 0·2 | 1·6 | 1·4 | 0·5 | 0 |
| CD4/CD8 ratio | 1·9 | 1·4 | 2·3 | 2·2 | 0·6 | 4·7 |
| Absolute number of lymphocytes* | 1·9 | 1·8 | 3·2 | 0·3 | 0·9 | 0·6 |
Diagnosis of IPF was based on clinical criteria for IPF defined by The Ministry of Health and Welfare research group in Japan.
× 104/ml.
Bronchoalveolar lavage and peripheral blood lymphocytes
BAL was performed on the involved lung segment (right lower lobe and posterior segment) of the three patients with IPF and three healthy control subjects. After topical anaesthesia, the fiberoptic bronchoscope (Olympus type BF20, Olympus Co., Tokyo, Japan) was advanced into the described segment and wedged, and 50 ml sterile 0·9% saline at 37°C was injected through the bronchoscope. The latter process was performed three times. The volume of BALF recovered from the involved segment was approximately 100 ml. Cells in BALF were passed through sterile gauze to remove debris. BALF from patients was centrifuged at 20 g and 4°C for 10 min, and washed twice with PBS. Following cell count, part of the cell mass was subjected to flow cytometric analysis. A number of cells (2 × 105) were stained with MoAb against Leu 4 (anti-CD3), Leu 3a (anti-CD4) and Leu 2a (anti-CD8) (Becton Dickinson, Mountain View, CA, USA). After flow cytometry, PBLs from patients with IPF were obtained by Ficoll-Hypaque density gradient centrifugation, and analysed immediately.
PCR, Southern blot analysis and SSCP
Total RNA from BALF cells was prepared with Isogen (Nippon Gene Co., Tokyo, Japan). PCR and cDNA synthesis were performed as described previously by Sumida et al.[11]. Briefly, first-strand cDNA was synthesized from 1 μg total RNA in a 20-μl reaction mixture containing an oligo(dT) primer by avian myeloblastosis virus reverse transcriptase. Amplification was performed with Taq polymerase in 50 μl standard buffer, using 0·2 μl cDNA (corresponding to 10 ng total RNA), with primers specific for 20 different TCR BV genes and BC gene. Sequences of the primers were obtained from previously published data [10]. Denaturing was performed at 95°C for 1·5 min, annealing at 60°C for 1·0 min and extension at 72°C for 1·0 min for 30 cycles in a DNA Thermal Cycler (Perkin-Elmer Corporation, Norwork, CT, USA). One-tenth of each amplified PCR product was subjected to 2% agarose gel electrophoresis and transferred to a nylon membrane. Membranes were hybridized further with digoxygenin-labelled TCR BC probe, and visualized using the DIG luminescent detection kit (Boehringer Mannheim, Mannheim, Germany). The digoxygenin-labelled TCR BC probe was synthesized employing the PCR DIG probe synthesis kit (Boehringer Mannheim), with 5′-TCR BC (5′-GAGGATCTGA GAAATGTGACT-3′) and 3′-TCR BC (5′-CAAGCACACAC GAGGGTAGCCT-3′) primers. For individual single-strand conformation polymorphism (SSCP) assays, amplified DNA was diluted (1:20) in a denaturing solution (95% formamide, 10mm EDTA, 0·1% bromophenol blue, 0·1% xylene cyanol) at 90°C for 2 min. Diluted samples (2:1) were subjected to electrophoresis in non-denaturing 5% polyacrylamide gels containing 10% glycerol [19]. Gels were run at 35W constant power for 2 h. Following electrophoresis, DNA was transferred to Immobilon-S (Millipore Intertech, Bedford, MA, USA) and hybridized with biotinylated TCR BC probe (5′-A (AC) AA (GC) GTGTTCCCACCCGAG GTCGCTGTGTT-3′), streptavidin, biotinylated alkaline phosphatase and a chemiluminescent substrate system (Plex™ Luminescence kit, Millipore).
Sequencing of cDNA encoding TCR BV genes
Complementary DNA, encoding TCR BV genes from BALF and PBLs, was purified from polyacrylamide gels for subjection to SSCP and amplified by PCR using the primers described above. PCR products were ligated to plasmids using the TA cloning kit (Invitrogen, San Diego, CA, USA), transformed into competent INVαF′Escherichia coli cells and grown under appropriate conditions. After selection of TCR BC-positive colonies, plasmid DNA was purified by alkaline lysis for DNA sequencing. Sequencing reactions were performed using an automated DNA sequencer (model 377 A, Applied Biosystems, Foster City, CA, USA).
RESULTS
Heterogeneous TCR BV repertoire of BALF T cells in patients with IPF
To analyse the pathogenesis of IPF, we examined the TCR repertoire of BALF T cells from three patients with IPF (IPF-1, -2, and -3) using the family PCR method. PBLs from identical patients were used as a control. Clinical profiles of IPF patients are summarized in Table 1. Chest CT revealed diffuse reticulonodular opacities, honeycombing and ground-glass attenuation. Histological examination of lung biopsies from patients with IPF showed a large number of mononuclear cells in the inflamed alveolar septa. The pathological changes in the interstitial septa, alveolar spaces, bronchial mucosa and pleura were similar to those in lungs with UIP. Infectious agents or parasites were not observed, and there was no evidence of vasculitis in biopsy specimens. TCR analysis showed expression of the majority of TCR BV family genes in both BALF T cells and PBL in all patients (Fig. 1). These results suggest that the TCR BV repertoire of T cells in the lung is heterogeneous and that there is no restricted predominant usage of TCR BV genes. HLA typing of two patients with IPF showed that one was the DR B1*0405, 0405 allele and the other was the DR B1*0901, 1502 allele.
Fig. 1.
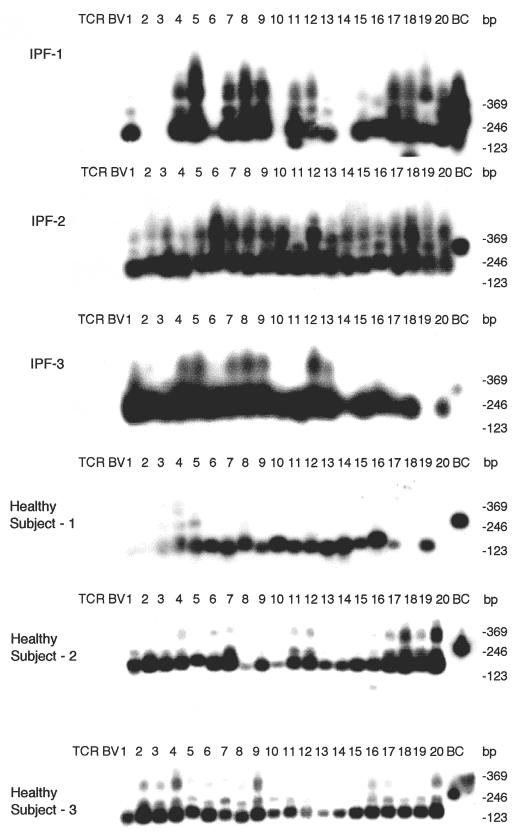
TCR BV gene repertoire of BALF T cells from three patients with IPF and three healthy subjects. PCR, Southern blot analysis and SSCP methods are described in Patients and methods. The numbers indicate the family of TCR BV genes.
Lung-specific T cell clones in patients with IPF
TCR BV genes in BALF and peripheral T cells were examined by PCR-SSCP in order to investigate the clonality of pulmonary T cells in patients with IPF. Lung-specific bands that were found in several TCR BV genes of the three IPF patients. These bands were detected mainly in TCR BV 3, 11 and 15 genes. The number of bands encoding TCR BV genes in the lung is summarized in Table 2. We observed a significant increase in the number of expanded clones in BALF from IPF-1 and -3 patients (26 and 24 clones), compared with the healthy subjects (six clones) (P < 0·05). In contrast, there was no difference in the number of T cell clones in the lung between IPF-2 patients and healthy controls. These results suggest that some T cells accumulate in the lungs of patients with IPF, suggesting that these cells proliferate by antigen stimulation.
Table 2.
Accumulated T cell clones in BALF of patients with IPF
| TCR BV gene | 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 | 9 | 10 | 11 | 12 | 13 | 14 | 15 | 16 | 17 | 18 | 19 | 20 | Total |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| IPF -1 | 0 | 0 | 2 | 0 | 0 | 3 | 0 | 0 | 3 | 1 | 7 | 0 | 0 | 0 | 2 | 0 | 2 | 2 | 4 | 0 | 26 |
| IPF -2 | 1 | 1 | 1 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 1 | 0 | 0 | 0 | 1 | 1 | 0 | 0 | 0 | 1 | 8 |
| IPF -3 | 1 | 1 | 3 | 0 | 1 | 1 | 1 | 0 | 0 | 2 | 1 | 0 | 3 | 3 | 3 | 1 | 0 | 2 | 0 | 1 | 24 |
| Healthy subject 1 | 1 | 2 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 2 | 6 |
| Healthy subject 2 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 1 | 0 | 0 | 1 | 0 | 0 | 1 | 1 | 0 | 0 | 1 | 0 | 0 | 6 |
| Healthy subject 3 | 0 | 0 | 1 | 0 | 0 | 0 | 1 | 0 | 0 | 1 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 1 | 0 | 1 | 6 |
Numbers represent T cell clones accumulated in BAL from patients with IPF. The distinct bands encoding TCR BV genes on SSCP were described as the number of BAL-specific T cell clones.
Conserved amino acid sequence motifs in the CDR3 region of TCR BV genes from BALF-specific T cells of IPF patients
To examine the amino acid sequences of the CDR3 region in the TCR BV gene, we focused on the lung-specific T cell clones on SSCP analysis. DNAs encoding the TCR BV genes from BALF-specific bands were eluted from gels, following which the corresponding CDR3 regions were sequenced. As shown in Table 3, the CDR3 region of the lung-specific accumulated T cell clones contained conserved amino acid motifs. A LAxG motif was found in BV3-2 and BV6-1 clones from IPF-1 and BV8-2 and BV16-1 clones from the IPF-2 patient. Furthermore, in the IPF-2 patient, QGQ, GxQP, GRxG and EIGRSG motifs were found in BV6-1 and BV9-1, BV11-1 and BV11-2, BV15-2, BV 20-1 and BV20-2, and BV20-1 and BV20-2, respectively. For the IPF-3 patient, VAR, PGT, TGR, SGQ, LxLxQ, GGT, PGT, GTI sequences were observed in BV1-1 and BV1-2, BV2-1 and BV15-1, BV2-2 and BV18-2, BV3-1 and BV3-2, BV3-1 and BV11-1, BV10-2 and BV20-1, BV10-2 and BV15-1, and BV14-2 and BV16-1, respectively. In contrast, BALF-specific bands of healthy subjects revealed only two motifs, VTGG and GGE. A VTGG motif was found in BV1-2 clone from healthy subject 1 and BV11-2 clone from healthy subject 2 and a GGE motif was found in BV2-1 clone from healthy subject 1 and BV18-2 clone from healthy subject 2 (Table 3). These amino acid motifs were not detected in patients with IPF. The conserved amino acids motifs in TCR BV CDR3 region of BAL T cells in patients with IPF was summarized in Table 4. These findings suggest that accumulated T cells in the lungs of patients with IPF recognize a limited epitope on antigens.
Table 3.
TCR BV CDR3 region of BAL T cells in patients with IPF
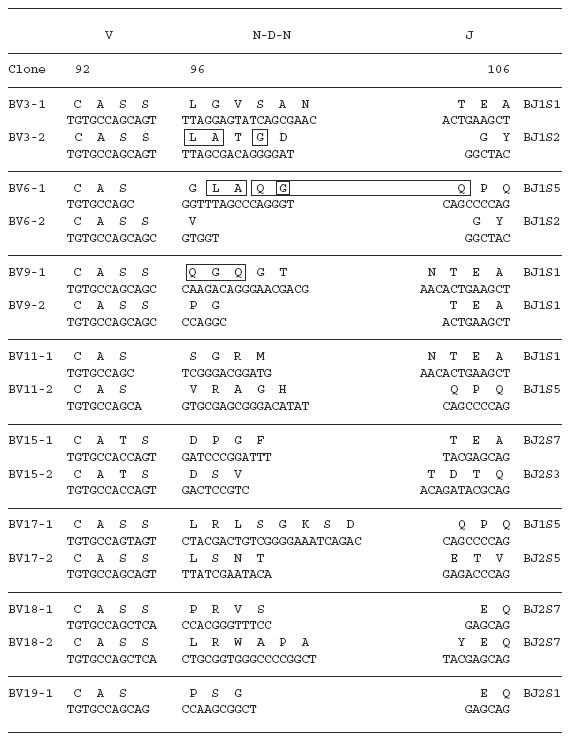
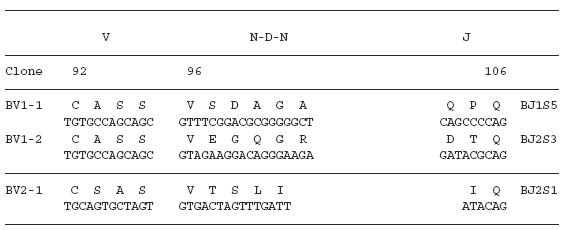
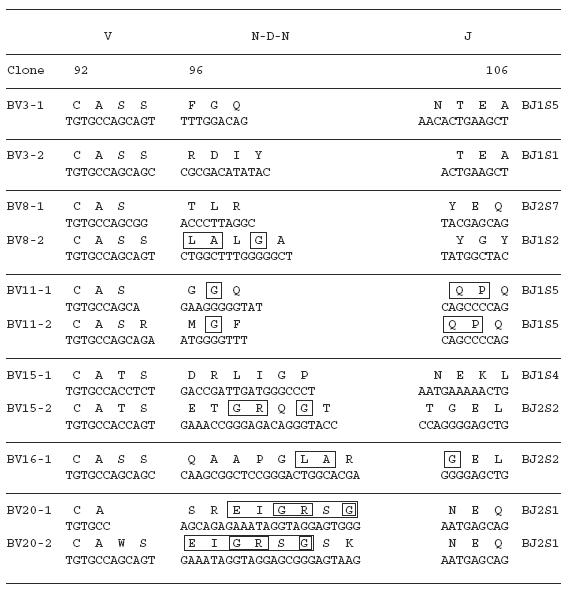
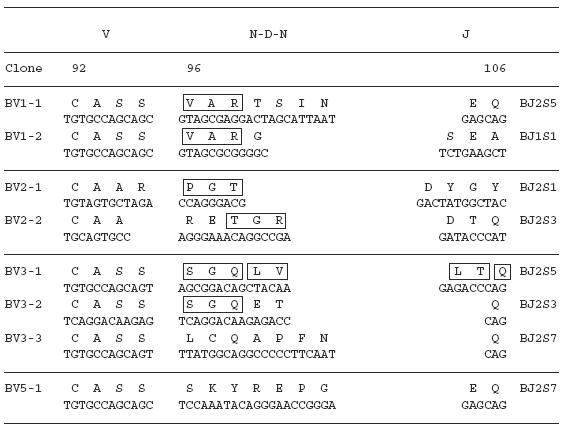
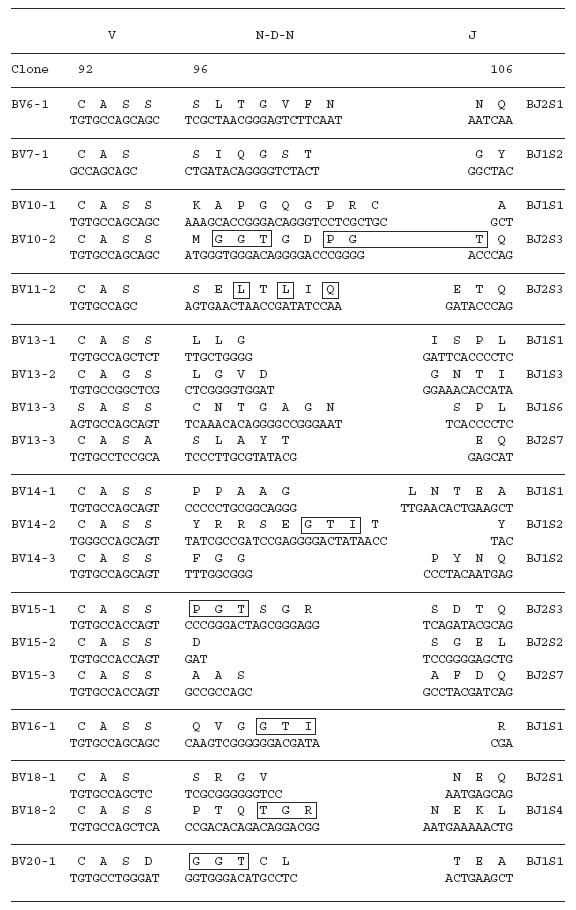
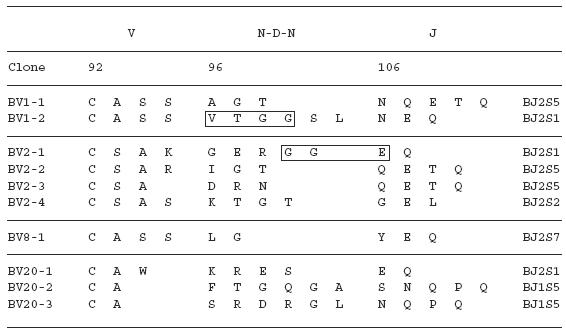
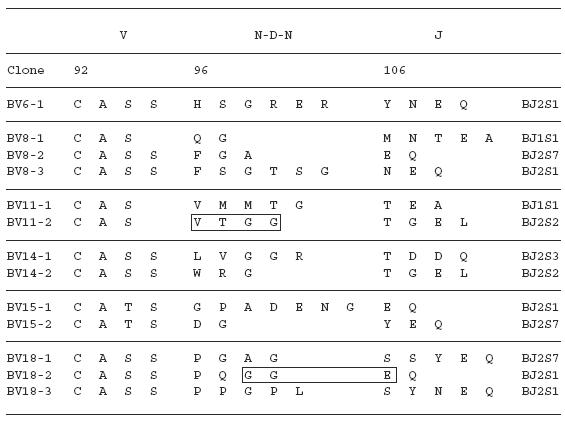
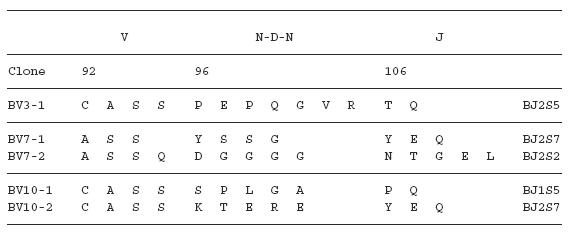
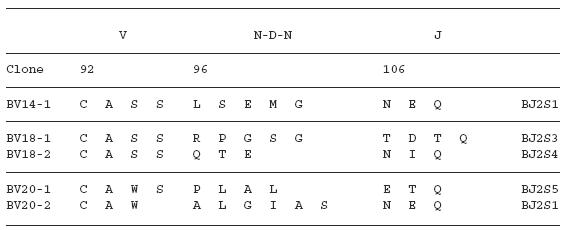
Table 4.
Conserved amino acid motifs in TCR BV CDR3 region of BAL T cells in patients with IPF
| (1) IPF | |
| LAxG | (IPF-1; BV3-2, BV6-1, IPF-2; BV8-2, BV16-1) |
| QGQ | (IPF-1; BV6-1, BV9-1) |
| GxQP | (IPF-2; BV11-1, BV11–2) |
| GRxG | (IPF-2; BV15-2, BV 20–1, BV20-2) |
| EIGRSG | (IPF-2; BV20-1, BV20-2) |
| VAR | (IPF-3, BV1-1, BV1-2) |
| PGT | (IPF-3, BV2-1, BV15-1) |
| TGR | (IPF-3, BV2-2, BV18-2) |
| SGQ | (IPF-3, BV3-1, BV3-2) |
| LxLxQ | (IPF-3, BV3-1, BV11-1) |
| GGT | (IPF-3, BV10-2, BV20-1) |
| PGT | (IPF-3, BV10-2, BV15-1) |
| GTI | (IPF-3, BV14-2, BV16-1) |
| (2) Healthy subject | |
| VTGG | (healthy subject 1; BV1-2, healthy subject 2; BV11-2) |
| GGE | (healthy subject 1; BV2-1, healthy subject 2; BV18-2) |
DISCUSSION
IPF is an inflammatory lung disease characterized by the accumulation of lymphocytes and other inflammatory cells, resulting in lung remodeling. T cells comprise the major population of lymphocytes in pulmonary tissue and BALF [15,16,21]. In the present study of TCR genes, we provide evidence that some T cells accumulate in BALF, suggesting that cells in pulmonary lesions might expand by antigen stimulation in the context of HLA, rather than stimulation by superantigen, as suggested previously [20,21]. This conclusion is in agreement with the results of Lympany et al.[22], where the TCR repertoire of T cells in lung biopsy samples and BALF was analysed using RT-PCR, and individual TCR AV and BV expression bias was observed in subjects with fibrosing alveolitis. The findings are also similar to those observed in T cells from the lungs of patients with sarcoidosis [14–16] and atopic bronchial asthma [17]. Considered together, these findings suggest that the pathogenesis of IPF occurs in two steps. The first step includes the presentation of antigens on the HLA molecule. This is followed by accumulation of reactive T cells in the pulmonary lesion, which in turn induces inflammation, resulting in pulmonary fibrosis.
Using flow cytometry, Gruber et al.[23] showed a high proportion of CD8+ T cells, low proportion of CD4+ T cells and no change in γδ T cells in IPF, compared with sarcoidosis. However, we observed a greater increase in the number of CD4+ T cells compared to CD8+ T cells, while γδ T cells (data not shown) were not detected in BALF of all three patients (data not shown). The discrepancy in these results may be due either to individual observations during different stages of IPF or the presence of distinct pathogenic antigens.
Several conserved amino acid motifs were found in the CDR3 region of the TCR BV gene in clonally expanded BALF T cells of patients with IPF, suggesting that these T cells recognize highly limited epitopes on the antigen. The phenomenon appears to be disease-specific, because there were no corresponding conserved amino acids in BALF T cells of the control subjects. The restricted T cell epitope on the antigen was not dependent on HLA, because HLA-DR of the two IPF patients varies widely among the two patients. This finding suggests the presence of dominant and common T cell epitopes on the antigen. Although this study is dependent on small numbers of IPF patients, a further large-scale study would be necessary to confirm our conclusion.
The identity of antigens recognized by accumulated T cells in BALF of patients with IPF remains to be established. While previous studies have recognized candidate antigens in IPF, no direct evidence has been presented to date [24,25]. Recently, viral proteins such as cytomegalovirus and Epstein–Barr virus (EBV) proteins were considered [26–30]. Antibodies against cytomegalovirus and EBV were detected in patients with pulmonary fibrosis [27,28] and their viral products were expressed in immunostained pulmonary tissues [26,30]. Moreover, DNA and mRNA encoding cytomegalovirus and EBV were detected in patients with pulmonary fibrosis by RT-PCR and ISH [29,30]. These observations suggest that particular viral products are antigens recognized by T cells in pulmonary lesions, and may therefore play a crucial role in the pathogenesis of pulmonary fibrosis.
Therefore, determination of the amino acid sequences of T cell epitopes of the antigen is essential before any vaccine using an analogue peptide of the antigen is used for antigen-specific regulation of IPF.
Acknowledgments
We thank Miss Miyuki Nishihara for technical assistance, and Dr F. G. Issa (www.word-medex.com.au) for critical reading of the manuscript.
REFERENCES
- 1.Crystal RG, Bitterman PB, Rennard SI, Hance AJ, Keogh BA. Interstitial lung diseases of unknown cause. N Engl J Med. 1984;310:235–44. doi: 10.1056/NEJM198401263100406. [DOI] [PubMed] [Google Scholar]
- 2.Crystal RG, Gedek JE, Ferrans VJ, Fulmer JD, Line BR, Hunninghaku GW. Interstitial lung disease. current concepts of pathogenesis, staging, and therapy. Am J Med. 1981;70:542–68. doi: 10.1016/0002-9343(81)90577-5. [DOI] [PubMed] [Google Scholar]
- 3.Rudd RM, Haslam PL, Turner-Warwick M. Cryptogenic fibrosing alveolitis. Relationship of pulmonary physiology and bronchoalveolar lavage to response to treatment and prognosis. Am Rev Respir Dis. 1981;124:1–8. doi: 10.1164/arrd.1981.124.1.1. [DOI] [PubMed] [Google Scholar]
- 4.Haslam PL, Turton CW, Lukoszik A, et al. Bronchoalveolar lavage fluid cell counts in cryptogenic fibrosing alveolitis and relation to therapy. Thorax. 1980;35:328–39. doi: 10.1136/thx.35.5.328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Haslam PL, Turton CW, Heard B, et al. Bronchoalveolar lavage in pulmonary fibrosis: comparison of cells obtained with lung biopsy and clinical features. Thorax. 1980;35:9–18. doi: 10.1136/thx.35.1.9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Katzenstein ALA, Myers JL. Idiopathic pulmonary fibrosis. Clinical relevance and pathologic classification. Am J Respir Crit Care Med. 1998;157:130–15. doi: 10.1164/ajrccm.157.4.9707039. [DOI] [PubMed] [Google Scholar]
- 7.Bjoraker JA, Ryu JH, Edwin MK, et al. Prognostic significance of histopathological subsets in idiopathic pulmonary fibrosis. Am J Respir Crit Care Med. 1998;157:199–203. doi: 10.1164/ajrccm.157.1.9704130. [DOI] [PubMed] [Google Scholar]
- 8.Ryu JH, Colby TV, Hartmann TE. Idiopathic pulmonary fibrosis: current concepts. Mayo Clin Proc. 1998;11:1085–101. doi: 10.4065/73.11.1085. [DOI] [PubMed] [Google Scholar]
- 9.Paliard X, West SG, Lafferty JA, et al. Evidence for the effects of a super antigen in rheumatoid arthritis. Science. 1991;253:325–8. doi: 10.1126/science.1857971. [DOI] [PubMed] [Google Scholar]
- 10.Sumida T, Sakamaki T, Yonaha F, et al. HLA-DR alleles in patients with Sjögren's syndrome over-representing Sjögren's syndrome Vb2 and Vb13 genes in the labial salivary glands. Br J Rheumatol. 1994;33:42014. doi: 10.1093/rheumatology/33.5.420. [DOI] [PubMed] [Google Scholar]
- 11.Sumida T, Yonaha F, Maeda T, et al. T cell receptor repertoire of infiltrating T cells in lips of Sjögren's syndrome patients. J Clin Invest. 1992;89:681–5. doi: 10.1172/JCI115635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kotzin BL, Karuturi S, Chou YK, et al. Preferential T-cell receptor beta-chain variable gene use in myelin basic protein-reactive T-cell clones from patients with multiple sclerosis. Proc Natl Acad Sci USA. 1991;88:9161–5. doi: 10.1073/pnas.88.20.9161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wucherpfening KW, Ota K, Endo N, et al. Shared human T cell receptor V beta usage to immunodominant regions of myelin basic protein. Science. 1990;248:1016–9. doi: 10.1126/science.1693015. [DOI] [PubMed] [Google Scholar]
- 14.Moller DR, Konishi K, Kirby M, Balbi B, Crystal RG. Bias toward use of specific T cell receptor beta-chain variable region in a subgroup of individuals with sarcoidosis. J Clin Invest. 1988;82:1183–91. doi: 10.1172/JCI113715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zissel G, Baumer I, Fleischer B, Schlaak M, Muller-Quernheim J. TCR V beta families in T cell clones from sarcoid lung parenchyma, BAL, and blood. Am J Respir Crit Care Med. 1997;156:1593–600. doi: 10.1164/ajrccm.156.5.97-01037. [DOI] [PubMed] [Google Scholar]
- 16.Bellocq A, Lecossier D, Pierre-Audigier C, Tazi A, Valeyre D, Hance AJ. T cell receptor repertoire of T lymphocytes recovered from the lung and blood of patients with sarcoidosis. Am J Respir Crit Care Med. 1994;149:646–54. doi: 10.1164/ajrccm.149.3.7906994. [DOI] [PubMed] [Google Scholar]
- 17.Hodges E, Dasmahapatra J, Smith JL, et al. T cell receptor Vβ gene usage in bronchoalveolar lavage and peripheral blood T cells from asthmatic and normal subjects. Clinical & Experimental Immunology. 1998;112:363–74. doi: 10.1046/j.1365-2249.1998.00611.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Umibe T, Kita Y, Nakao A, et al. Clonal expansion of T cells infiltrating in the airways of non-atopic asthmatics. Clinical & Experimental Immunology. 2000;119:390–7. doi: 10.1046/j.1365-2249.2000.01148.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Yamamoto K, Sakoda H, Nakajima T, et al. Accumulation of multiple T cell clonotypes in the synovial lesions of patients with rheumatoid arthritis revealed by a novel clonality analysis. Int Immunol. 1992;4:1219–23. doi: 10.1093/intimm/4.11.1219. [DOI] [PubMed] [Google Scholar]
- 20.Jones CM, Lake RA, Wijeyekoon JB, Mitchell DM, du Bois RM, O'Hehir RE. Oligoclonal V gene usage by T lymphocytes in bronchoalveolar lavage fluid from sarcoidosis patients. Am J Cell Mol Biol. 1996;14:470–7. doi: 10.1165/ajrcmb.14.5.8624252. [DOI] [PubMed] [Google Scholar]
- 21.Vissinga C, Springmeyer SC, Concannon P. TCR expression and clonality analysis in pulmonary sarcoidosis. Hum Immunol. 1996;48:98–106. doi: 10.1016/0198-8859(96)00078-x. [DOI] [PubMed] [Google Scholar]
- 22.Lympany PA, Southcott AM, Welsh KI, Boylston AW, du Bois RM. T-cell receptor gene usage in patients with fibrosing alveolitis and control subjects. Eur J Clin Invest. 1999;29:173–81. doi: 10.1046/j.1365-2362.1999.00434.x. [DOI] [PubMed] [Google Scholar]
- 23.Gruber R, Pforte A, Beer B, Riethmuller G. Determination of gamma/delta and other T-lymphocyte subsets in bronchoalveolar lavage fluid and peripheral blood from patients with sarcoidosis and idiopathic fibrosis of the lung. APMIS. 1995;104:199–203. doi: 10.1111/j.1699-0463.1996.tb00708.x. [DOI] [PubMed] [Google Scholar]
- 24.Ueda T, Ohta K, Suzuki N, et al. Idiopathic pulmonary fibrosis and high prevalence of serum antibodies to hepatitis C virus. Am Rev Respir Dis. 1992;146:266–8. doi: 10.1164/ajrccm/146.1.266. [DOI] [PubMed] [Google Scholar]
- 25.Kuwano K, Nomoto Y, Kunitake R, et al. Detection of adenovirus EIA DNA in pulmonary fibrosis using nested polymerase chain reaction. Eur Respir J. 1997;10:1445–9. doi: 10.1183/09031936.97.10071445. [DOI] [PubMed] [Google Scholar]
- 26.Egan JJ, Stewart JP, Hasleton PS, Arrand JR, Carrol KB, Woodcock AA. Epstein–Barr virus replication within pulmonary epithelial cells, in cryptogenic fibrosing alveolitis. Thorax. 1995;50:1234–9. doi: 10.1136/thx.50.12.1234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Yonemaru M, Kasuga H, Kusumoto H, et al. Elevation of antibodies to cytomegalovirus and other herpes viruses in pulmonary fibrosis. Eur Respir J. 1997;10:2040–5. doi: 10.1183/09031936.97.10092040. [DOI] [PubMed] [Google Scholar]
- 28.Vergnon JM, Vincent M, de The G, Mornex JF, Weynants P, Brune J. Cryptogenic fibrosing alveolitis and Epstein–Barr virus: an association. Lancet. 1984;ii:768–71. doi: 10.1016/s0140-6736(84)90702-5. [DOI] [PubMed] [Google Scholar]
- 29.Wangoo A, Shaw RJ, Diss TC, Farrell PJ, du Bois RM, Nicholson AG. Cryptogenic fibrosing alveolitis. Lack of association with Epstein– Barr virus infection. Thorax. 1997;52:888–91. doi: 10.1136/thx.52.10.888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Jiwa M, Steenbergen RD, Zwaan FE, Kluin PM, Raap AK, Ploeg M. Three sensitive methods for the detection of cytomegalovirus in lung tissue of patients with interstitial pneumonia. Am J Clin Pathol. 1990;93:491–4. doi: 10.1093/ajcp/93.4.491. [DOI] [PubMed] [Google Scholar]


