Abstract
A recombinant Fab antibody, designated 1E8–4b, which reacts with the Alzheimer's disease (AD)-related Aβ peptides, Aβ[1–40], Aβ[1–42] and Aβ[1–43] has been developed. The 1E8–4b Fab was constructed by cloning the VHCH1 and VLCL domains from the parent hybridoma 1E8 antibody, reported previously to recognize these Aβ peptides. Briefly, a C-terminal Flag tag sequence was incorporated into this construct, which was ligated into the vector pHFA2 and expressed in Escherichia coli. Following purification on an M2 anti-Flag affinity column, the 1E8–4b recombinant Fab antibody was shown to bind plaques within sections of brain tissue from CERAD-defined AD patients by immunohistochemistry. ELISA, epitope mapping and immunoblotting confirmed the recognition of the Aβ1–40/42/43] peptides by the 1E8–4b Fab. The 1E8–4b Fab did not recognize APP695 or APP770 which contain the Aβ sequence. The Aβ specificity of the recombinant 1E8–4b Fab antibody was identical to the parent 1E8 monoclonal antibody.
Keywords: recombinant Fab antibody, monoclonal antibody, Alzheimer's disease, Aβ peptide
INTRODUCTION
Alzheimer's disease (AD), a central nervous system (CNS) neurodegenerative disorder, is the predominant cause of dementia in elderly people and is seen in Down's syndrome individuals [1,2]. Early-onset familial AD (FAD) patients carrying mutations in APP, PS-1 or PS-2 encoding genes show increased β-amyloid (Aβ)1–42/43] and x-42] peptide levels by altered amyloid precursor protein (APP) processing, suggesting a major role for Aβ in AD pathogenesis [3]. Aggregated and polymerized Aβ deposits extracellularly in AD brain as highly insoluble neuritic plaques of β-pleated amyloid fibrils (predominantly Aβ1–42/43] with less Aβ[1–40]) and in cerebrovasculature as Aβ[1–40] with less Aβ[1–42], [45]. Toxicity of insoluble fibril Aβ aggregates which form at high concentrations [6] and of soluble oligomers at nanomolar concentrations [7] has been proposed. Peptides Aβ1–42/43] display higher fibrillogenicity compared to Aβ[1–40] and exhibit earlier and selective deposition in AD, which is implicated in neurotoxicity and Aβ deposition in FAD plaques [8,9].
Decreased total Aβ or Aβ[1–42] levels in AD CSF relative to control patients, correlating with increased dementia severity, have been reported [10] but measuring Aβ levels in biological fluids to diagnose AD is controversial. Diagnostic sensitivity and specificity remain dependent on social, genetic, medical, psychiatric and neurological examinations with confirmation by post mortem immunohistochemistry [11,12].
Using highly specific antibodies reactive with AD biological markers present in accessible bodily fluids, immunodiagnostics offer the opportunity for specific and sensitive detection of AD during life. Recombinant techniques provide tools for the generation of antibodies of enhanced specificity and sensitivity for AD markers, such as Aβ in CSF/plasma, by incorporating tags, affinity enhancement, enzyme linkage, bispecificity and increased avidity [13]. Although Aβ[1–42]:Aβ[1–40] plasma levels have been reported to be increased in Down syndrome and FAD cases [14,15], increased assay sensitivity may minimize the existing controversy in sporadic AD [16]. Recombinant antibodies also have importance in AD therapy. Immunization of mice with human Aβ has been shown recently to inhibit amyloid plaque formation [17]. ‘Specific anti-Aβ antibody-directed mopping-up’ of excess Aβ may be therapeutic in AD. With these considerations in mind, the novel Aβ-specific recombinant 1E8–4b Fab fragment was generated from Escherichia coli and compared with the parent antibody.
MATERIALS AND METHODS
Generation of the immunoglobulin heavy and light chain constructs
The mRNA was extracted from 4·0 × 106 hybridoma cells by Dynabeads mRNA Direct Kit (Dynal, Oslo, Norway). The hybrid-oma used was 1E8 [17–22] (SmithKlein Beecham Laboratories, Harlow, UK) [18]. The cDNA was generated by the Promega (Madison, WI, USA) reverse transcription system kit in a 20-μl reaction containing 1 × RT buffer, 5 mm MgCl2, 5 mm DTT, 1 U/μl RNase inhibitor, 1 mm dNTP mix, 0·5 μg oligo (dT)15 primer/μg mRNA, 15 U AMV reverse transcriptase/μg mRNA, 1 μg mRNA and RNase-free ddH2O. The reaction was incubated at 42°C for 1h and inhibited at 99°C for 5 min. PCR was performed in a 100-μl reaction containing 0·50 pm each forward and reverse primer (section 1·2), 0·20 mm dNTP (Amresco), 1 × ThermoPol reaction buffer (New England BioLabs, Beverly, MA, USA), 100 μg/ml BSA, 10 μl of cDNA and ddH2O. Samples were ‘hot started’ by heating to 94°C for 1 min and adding 2 Units of Vent DNA polymerase (New England BioLabs, Beverly, MA, USA) per 100 μl reaction. Reactions were overlaid with 50 μl of mineral oil (Sigma, St Louis, MO, USA) and incubated at 94°C 1 min, 55°C 1 min and 72°C 1 min for three cycles, followed by 94°C 1 min, 55°C 1 min and 72°C 2 min, for 30 cycles on a FTS-1 thermal sequencer (Corbett Research, Mortlake, NSW, Australia). DNA was purified by the BIO101 GeneClean (GC) Spin Kit (Integrated Sciences, NSW, Australia).
Primers used to generate heavy and light chain constructs
The VHCH1 construct was generated using the forward primer CH1 degen(212–223)] (5′ at taa gtc gac T/G/AAT T/CTT T/CTT GTC CAC C/TG/TC GGT G/CC/TT GCT GGC C/TGG GTG 3′) modified from Kettleborough et al.[19] with data from Kabat et al.[20], containing a SalI site and the reverse primer VH back 3350] containing a SfiI site (5′ tta tta ctc gcg gcc cag ccg gcc atg gcc GAG GTC CAG CTG CAG CAG TC 3′). The VLCL construct was generated using CL forward 3964] containing a NotI site (5′ at gag ttt ttg ttc tgc ggc cgc ggc ACA CTC ATT CCT GTT GAA GCT CTT 3′) and the reverse primer VL back 3959] containing a NcoI site (5′ c cgg gtg tct gcc atg gcc GAC ATT GTG ATG ACC CAG TCT 3′). All restriction sites are underlined.
Cloning the immunoglobulin heavy and light chain constructs
Purified PCR fragments were cloned into pCR-Script Amp SK(+) vector and transformed into E. coli XL1-Blue MRF′ Kan cells using pCR-Script Amp SK(+) an electroporation-competent cell cloning kit (Stratagene, CA, USA). Positive colonies were screened by restriction enzyme digests of Miniprep DNA or colony PCR. Following sequencing using the ABI PRISM dye terminator cycle sequencing ready reaction kit (Perkin Elmer, Norwalk, CT, USA), positive clones were digested with appropriate enzymes, purified from 1% (w/v) low melt agarose gels and ligated into a similarly digested and purified pHFA2 expression vector [21]. The ligation mixture was transformed into E. coli HB2151 cells and plated onto 1·5% (w/v) agar plates containing 2 × YT, 100 μg/ml ampicillin and 1% (v/v) glucose, overnight at 37°C. Positive colonies were screened as above and resequenced for affirmation using pUC/M13 22-mer reverse sequencing primer (Promega, Madison, WI, USA) (5′TCACACAGGAAACAGC TATGAC 3′) and Fd gene III primer (Beckman, NSW, Australia) (5′ACTTTCAACAGTCTATGCCGCG 3′).
Expression and purification of the antibody Fab fragments
E. coli HB2151 cells [22] containing the recombinant plasmids were grown overnight at 37°C in 2 × YT containing 100 μg/ml Ampicillin and 1% (v/v) glucose (2 ×YT/AMP/Glu). The following day, cells were subcultured to an O.D. 600nm value of 0·1 Unit into 2 × YT/AMP. The cells were grown at 30°C, 125r.p.m. in an orbital incubator to an O.D. 600nm value of 0·8–1·0Units (about 3h). IPTG was added to a final concentration of 1 mm and the cultures allowed to shake at 125 r.p.m., 25°C overnight.
Overnight culture medium was clarified by two successive centrifugations at 10 000 gmax for 30 min, 4°C. The supernatant was treated with 2 mm EDTA, 0·1 μg/ml leupeptin, 0·1 μg/ml pepstatin, 1 mm PMSF and 0·02% (w/v) NaN3. The supernatant was treated with 100 μg DNAaseI/l supernatant (SIGMA, St Louis, MO, USA) for 30 min at RT with stirring and then filtered through a 0·45-μm filter (Nalgene). The supernatant was concentrated by a 60% (v/v) saturated ammonium sulphate (SAS) precipitation as described in Coppola et al.[23]. The pellet was resuspended in 1/40th of the original volume in TBS/NaN3 and dialysed against 100 volumes of TBS/NaN3 over 48h with three changes of buffer at 4°C. Dialysed samples were centrifuged at 10 000gmax for 30 min at 4°C and the supernatant collected. The M2 anti-Flag antibody affinity column (Sigma, St Louis, MO, USA) was equilibrated with four column volumes of TBS/NaN3/I (TBS/NaN3 containing 2 mm EDTA, 0·1 μg/ml leupeptin, 0·1 μg/ml Pepstatin, 2·5 μg/ml Aprotinin) at 1 ml/min at 4°C. The concentrated supernatant was treated with inhibitors (2 mm EDTA, 0·1 μg/ml leupeptin, 0·1 μg/ml Pepstatin, 5 μg/ml Aprotinin), ultracentrifuged at 100 000 gmax 4°C for 1 h, filtered through a 0·22-μm filter (Millipore, Bedford, MA, USA) and applied at 1 ml/min 4°C to the affinity column. The sample was recirculated over the column three times or overnight. The affinity column was then washed with four column volumes of TBS/NaN3/I at 1 ml/min 4°C. The bound antibody was eluted at 4°C with 0·1m glycine-HCl/0·02% (w/v) NaN3 pH 3·5 at 1 ml/min. One-millilitre fractions were collected and immediately neutralized with 20–40 μl of 1m Tris-HCl, pH 8·0. Fractions were examined by immunoblot for the presence of antibody and by ELISA for antibody reactivity. Eluates containing active antibody were pooled, concentrated in 2 ml Centricon concentrators (Millipore, Bedford, MA, USA) and buffer exchanged into PBS/NaN3.
SDS-PAGE and immunoblotting analysis
SDS-PAGE and immunoblotting were carried out under standard conditions [24]. Samples were boiled for 5 min in sample buffer and electrophoresed on 10% SDS-polyacrylamide gels. For reducing conditions, the sample buffer contained 20% (v/v) 2-mercaptoethanol (Sigma, St. Louis, MO, USA). The proteins were transferred onto nitrocellulose (Biorad, Hercules, CA, USA) in a Biorad Mini Trans-Blot apparatus at 300 mA for 1 h. The recombinant antibody, containing C-terminal flag and tubulin tags, was detected with mouse M2 anti-Flag (Sigma, St Louis, MO, USA) or rat antitubulin antibodies (Serotec, Oxford, UK), respectively, followed by goat anti-mouse immunoglobulin coupled to alkaline phosphatase (Promega, Madison, WI, USA) or goat anti-rat immunoglobulin coupled to alkaline phosphatase (Southern Biotechnology Associates), respectively, and developed with a substrate solution containing fast red and naphthol phosphate AS-MX reagents (Sigma, St Louis, MO, USA).
ELISA analysis of the 1E8–4b Fab fragment
In order to analyse the specificity of the 1E8–4b recombinant Fab, 96-well microtitre Cliniplates EB (Labsystems, Helsinki, Finland) were coated with 50 ng/well of the peptides Aβ[1–40], Aβ[1–42], Aβ[1–43], APP 695 or APP 770 with or without serial dilutions in carbonate-bicarbonate buffer pH 9·6 for 4h. After two washes with PBS 100 μl/well of 1% (w/v) BSA was added for 2h at 37°C. After two PBS washes, antibodies were incubated overnight at 4°C at 10 μg/ml with serial dilutions or 5 μg/ml with titrated peptide plates. Plates had three PBST washes and three PBS washes. To detect the Fab and isotype control, mouse M2 anti-Flag antibody (Sigma, St Louis, MO, USA) was applied at 100 ng/well for 1 h, followed by three PBST washes and three PBS washes, then rabbit anti-mouse immunoglobulins coupled to horseradish peroxidase (Dako, Carpinteria, CA, USA) at 1/500 dilution was added at 50 μl/well for 1 h. To detect the parent, anti-mouse immunoglobulins couple to horseradish peroxidase (Dako, Carpinteria, CA, USA) at 1/500 dilution was added at 50 μl/well for 1 h. ELISA plates were washed three times with PBST and three times with PBS followed by 100 μl/well ‘o’-phenylenediamine (Sigma, St Louis, MO, USA) developing solution for 1 h. The development reaction was stopped with 50μl/well 8m H2SO4 and the colour reaction determined at O.D. 490nm on a Model 450 microplate reader (Biorad, Hercules, CA, USA).
Detection of Aβ and APP using immunoblotting
The following procedure was derived from Ida et al.[16]. Samples containing 100 ng of human Aβ or recombinant APP were denatured, electrophoresed in 14% Tris-tricine gels and transferred onto nitrocellulose (Biorad, Hercules, CA, USA). Blots were boiled in PBS for 5 min and blocked for 1·5h with 1% (w/v) BSA. The Fab, isotype and parent antibodies were incubated overnight at 4°C (5 μg/ml of Fab or isotype and 50 μg/ml of parent). The blots were washed once for 5 min then twice for 20 min with TBST. To detect the Fab and isotype antibodies, mouse M2 anti-Flag (Sigma, St Louis, MO, USA) was applied at 2 μg/ml, followed by one 5-min then two 20-min washes with TBST, followed by anti-mouse immunoglobulins coupled to horseradish peroxidase (Dako, Carpinteria, CA, USA) at 1/2000 for 1 h. To detect the parent, anti-mouse immunoglobulins coupled to horseradish peroxidase (Dako, Carpinteria, CA, USA) was applied at 1/2000 for 1 h. All blots were washed with TBST once for 5 min and twice for 20 min Blots were rinsed with PBST and developed with the ECL development reagent system (Amersham Pharmacia Biotech, Buckinghamshire, UK).
Epitope mapping analysis
To determine the specificity of the Aβ parent and Fab antibodies, Pepset analysis was undertaken comprising a set of 15 mer overlapping biotinylated peptides incrementing by three amino acids per peptide and encompassing the human [695–652] sequence (Chiron Mimotopes Pty Ltd, Melbourne, Australia) [18]. The 96-well microtitre Cliniplates EB (Labsystems, Helsinki, Finland) were incubated with 500 ng/well streptavidin at 37°C overnight and evaporated to dryness. Then 50 ng/well of biotinylated peptides were incubated for 30 min followed by three washes in PBS. BSA 1% (w/v)] (Sigma, St Louis, MO, USA) at 100 μl/well was incubated at 37°C for 2h and plates washed three times with PBS. The parent and Fab antibodies at 500 ng/well were incubated for 30 min and the plates washed three times with PBST and three times with PBS. To detect the parent antibody, 50 μl/well anti-mouse immunoglobulins coupled to horseradish peroxidase (Dako, Carpinteria, CA, USA) at 1/500 was incubated for 30 min. To detect the Fab antibody 100 ng/well M2 anti-Flag monoclonal antibody (Sigma, St Louis, MO, USA) was applied for 30 min, plates were washed three times with PBST and three times with PBS, and 50 μl/well anti-mouse immunoglobulins coupled to horseradish peroxidase (Dako, Carpinteria, CA, USA) at 1/500 was incubated for 30 min. Plates were washed three times with PBST, three times with PBS and bound mouse immunoglobulin or Fab was detected by the addition of 100 μl/well of ‘o’-phenylenediamine (Sigma, St Louis, MO, USA) development solution for 1 h. The development reaction was stopped by adding 25 μl/well of 8m H2SO4. The O.D. 490nm was determined using a Model 450 microplate reader (Biorad, Hercules, CA, USA).
Immunohistochemistry of human AD (certified by CERAD criteria) and normal, non-AD control brain
Paraffin-embedded, formalin fixed, 4 μm tissue sections were incubated over three changes of Shellex for 2 min each, followed by 2 min in 90% (v/v) ethanol and 70% (v/v) ethanol. Sections were washed with ddH2O and treated for 5 min in 80% (v/v) formic acid. After a ddH2O wash, sections were treated with 3% (v/v) H2O2 (BDH Chemicals, Poole, Dorset, UK) for 5 min, washed in ddH2O and TBS pH 7·4. Sections were blocked with 20% (v/v) rabbit serum in TBS pH 7·4 for 30 min and 100 μl/section of the parent, Fab and isotype control antibodies were applied at either 20 μg/ml or 10 μg/ml for 1 h. Slides were washed twice for 5 min with TBS pH 7·4. To detect the parent antibody 100 μl/section of anti-mouse immunoglobulins coupled to horseradish peroxidase (Dako, Carpinteria, CA, USA) was applied at 6·5 μg/ml for 1 h. To detect the Fab and isotype control, 100 μl/section of M2 anti-Flag monoclonal antibody (Sigma, St Louis, MO, USA) was applied at 10 μg/ml for 1h followed by two 5-min washes with TBS pH 7·4, then 100 μl/section of anti-mouse HRP (Dako, Carpinteria, CA, USA) was applied at 6·5 μg/ml for 1 h. Sections were washed twice for 5 min with TBS pH 7·4 and developed with diaminobenzidine solution (BDH Chemicals, Poole, Dorset, UK) for 5 min. Sections were washed in TBS pH 7·4 and ddH2O, then counterstained in Mayer's haematoxylin for 1min, washed in ddH2O, immersed in Scott's tap water for 30 s, washed in ddH2O, dehydrated in two changes of absolute alcohol, cleared in three changes of shellex (Shell Chemicals, Victoria, Australia) and mounted in DPX (BDH Chemicals, Poole, Dorset, UK).
Purification of monoclonal antibodies
The mouse monoclonal antibody 1E8 was purified from hybridoma culture supernatant by affinity chromatography on a Protein-G Hi-trap affinity column (Pharmacia Biotech, Sweden). Briefly, a 100-ml aliquot of hybridoma supernatant was recirculated overnight at 4°C using a peristaltic pump. The Protein G column containing bound antibody was washed with 30 ml of 0·1m sodium phosphate buffer pH 7·4. Bound antibody was eluted with 0·1 m glycine-HCl pH 2·7 into 1 ml fractions containing 42 μl/ml of 1·0m Tris-HCl pH 9·0. Eluted fractions were analysed spectrophotometrically at A280 nm to determine the antibody-containing fractions. Peak fractions were pooled and dialysed against 100 volumes of PBS at 4°C. Following dialysis, protein concentrations were measured spectrophotometrically and the antibody stored at −20°C.
Preparation of the FAD PS-1 mutation brain sample
The FAD brain sample containing the Australian PS-1 mutation (Leu219Pro) [25] was prepared from total brain tissue extract, homogenized in 1 ml TRIZOL (Life Technologies), incubated for 5 min at RT and 2 ml of chloroform was added. The sample was shaken for 15 s, incubated for 2min at RT and centrifuged at 12 000 gmax for 15 min, 4°C. Isopropyl alcohol (1·5 ml) was added to the lower interphase, incubated 10 min at RT and centrifuged at 12 000 gmax for 10 min at 4°C. The pellet was washed three times with 2 ml of 0·3m guanidine-HCl in 95% (v/v) ethanol for 20 min at RT and centrifuged at 7500 gmax for 5 min at 4°C after each wash. The pellet was vortexed in absolute ethanol (2 ml), incubated 20 min at RT, centrifuged at 7500 gmax for 5 min at 4°C, air-dried and redissolved in 0·5 ml of 2% (w/v) SDS, by heating and sonification. Insoluble material was centrifuged at 10 000 gmax for 10 min at 4°C. The supernatant was aliquoted and frozen at −80°C.
RESULTS
Generation, expression and purification of 1E8–4b
Fab antibody gene fragments from the mouse monoclonal hybridoma cell line 1E8 [17–22] were generated by PCR using primers generated from immunological sequences in Kabat et al.[20]. The VHCH1 construct was created with primers flanking amino acids 1–6/7 of the mouse variable heavy chain (VH) framework 1 region and amino acids 212–223 of the mouse first constant heavy chain region (CH1). The VLCL construct was created with primers flanking amino acids 1–7 of the variable light chain (VL) framework 1 region and amino acids 207–214 of the constant light chain region (CL).
Constructs were cloned into pCR-Script Amp SK(+) vectors (Stratagene, CA, USA), derived from pBluescript® II SK(+) vectors (Stratagene, CA, USA). Six independent clones for both VHCH1 and VLCL were sequenced, generating a consensus sequence. Sequence analysis in the Kabat and Wu database [20] suggested the heavy and light chain constructs had not previously been published. The entire VHCH1 chain spanned 213 amino acids and the VH region belonged to the immunoglobulin mouse heavy chain subgroup IIA. The VH region exhibited highest homology with heavy chains from the mouse immunoglobulins DBF1-386·5, DBF1-235·4 and DB2-101·1 [20]. The CH1 region was slightly abridged and most homologous with the heavy constant chain CH1 region of the mouse immunoglogulin IgG1CL[20]. The VLCL chain spanned 219 amino acids and the VL region was found to belong to the mouse kappa light chain subgroup II. The VL region displayed highest homology with kappa light chains from eight mouse immunoglobulins [20]. The CL chain was most homologous with the kappa light constant chains of the mouse immunoglobulins 17/9′ CL, C.C58M75′CL and MOPC21 [20].
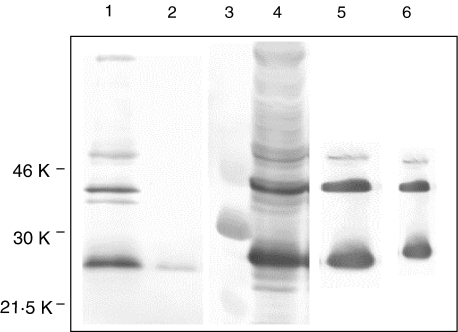
The six VHCH1 and VLCL constructs were subcloned into the expression vector pHFA2 and expressed the 1E8 Fab fragments equally well at 8h post-induction, with identical reactivity to the Aβ[1–42] peptide by ELISA. The VHCH1 chain migrated at 26 kDa and the VLCL chain at 28kDa. Clone 1E8–4b was selected and sequenced (Fig. 1). Time-course expression experiments conveyed highest Fab antibody expression at 24h post induction and greatest Fab antibody reactivity to Aβ[1–42] peptide by ELISA. The intact Fab fragment resolved at 48kDa with N-terminal fragments present on the immunoblot. At 24h post induction, 0·5 mg of 1E8–4b/l culture supernatant was produced. 1E8–4b was extracted most fully from culture supernatant by a 60% (v/v) saturated ammonium sulphate precipitation [23] (Fig. 2). After dialysing into PBS, 1E8–4b was purified on a M2 anti-Flag affinity column. Reactive eluates were pooled, concentrated and buffer exchanged into PBS (Fig. 2). Purity was assessed by Coomassie Blue stain (Fig. 2).
Fig. 1.
DNA and protein sequences of 1E8-4b Fab in the pHFA2 cloning vector. Both the heavy (VHCH1) and light (VLCL) chains of the 1E8-4b Fab are preceeded by N-terminal pelB signal sequences (italicised). Non-coding regions located 5′ to the pelB signal sequences are shown in lower case. C-terminal peptide tag sequences (tubulin (Glu Glu Phe) and Flag) are shown in bold. The framework, CDR and constant regions are outlined above the sequence, according to Kabat et al. [20]. Important restriction sites are underlined.
Fig. 2.
Purification of recombinant 1E8-4b Fab-EEF/FLAG conjugate from E. coli culture supernatant as analysed by SDS-PAGE under non-reducing conditions and immunoblotting (lanes 1–5) or Coomassie Brilliant Blue staining (lane 6). Immunoreactive protein bands were detected using M2 anti-Flag antibody followed by goat anti-mouse alkaline phosphatase conjugated antibody and developed with Fast Red/AS-MX Naphthol reagents. Lane 1: Culture supernatant; lane 2: Supernatant following 60% (v/v) SAS precipitation; lane 3: Low-molecular-weight-protein markers (with numbers to the left in kDa); lane 4: precipitated 1E8-4b Fab protein following 60% (v/v) SAS precipitation; lane 5: pooled and concentrated M2 anti-Flag affinity gel purified 1E8-4b Fab eluates; lane 6: Coomassie stain of lane 5.
Specificity comparison of the recombinant Fab and parent 1E8 monoclonal antibodies
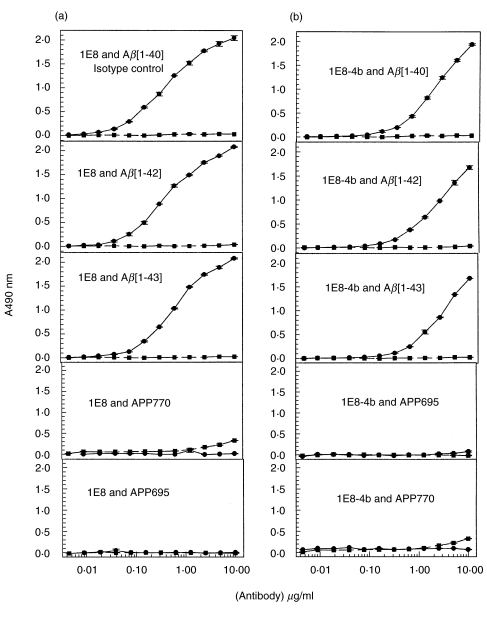
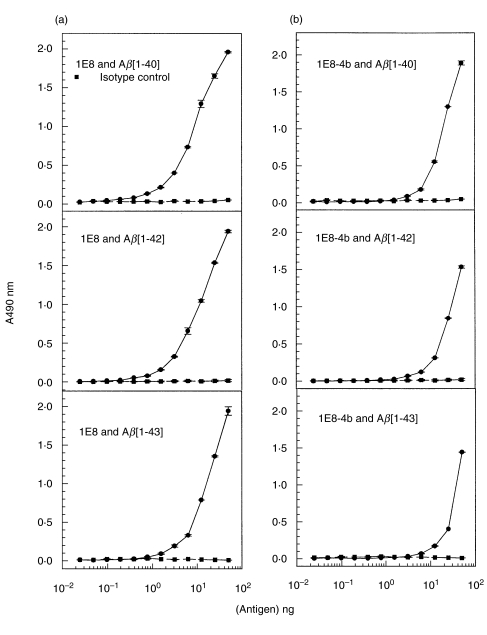
By ELISA, the recombinant Fab 1E8–4b and the parent 1E8 antibody were titrated against Aβ1–40/42/43] peptides and against APP 695 and 770. Both antibodies displayed binding to Aβ1–40/42/43] but not to APP 695 and 770. The antibody 1E8 exhibited binding at lower concentrations than the monovalent 1E8–4b recombinant Fab fragment (Fig. 3). With peptide titration, 1E8–4b detected Aβ1–40/42/43] peptides down to 6 ng, suggesting that 1E8–4b had a fast on–off rate compared to the parent antibody, 1E8. The reactivity of 1E8 to the titrated peptides demonstrated detection of Aβ1–40/42/43] down to 1 ng, producing higher O.D. 490 nm Units per peptide concentration than the recombinant 1E8–4b fragment (Fig. 4).
Fig. 3.
ELISA depicting the reactivity of the parent 1E8 antibody (a) and the recombinant 1E8-4b Fab fragment (b) to Aβ[1–40], Aβ [1–42], Aβ[1–43], APP695 and APP770 peptides. The antibody is titrated from 10 μg/ml with serial doubling dilutions, while the peptide is at a constant concentration of 50 ng/well. 1E8 was probed with antimouse-HRP while 1E8-4b was probed with M2 anti-Flag antibody and then with antimouse-HRP. Detection of antibody reactivity to the peptides was via an ‘o’-phenylenediamine development mixture. The isotype control used was M2 anti-Flag antibody which is a mouse IgG1.
Fig. 4.
Determination of the sensitivity of the parent antibody 1E8 (a) and the recombinant Fab antibody 1E8-4b (b) to the amyloid peptides Aβ[1–40], Aβ[1–42] and Aβ[1–43] by ELISA. The peptides were serially diluted with doubling dilutions starting from 50 ng/ml while the concentrations of 1E8 and 1E8-4b were kept constant at 5 μg/ml. 1E8 was detected with antimouse-HRP conjugated antibody while 1E8-4b was probed with M2 anti-Flag antibody and then detected with antimouse-HRP conjugated antibody. Detection was via an ‘o’-phenylenediamine development mixture. The isotype control used was M2 anti-Flag antibody which is a mouse IgG1.
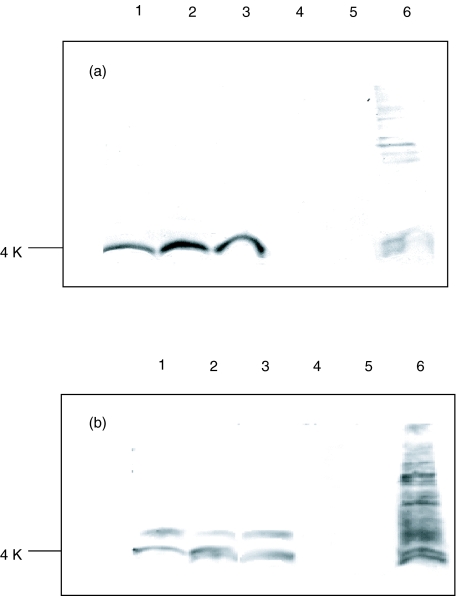
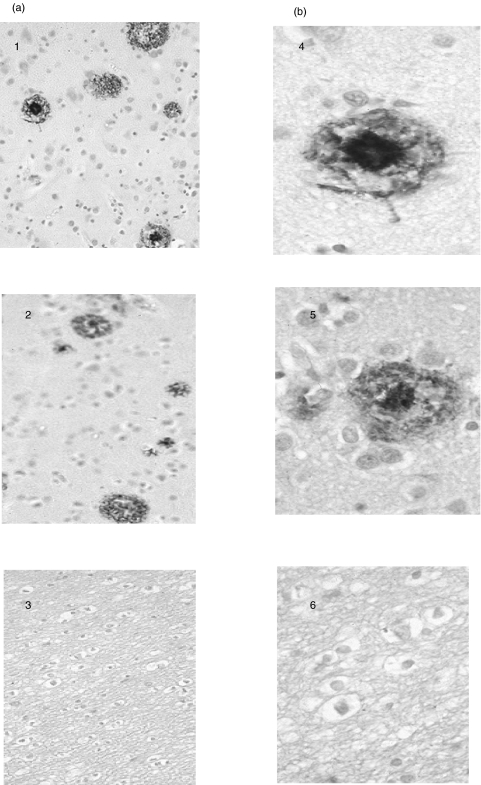
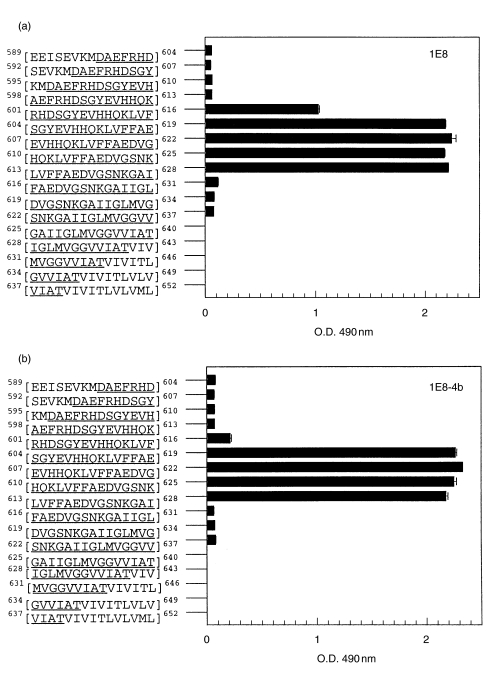
The 1E8 (50 μg/ml) antibody and 1E8–4b (5 μg/ml) antibody also bound to Aβ1–40/42/43] peptides but not to APP 695 and 770 by immunoblotting. Figure 5 shows the detection of an approximately 4kDa band by both antibodies, representing the Aβ1–40/42/43] peptides and Aβ from the PS-1 mutation FAD brain sample [25]. 1E8 at 50 μg/ml also detects an Aβ dimer. In addition, by immunohistochemistry, 1E8–4b and 1E8 at 20 μg/ml were able to detect Aβ in AD brain sections. Figure 6 shows the detection of amyloid associated with plaques. Epitope mapping (Fig. 7) showed that the 1E8–4b recombinant fragment and the 1E8 antibody recognized the following Aβ peptides: Aβ[5–19], Aβ[8–22], Aβ[11–25], Aβ[14–28] and Aβ[17–31], with 1E8 exhibiting much stronger binding than 1E8–4b against theAβ[5–19] peptide.
Fig. 5.
Detection of Aβ peptides [1–40], [1–42] and [1–43] and total Aβ in FAD brain by 1E8-4b (a) and 1E8 (b) antibodies using immunoblotting followed by ECL development. Samples were electrophoresed on 14% Tris-tricine gels with 100 ng/well of Aβ peptides [1–40], [1–42] and [1–43] and APP peptides 695 and 770. A total of 5 μl of FAD brain sample was used. The recombinant Fab 1E8-4b and the isotype control M2 anti-Flag monoclonal antibody were applied to the immunoblots at 5 μg/ml, while 1E8 was used at 50 μg/ml. The isotype control displayed no reactivity to the peptides or FAD brain sample. Lane 1: Aβ peptide [1–40]; lane 2: Aβ peptide [1–42]; lane 3: Aβ peptide [1–43]; lane 4: APP peptide 695; lane 5: APP peptide 770; lane 6: FAD (PS-1mut) brain sample.
Fig. 6.
Immunohistochemistry of AD brain sections. Brain sections were taken from the same AD brain patient for accurate comparison. The 1E8-4b recombinant Fab fragment, the 1E8 antibody and the isotype control M2 anti-Flag antibody were applied to the sections at a concentration of 20 μg/ml at x20 (a) and x60 (b) magnification. At 10 μg/ml and 20 μg/ml the 1E8-4b and 1E8 antibodies showed reactivity to amyloid associated with plaques, while the isotype control showed no reactivity. Sections 1 and 4: 1E8; sections 2 and 5: 1E8-4b; sections 3 and 6: isotype control M2 anti-Flag monoclonal antibody.
Fig. 7.
Epitope mapping of 1E8 (a) and 1E8-4b (b). The pepset encompasses the human APP695[589–652] sequence and consists of an overlapping set of 15-mer linear peptides, incrementing by 3 amino acids per peptide. APP[597–640] amino acid residues represent the Aβ[1–43] peptide sequence and are underlined.
DISCUSSION
Despite procedures for pre-mortem AD diagnosis (i.e. neurological examination, ELISA-based tests, risk assessment by family history/genetic predisposition to AD) post-mortem immunohistological brain tissue examination is the definitive AD diagnostic [26,27]. Generating antibodies specific for key AD markers is important for ante-mortem reagent development to diagnose AD rapidly and accurately, eliminating stringent tests and permitting therapy prior to neurodegeneration [26]. There is currently no cure for AD and although therapies such as cholinesterase inhibitors are reported to improve cognitive function, disease progression continues [28]. Ideal therapeutics should prevent AD progression and restore normal function. Antibodies may clear amyloid from brain [17] and combined with 5 mer β-sheet breaker peptide tags [29], neurotrophic tags [30] and Aβ solubilizing substances [31], therapeutic ‘magic bullets’ could ensue. We have produced the recombinant Fab antibody, 1E8–4b, and performed comparitive binding studies with the parent antibody, 1E8.
To correct for the differences in size and number of valencies between 1E8 and 1E8–4b, the antibodies should be compared on a 1:1 mole ratio. As 1E8 is an intact, divalent IgG1 of 160kDa and 1E8–4b is a monovalent Fab of 48 kDa, the amount of 1E8 needed relative to 1E8–4b is: (160 kDa/48 kDa)/2 valencies = 1·67 times more 1E8 than 1E8–4b to give a 1:1 mole ratio. Hence 10μg/ml of 1E8 is equivalent to 6 μg/ml of 1E8–4b to have the same number of moles. In ELISA experiments using a high epitope density of 50 ng/well Aβ1–40/42/43] peptides and titrated antibody (Fig. 3), 1E8–4b did not perform as well as 1E8 on a mole to mole ratio, although both antibodies are quite sensitive. 1E8 produced a signal down to 20 ng/ml and produced a curve with a steep linear slope of 1·5 from low to high antibody dilutions. In contrast, 1E8–4b produced a signal down to 200 ng/ml with a decreased linear slope of 0·4. As bivalently bound antibody dissociates slower than univalently bound antibody, apparent during ELISA washing steps [32], the characteristic curve for 1E8 may have arisen (Fig. 3). High-affinity antibodies generally produce steeper-sloped titration curves, suggesting a lower affinity for 1E8–4b [33,34]. At the same antibody concentration, with 1·7 times less moles of 1E8 than 1E8–4b, 1E8 detected lower levels of Aβ (1·5 ng/well Aβ) than 1E8–4b (6 ng/well Aβ). The slower titration of 1E8 suggested stronger binding of 1E8 to the Aβ peptides than 1E8–4b and perhaps a slower dissociation rate due to divalency or higher affinity [32]. Presence of the Flag tag on 1E8–4b was expected to produce an enhanced signal for the 1E8–4b ELISA due to the additional M2 anti-Flag monoclonal antibody detection step [13].
By immunohistochemistry, 1E8–4b and 1E8 detected Aβ in AD brain sections, which was localized to plaques [4]. Immunoblotting depicted reactivity to Aβ1–40/42/43] peptides by 1E8 and 1E8–4b. In addition, in FAD brain characterized with a novel PS-1 mutation [25] we detected a 4 kDa fragment corresponding to Aβ. Immunoblotting and ELISA results conveyed that 1E8 and 1E8–4b did not react with APP 695 or APP 770, suggesting Aβ specificity by these antibodies. Pepset epitope mapping showed specific binding of 1E8–4b and 1E8 to Aβ amino acid residues 17–22. Removal of Aβ[17–19] obliterated binding despite the presence of [20–31]. Stronger binding was seen when Aβ[20–22] was included with Aβ[17–19]. 1E8–4b showed a weaker response to Aβ[17–19] than 1E8 due perhaps to altered conformation or unique requirements of only Aβ[18,19/19] plus [20–22]. Overall, 1E8 and 1E8–4b contained a specific epitope encompassing Aβ[17,18,18–22].
On a mole:mole ratio, 1E8 showed greater reactivity than 1E8–4b with Aβ[1–40/42/43] but the specificity of 1E8–4b and 1E8 was nearly identical. Improved detection of Aβ in CSF and plasma requires an antibody of higher affinity than 1E8 or 1E8–4b. With several rounds of affinity selection, 1E8–4b may attain this characteristic [13] with diagnostic benefits for AD.
The use of recombinant technology for antibody production in the diagnosis and therapy of AD is relatively unexplored and the need to examine recombinant antibody design and application should be pursued. The recombinant Fab fragment 1E8–4b produced in this study, is the first recombinant Fab fragment to be described that is reactive with the AD-related Aβ protein.
Acknowledgments
The authors thank Drs Steven Holmes and Carol Gray of SmithKline Beecham for the 1E8 hybridoma cell line which was provided in 1995 for the development of the double-antibody capture ELISA as reported in Jayasena et al.[18]. The mRNA and purified 1E8 antibodies from the 1E8 hybridoma, cultured in Dr Underwood's laboratory in the Department of Pathology at the University of Melbourne, were used for this study. The authors also thank Ms Tina Cardamone for her assistance with immunohistochemical staining, Dr Genevieve Evin for supplying the PS-1 mutation brain sample, Ms Denise Galatis for her advice and Dr Bob Irving for providing reagents and advice.
REFERENCES
- 1.Wisniewski KE, Dalton AJ, McLachlan C, Wen GY, Wisniewski HM. Alzheimer's disease in Down's syndrome: clinicopathologic studies. Neurology. 1985;35:957–61. doi: 10.1212/wnl.35.7.957. [DOI] [PubMed] [Google Scholar]
- 2.Ebly EM, Parhad IM, Hogan DB, Fung TS. Prevalence and types of dementia in the very old: results from the Canadian study of Health and Aging. Neurology. 1994;44:1593–600. doi: 10.1212/wnl.44.9.1593. [DOI] [PubMed] [Google Scholar]
- 3.Hardy J. Amyloid, the presenilins and Alzheimer's disease. Trends Neurosci. 1997;20:154–9. doi: 10.1016/s0166-2236(96)01030-2. [DOI] [PubMed] [Google Scholar]
- 4.Gravina SA, Ho L, Eckman CB, et al. Amyloid β protein (Aβ) in Alzheimer's disease brain. Biochemical and immunocytochemical analysis with antibodies specific for forms ending at Abeta 40 or A beta 42 (43) J Biol Chem. 1995;270:7013–6. doi: 10.1074/jbc.270.13.7013. [DOI] [PubMed] [Google Scholar]
- 5.Roher AE, Lowenson JD, Clarke S, et al. Structural alterations in the peptide backbone of β-amyloid core protein may account for its deposition and stability in Alzheimer's disease. J Biol Chem. 1993;268:3072–83. [PubMed] [Google Scholar]
- 6.Mattson MP. Cellular actions of beta-amyloid precursor protein and its soluble and fibrillogenic derivatives. Physiol Rev. 1997;77:1081–132. doi: 10.1152/physrev.1997.77.4.1081. [DOI] [PubMed] [Google Scholar]
- 7.Roher AE, Kuo YM, Kokjohn KM, Emmerling MR, Gracon S. Amyloid and lipids in the pathology of Alzheimer disease. Amyloid. 1999;6:136–45. doi: 10.3109/13506129909007315. [DOI] [PubMed] [Google Scholar]
- 8.Jarrett JT, Lansbury PT. Seeding ‘one-dimensional crystallization’ of amyloid. Cell. 1993;73:1055–8. doi: 10.1016/0092-8674(93)90635-4. [DOI] [PubMed] [Google Scholar]
- 9.Mann DM, Iwatsubo T, Cairns NJ, et al. Amyloid beta protein (Abeta) deposition in Chromosome 14 linked Alzheimer's disease. Predominance of Abeta42 (43) Ann Neurol. 1996;40:149–56. doi: 10.1002/ana.410400205. [DOI] [PubMed] [Google Scholar]
- 10.Hock C, Golombowski S, Muller-Spahn F, et al. Histological markers in nasal mucosa of patients with Alzheimer's disease. Eur Neurol. 1998;39:111–8. doi: 10.1159/000007953. [DOI] [PubMed] [Google Scholar]
- 11.Shoji M, Matsubara E, Kanai M, et al. Combination assay of CSF Tau, Aβ1–40 and Aβ1-42 (43) as a biochemical marker of Alzheimer's disease. J Neurol Sci. 1998;158:134–40. doi: 10.1016/s0022-510x(98)00122-1. [DOI] [PubMed] [Google Scholar]
- 12.Vanmechelen E, Vanderstichele H. Alzheimer tau test and detergent cellulase made by genetic engineering. J Biotechnol. 1998;66:229–33. doi: 10.1016/s0168-1656(98)00169-2. [DOI] [PubMed] [Google Scholar]
- 13.Hudson PJ. Recombinant antibody fragments. Curr Opin Biotech. 1998;9:395–402. doi: 10.1016/s0958-1669(98)80014-1. [DOI] [PubMed] [Google Scholar]
- 14.Scheuner D, Eckman C, Jensen M, et al. Secreted amyloid beta-protein similar to that in the senile plaques of Alzheimer's disease is increased in vivo by the presenilin 1 and 2 and APP mutations linked to familial Alzheimer's disease. Nat Med. 1996;2:864–70. doi: 10.1038/nm0896-864. [DOI] [PubMed] [Google Scholar]
- 15.Mehta PD, Dalton AJ, Mehta SP, Kim KS, Sersen EA, Wisniewski HM. Increased plasma amyloid β protein 1–42 levels in Down syndrome. Neurosci Lett. 1998;241:13–6. doi: 10.1016/s0304-3940(97)00966-x. [DOI] [PubMed] [Google Scholar]
- 16.Ida N, Hartmann T, Pantel J, et al. Analysis of heterogeneous βA4 peptides in human cerebrospinal fluid and blood by a newly developed sensitive Western blot assay. J Biol Chem. 1996;271:22908–14. doi: 10.1074/jbc.271.37.22908. [DOI] [PubMed] [Google Scholar]
- 17.Schenk D, Barbour R, Dunn W, et al. Immunization with amyloid-beta attenuates Alzheimer-disease-like pathology in the PDAPP mouse. Nature. 1999;400:173–7. doi: 10.1038/22124. [DOI] [PubMed] [Google Scholar]
- 18.Jayasena ULHR, Gribble SK, McKenzie A, Beyreuther K, Masters CL, Underwood JR. Identification of structural variations in the carboxyl terminus of Alzheimer's disease-associated betaA4. Clin Exp Immunol. 2001;124:297–305. doi: 10.1046/j.1365-2249.2001.01209.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kettleborough CA, Saldanha J, Ansell KH, Bendig MM. Optimization of primers for cloning libraries of mouse immunoglobulin genes using the polymerase reaction. Eur J Immunol. 1993;23:206–11. doi: 10.1002/eji.1830230132. [DOI] [PubMed] [Google Scholar]
- 20.Kabat EA, Wu TT, Perry HM, Gottesman KS, Foeller C. Sequences of proteins of immunological interest. 5. Bethesda, MD: National Institutes of Health; 1991. [Google Scholar]
- 21.Dolezal O, Coia G, Guthrie RE, Lilley GG, Hudson PJ. Escherichia coli expression of a bifunctional Fab peptide epitope reagent for the rapid diagnosis of HIV-1 and HIV-2. Immunotechnology. 1995;1:197–209. doi: 10.1016/1380-2933(95)00021-6. [DOI] [PubMed] [Google Scholar]
- 22.Hoogenboom HR, Griffiths AD, Johnson KS, Chiswell DJ, Hudson P, Winter G. Multi-subunit proteins on the surface of filamentous phage: methodologies for displaying antibody (Fab) heavy and light chains. Nuc Acids Res. 1991;19:4133–7. doi: 10.1093/nar/19.15.4133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Coppola G, Underwood J, Cartwright G, Hearn MTW. Comparison of methods for the purification of mouse monoclonal immunoglobulin M autoantibodies. J Chromatogr. 1989;426:269. doi: 10.1016/s0021-9673(01)93875-0. [DOI] [PubMed] [Google Scholar]
- 24.Sambrook J, Fritsch EF, Maniatis T. Molecular cloning, a laboratory manual. 2. New York: Nolan C; 1989. [Google Scholar]
- 25.Smith MJ, Gardner RJ, Knight MA, et al. Early-onset Alzheimer's disease caused by a novel mutation at codon 219 of the presenilin-gene. Neuroreport. 1999;10:503–7. doi: 10.1097/00001756-199902250-00011. [DOI] [PubMed] [Google Scholar]
- 26.Arai H. Biological markers for the clinical diagnosis of Alzheimer's disease. Tohoku J Exp Med. 1996;179:65–79. doi: 10.1620/tjem.179.65. [DOI] [PubMed] [Google Scholar]
- 27.Selkoe DJ. Translating cell biology into therapeutic advances in Alzheimer's disease. Nature. 1999;399:A23–A31. doi: 10.1038/399a023. [DOI] [PubMed] [Google Scholar]
- 28.Hecker J. Alzheimer's disease: the advent of effective therapy. Aust NZ J Med. 1998;28:765–71. doi: 10.1111/j.1445-5994.1998.tb01551.x. [DOI] [PubMed] [Google Scholar]
- 29.Soto C. Alzheimer's and prion disease as disorders of protein conformation: implications for the design of novel therapeutic approaches. J Mol Med. 1999;77:412–8. doi: 10.1007/s001090050371. [DOI] [PubMed] [Google Scholar]
- 30.Birkhauser MH, Strnad J, Kampf C, Bahro M. Oestrogens and Alzheimer's disease. Int J Geriatr Psychiatry. 2000;15:600–9. doi: 10.1002/1099-1166(200007)15:7<600::aid-gps155>3.0.co;2-7. [DOI] [PubMed] [Google Scholar]
- 31.Janciauskiene S, de Frutos PG, Carlemalm E, Dahlback B, Eriksson S. Inhibition of Alzheimer β-peptide fibril formation by serum amyloid P component. J Biol Chem. 1995;270:26041–4. doi: 10.1074/jbc.270.44.26041. [DOI] [PubMed] [Google Scholar]
- 32.Mason DW, Williams AF. The kinetics of antibody binding to membrane antigens in solution and at the cell surface. Biochem. 1980;187:1–20. doi: 10.1042/bj1870001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lew AM. The effect of epitope density and antibody affinity on ELISA as analysed by monoclonal antibodies. J Immunol Meth. 1984;72:171. doi: 10.1016/0022-1759(84)90445-9. [DOI] [PubMed] [Google Scholar]
- 34.Nimmo GR, Lew AM, Stanley CM, Steward MW. Influence of antibody affinity on the performance of different antibody assays. J Immunol Meth. 1984;72:177. doi: 10.1016/0022-1759(84)90446-0. [DOI] [PubMed] [Google Scholar]