Abstract
Autoantibodies to 65 kDa glutamic acid decarboxylase (GAD65) are produced in many patients with autoimmune polyendocrine syndrome type II (APS-II) or stiff-man syndrome (SMS) and are heterogeneous in their epitope specificities, recognizing both conformational and linear determinants. Major linear epitopes of GAD, which are recognized by autoantibodies in a minority of these patients, occur in the N-terminal and C-terminal regions. We have investigated antibody recognition of the N- and C-termini of GAD65 in relation to their structural features as an approach to understanding what modifications to the native GAD structure may occur that facilitate the generation of antibodies specific to linear epitopes in these regions during the autoimmune pathogenesis. A monoclonal antibody specific to the N-terminus of GAD65 bound both native and denatured GAD in ELISA, whereas monoclonal and polyclonal antibodies specific to the C-terminus of GAD bound only denatured GAD. These antibodies were epitope mapped using random peptide phage-display libraries and the epitopes related to a previously proposed structural model of GAD65. This has led us to propose that the α-helical secondary structure of the C-terminus of GAD65 must be denatured to generate linear epitopes. In contrast, the N-terminus is both surface exposed and linear in the native structure, but may be masked by membrane interactions, which must be broken to facilitate recognition by B cells.
Keywords: autoimmune polyendocrine syndrome type II, GAD-65, linear epitopes, phage peptide-display libraries, stiff-man syndrome
INTRODUCTION
Autoantibodies to glutamic acid decarboxylase (GAD) are produced in several autoimmune disorders that exhibit both distinct and overlapping features in their pathogenesis and clinical manifestations [1]. GAD occurs in the central nervous system and in the pancreatic islet β-cells; it exists as a 65-kDa membrane-associated form (GAD65) and a 67-kDa soluble form (GAD67) [2]. These isoforms are homologous in their middle and carboxy (C)-terminal regions, but differ in their amino (N)-terminal regions. The autoantibodies are specific predominantly for GAD65, although some either cross-react with GAD67 or are GAD67-specific [3].
GADA were initially identified in a subset of patients with the central nervous system disease stiff-man syndrome (SMS) [4], about a third of whom develop type 1 diabetes mellitus (DM). Subsequently, GADA were also demonstrated in the majority of type 1 DM patients [5] and in patients with autoimmune polyendocrine syndrome type II (APS-II) [6], in whom different autoimmune endocrinopathies co-exist (particularly autoimmune thyroid disease, gastritis, Addison's disease and type 1 DM).
The pathogenetic mechanisms leading to the generation of GADA are unknown. However, analysis of the epitope specificities of GADA suggests common and distinct features of these mechanisms in type 1 Dm, APS-II and SMS. GADA recognize epitopes in the middle, C-terminal and N-terminal regions of GAD and show specificities for conformational/native determinants and linear/denatured determinants [3,7–22]. The serum GADA of many type 1 Dm patients have relatively low titres and recognize mainly conformational determinants. However, some type 1 Dm and APS-II patients are similar to many SMS patients in possessing high titre GADA which recognize linear/denatured determinants as well as conformational determinants.
The secondary and tertiary structures of the middle and C-terminal regions of GAD65 have been modelled using information from the epitope mapping of GAD65-specific human monoclonal antibodies (MoAbs) [19] and the known crystal structure of related decarboxylase enzymes [19,23]. Collectively, the human MoAbs to GAD recognize determinants scattered across virtually the whole exposed surface of these regions [19,23]. However, there is currently no model for the structure of the N-terminal region of GAD65 or the surface exposure of epitopes within this region.
Very little is known about how, and in what form, GAD is made available to the immune system during the autoimmune response. Responses to linear epitopes in the extreme N- and C-termini of GAD are relatively common in SMS, but also occur in some type 1 Dm and APS-II patients [7,11,13–16], and are probably characteristic of a relatively advanced stage of the autoimmune response to GAD. Indeed, the extreme termini of GAD appear to be main sites of linear (as opposed to conformational) epitopes of GAD65 [7]. This raises the question of how and why linear epitopes occur at these sites in particular. We have therefore investigated antibody recognition of the N- and C-termini of GAD65 in relation to their structural features as an approach to understanding what modifications to the native GAD structure may occur that facilitate the generation of antibodies specific to linear epitopes in these regions during the autoimmune pathogenesis. In this context, we have determined amino acid residues that contribute to epitopes in these terminal regions of GAD65 as defined by epitope mapping of monoclonal and polyclonal antibodies to these regions using random peptide phage-display libraries. This has further enabled us to infer some structural features of the N-terminus of GAD65 in relation to its antigenicity.
MATERIALS AND METHODS
Patients’ sera
Sera were used from four SMS patients, all of whom were positive for GADA. Only one of the patients (LB) had type 1 Dm. Thirty-one GADA+ patients with APS-II whose sera were used have been described in detail previously [17]: 19 did not have type 1 Dm; for five of the 12 that did have type 1 Dm, serum samples were available pre-onset and post-onset of diabetes.
Monoclonal and polyclonal antibodies
GAD-6, a murine IgG2a MoAb specific for GAD65 [24], was purchased as purified lyophilized antibody (Roche Diagnostics, Lewes, UK). GAD-6 reacts with an epitope between residues 475–585 of GAD65 [10] and requires residues 529–585 for binding [25]. The following antibodies were purchased from Affiniti Research Products (Exeter, UK): N-MoAb (GC3208, clone11) is a murine IgG1 MoAb that reacts specifically with a peptide representing residues 4–17 of GAD65 [26]; C-MoAb (GC3108, clone 111) is a murine IgG1 MoAb that recognizes the C-terminus of GAD65 and GAD67 (it reacts with a peptide representing residues 572–585 of GAD65) [27]; C-pc antibody (GC3008) is a rabbit antiserum that is also specific for the C-terminus of GAD65 and GAD67 (residues 572–585 of GAD65) and was provided as immunoglobulin purified partially from serum by precipitation with caprylic acid and ammonium sulphate. Affinity purification of the C-pc antibody was performed by absorption to, and elution from, rbGAD (see below) coupled to Sepharose 4B.
Synthetic peptides
A synthetic peptide representing amino acids 1–20 of the N-terminus of GAD65, plus a C-terminal cysteine residue (sequence MASPGSGFWSFGSEDGSGDSC) was synthesized commercially and supplied at>75% purity (Chiron Technologies, Clayton, Australia). A synthetic peptide representing amino acids 572–585 of the C-terminus of GAD65 (or 580–593 of GAD67), plus an N-terminal cysteine residue (sequence CDFLIEEIERLGQDL) was purchased at>95% purity (Affiniti Research Products, Exeter, UK). Peptides with the following irrelevant amino acid sequences were used as negative controls for antibody binding in the ELISAs for the detection of peptide-specific antibodies: CTIPLWFDDEIEVMIY in the ELISA for antibodies to the N-terminal peptide, and LTTEPDTTKFWPAPS in the ELISA for antibodies to the C-terminal peptide.
Partial purification of GAD from rat brain
Rat brain GAD (rbGAD) was prepared as described previously [17]. Briefly, whole brains of Wistar rats were homogenized in 10 mm Tris-HCl, pH 7·5 containing 100 µm phenyl methyl sulphonyl fluoride. The homogenate was ultracentrifuged and the supernatant fractionated by FPLC on a Q Sepharose Fast Flow column in 20 mm Tris-HCl pH 7·5, 1 m NaCl. The GAD-enriched fractions were identified in a dot immunoblot assay [17] and pooled.
Direct and capture ELISAs
The direct and capture ELISAs with rbGAD as the antigen (used at 40 µg/ml) were performed essentially as previously described [17,28]. For certain experiments, mild denaturation of rbGAD was achieved by incubation with 0·2% Triton X-100 at 4°C overnight prior to use in the capture ELISA. In the ELISAs for the detection of antibodies specific for the N-terminus or C-terminus of GAD65, the synthetic peptides (see above) were coated onto wells of ELISA plates at 5 µg/ml and the sera were assayed at dilutions of 1/40 or 1/100.
Screening the T7 linear or constrained 9-mer random peptide display libraries with anti-GAD antibodies
Two T7 libraries of linear or constrained 9-mer random peptides were synthesized in house. The random peptides were encoded by double-stranded DNA inserts assembled from synthetic degenerate oligonucleotides and cloned into gene X of the vector (T7select415–1) (Bioscience, Cambridge, UK). The mouse N-MoAb, at 10 µg/ml in 0·05 m sodium carbonate/sodium bicarbonate buffer pH 9·6, was coated onto Nunc immunotubes at 4°C overnight. The tubes were washed five times with 25 mm Tris-buffered saline (150 mm NaCl) pH 7·4 (TBS) containing 0·1% Tween-20 and then blocked with 5% BSA in TBS. The rabbit C-pc antibody was coated onto Nunc immunotubes at 10 µg/ml in TBS, and the tubes blocked with 5% milk powder (Marvel, Premier Beverages, Adbaston, UK) in TBS.
Either linear or constrained 9-mer T7 phage random peptide display library (about 1 × 1010 plaque-forming units per tube) in 1 ml of blocking solution, was added to antibody-coated tubes and incubated at 4°C for 20–30 min. The tubes were washed in TBS-0·1% Tween-20 and then 1 ml of a mid-log phase Escherichia coli BL 21 culture added and incubated at room temperature for 5 min for infection by the bound phage (termed the ‘eluate’). The eluate was added to 20–30 ml of mid-log phase E. coli BL 21 for amplification. The eluate phage were subjected to three further rounds of selection, as above.
The phage from the fourth round of selection were plated onto LB agar at 100–200 plaques per dish. A nitrocellulose membrane (0·45 µm pore size) (Millipore, UK) was placed onto the plate and incubated for 30 min at room temperature. The membrane was then blocked with 5% BSA/TBS or 5% milk powder/TBS. N-MoAb (10 µg/ml) or C-pc antibody (1/1000 dilution; preabsorbed with phage/E. coli lysate-treated nitrocellulose membrane) was added to the membrane and incubated on a rotator for 1–2 h at room temperature. The membrane was washed with TBS-0·1% Tween-20. Alkaline phosphatase-conjugated sheep antimouse IgG (whole molecule) or goat anti-rabbit IgG (1/1000 dilution; preabsorbed with phage/E. coli lysate-treated nitrocellulose membrane) was then added to the membrane and incubated at room temperature for 1 h. The membrane was washed and 5-bromo-4-chloro-3-indolyl phosphate/nitro-blue tetrazolium substrate (Sigma, Poole, UK) in deionized water with 5 mm levamisole was added to the membrane. Following the appearance of blue spots, the membrane was washed with TBS, then with water and dried.
Antibody-specific phage clones, which developed as blue spots on the membrane, were selected from the original plate and each of them mixed with 1 ml of mid-log phase E. coli BL 21 culture and incubated on a shaker at 37°C for 3 h for amplification.
Each clone of specific phage was further amplified by PCR and then sequenced in an ABI PRISM 310 Genetic Analyser (Applied Biosystems, Warrington, UK).
Screening the M13 pIII linear 12-mer random peptide display libraries with anti-GAD antibodies
The mouse N-MoAb or C-MoAb, and the rabbit C-pc antibody, were coated onto Nunc immunotubes essentially as described above. The tubes were blocked with BSA solution and washed. The M13 pIII linear 12-mer library was obtained commercially (New England Biolabs, UK). The phages were added to the antibody-coated immunotubes (about 2 × 1011 plaque-forming units per tube) in TBS-0·1% Tween-20 and incubated at 4°C for 30 min. The tubes were washed extensively and 1 ml of elution buffer (0·2 m glycine-HCl pH 2·2, 0·1% BSA) was added and incubated at room temperature for 10 min. The eluate was then neutralized with 1 m Tris-HCl pH 9·1 and added to early log phase E. coli 2537 and incubated at 37°C for 4·5 h for amplification. The ampified phages were concentrated and purified by repeated precipitation with one-sixth volume of 20% polyethylene glycol in 2·5 m NaCl (PEG/NaCl). The enriched phages were subjected to two further rounds of selection, as above.
Selected M13 phage clones were screened on immunoblots for specific reactivity with the selecting antibodies, essentially as described above for the T7 phage clones. Antibody-specific phage clones, which developed as blue spots on the membranes, were selected, mixed with 1 ml early log phase E. coli ER 2537 and incubated at 37°C for 4·5–5 h for amplification. Phage DNA was purified and then sequenced in an ABI PRISM 310 Genetic Analyser (Applied Biosystems).
Model of GAD65
The pdf files containing the coordinates of the three-dimensional model of GAD65 [16] were kindly provided by Professor S. Baekkeskov (University of California, San Francisco) and were viewed with the Swiss-PdbViewer version 3·7b2.
RESULTS
Binding of serum antibodies to N-terminal and C-terminal peptides of GAD65
Sera from GADA+ patients with SMS or APS-II were tested in ELISA for binding to linear synthetic peptides representing the 20 N-terminal amino acids or 14 C-terminal amino acids of GAD65 (described in Materials and Methods). In order to determine reactivity with the N-terminal peptide, patients’ sera were selected initially on the basis of giving an optical density (O.D.) reading for binding to the N-terminal peptide, which was at least three standard deviations above the mean O.D. given by sera of normal controls. Specific binding to either the N-terminal peptide or C-terminal peptide was determined from the difference in O.D. (δO.D.) given by the O.D. for binding to the relevant peptide minus the O.D. for binding to an irrelevant control peptide (described in Materials and Methods). Positive binding was taken as a δO.D. at least three standard deviations greater than the ΔO.D. of normal control sera. On this basis, a minority of SMS and APS II sera contained antibodies that bound specifically to the N-terminal and/or C-terminal peptides: these findings are summarized in Table 1 and exemplified in Fig. 1. Thus, a single non-diabetic SMS patient's serum showed specific binding with both peptides (Table 1; Fig. 1a,c). Four sera from non-diabetic APS-II patients that were positive for antibodies to the N-terminal peptide (Table 1; Fig. 1b) were not tested for reactivity with the C-terminal peptide, but the one APS-II serum positive for C-terminal peptide reactivity (Table 1; Fig. 1c) was not reactive with the N-terminal peptide. The diabetic APS-II patients were tested for reactivity with the N-terminal peptide only: two of these were positive (Table 1; Fig. 1b). Our finding of antibodies in some (but not all) patients specific to linear epitopes within the terminal amino acid sequences at either end of GAD65 is consistent with previous reports [7,10,11,13–15 ].
Table 1.
Binding of serum antibodies to N- and C-terminal peptides of GAD
| Patient group | Binding to N-term. Peptides: no. of + ve (no. of tested) | Binding to C-term. Peptides: no. of + ve (no. of tested) |
|---|---|---|
| SMS | 1a (3b) | 1a (4b) |
| APS II (non-DM) | 4 (19) | 1c (6) |
| APS II (DM) | 2 (12) | n.d.d |
The same non-diabetic SMS patient (DH) was positive for antibodies to N- and C-terminal peptides.
One of the SMS patients (LB) was diabetic.
This non-diabetic APS II patient, who was positive for antibodies to the C-terminal peptide, was not positive for antibodies to the N-terminal peptide.
Not done.
Fig. 1.
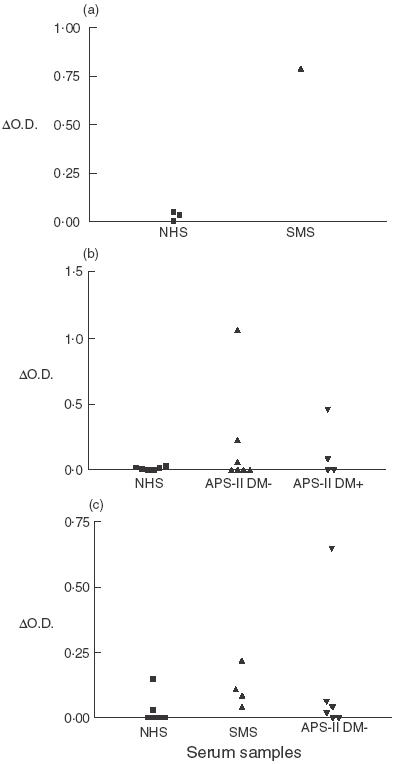
Examples of serum antibodies binding to N- and C-terminal peptides of GAD, expressed as O.D. of binding to the relevant peptide minus the O.D. of binding to an irrelevant peptide (δO.D.). (a) and (b) show reactivity with the N-terminal peptide, and (c) shows reactivity with the C-terminal peptide. (DM- and DM+ indicate non-diabetic and diabetic APS-II patients, respectively.)
Binding of monoclonal & polyclonal antibodies to the N- and C-termini of GAD
The antigenic nature of the terminal regions of GAD was probed with commercially available mouse monoclonal and rabbit polyclonal antibodies that react with N-terminal and C-terminal linear determinants (as described in Materials and Methods).
The results in Table 2a indicate that the three mouse MoAbs showed similar reactivities with a semipurified, detergent-free preparation of rat brain GAD (rbGAD) coated onto the wells of ELISA plates (direct ELISA). Isotype-matched MoAbs of irrelevant specificities did not react similarly with rat brain homogenate. The rabbit C-pc antibody also reacted strongly with rbGAD in direct ELISA (Table 2b), as did the GAD-specific antibodies of the C-pc antibody preparation which had been affinity-purified by binding to, and elution from, solid-phase rbGAD (Table 2b).
Table 2.
Interaction of monoclonal and polyclonal antibodies with solid-phase GAD
| Dilution | O.D.405 | |
|---|---|---|
| (a) Binding of mouse MoAbs to rbGAD by direct ELISA | ||
| MoAb | ||
| N-terma | 1 : 500 | 0·122 |
| C-terma | 1 : 1000 | 0·187 |
| GAD-6b | 1 : 1000 | 0·177 |
| (b) Binding of rabbit polyclonal antibodies (C-pc antibody) to rbGAD by direct ELISA | ||
| Preparation | ||
| Ig enriched fractionc | 1 : 800 | 0·514 |
| Affinity purified fractiond | 1 : 500 | 0·253 |
| Affinity depleted fractione | 1 : 500 | 0·090 |
Partially purified immunoglobulin preparation from hybridoma culture supernatant.
Purified immunoglobulin: stock concentration 1 mg/ml.
Partially purified immunoglobulin precipitated from serum with caprylic acid and ammonium sulphate.
Eluted after binding to rbGAD coupled to Sepharose 4B.
Effluent not bound by rbGAD coupled to Sepharose 4B.
In contrast to their similar interactions with GAD bound to plastic, the MoAbs showed differential reactivities with fluid-phase rbGAD in a capture ELISA (Table 3a). The MoAbs were bound to the wells of ELISA plates (at the concentrations which showed similar reactivities in the direct ELISA) and then exposed to rbGAD in solution. The GAD bound by the MoAbs was then detected by binding GADA of SMS sera followed by enzyme-conjugated anti-human immunoglobulins. The fluid-phase GAD was captured effectively by GAD-6 and the N-MoAb, but was bound relatively poorly by the C-MoAb (Table 3a). Although the poor reactivity of the C-MoAb under these conditions was unexpected, the rabbit C-pc antibody behaved similarly in the capture ELISA in that neither the original immunoglobulin preparation nor the GAD-specific affinity purified antibodies bound fluid-phase rbGAD (Table 3b).
Table 3.
Interaction of monoclonal and polyclonal antibodies with fluid-phase GAD
| O.D.405 for binding of detection antibodies | |||||
|---|---|---|---|---|---|
| Capture antibody | Diln | SMS (DH) | SMS (PM) | SMS (LB) | Normal control |
| Capture of rbGAD by mouse MoAb | |||||
| GAD-6 | 1 : 1000 | 0·652 | 0·696 | 0·364 | 0·000 |
| N-term | 1 : 500 | 1·677 | 1·598 | 1·477 | 0·008 |
| C-term | 1 : 1000 | 0·104 | 0·110 | 0·055 | 0·009 |
| Capture of rbGAD by rabbit polyclonal antibodies (C-pc antibody) | |||||
| GAD-6 | 1 : 1000 | 0·759 | 0·005 | ||
| Ig enr. rabb fr.a | 1 : 500 | 0·031 | 0·020 | ||
| Aff. pur. rabb. fr.b | 1 : 500 | 0·031 | 0·073 | ||
Immunoglobulin-enriched fraction of rabbit C-pc antibody.
Affinity purified rabbit C-pc antibody eluted after binding to rbGAD coupled to Sepharose 4B.
The N-MoAb and GAD-6 are specific for GAD65, whereas the C-MoAb and C-pc antibody recognize both GAD65 and GAD67 because the C-terminal sequences of these two isoforms of GAD are identical. The rbGAD preparation contains both GAD65 and GAD67. However, the poor performance of the C-MoAb and C-pc antibody in the capture ELISA cannot be explained by preferential capture of GAD67 rather than GAD65 because staining of the rbGAD preparation on Western blots with the C-MoAb or C-pc antibody showed the rbGAD to contain a higher proportion of GAD65 than GAD67 (data not shown).
As discussed above, the rbGAD was prepared and stored in detergent-free solution, and it is not bound by the C-MoAb or C-pc antibody in the capture ELISA; however, the rbGAD is recognized by these antibodies when it is bound to plastic in the direct ELISA (mildly denaturing conditions), or bound to nitrocellulose in the Western blotting assay (strongly denaturing conditions). We therefore investigated the effects of mild denaturation of the rbGAD with 0·2% Triton X-100 on its recognition by the C-pc antibody in a capture ELISA (Table 4). The N-MoAb was bound to the wells of ELISA plates to capture GAD65 from the Triton-treated or untreated rbGAD preparations. Antibodies in SMS serum reacted similarly with the untreated and Triton-treated captured GAD. The C-pc antibody showed negligible binding to the untreated captured GAD, but bound well to the Triton-treated captured GAD (Table 4). Thus, mild denaturation of GAD65 appeared to be necessary to expose the epitope(s) of the C-terminus-specific antibodies.
Table 4.
Binding of rabbit C-pc antibody to denatured GAD in capture ELISAa
| O.D.405 for binding of | ||
|---|---|---|
| GAD treatment | C-pc antibody | SMS (DH) |
| Untreated | 0·069 | 0·664 |
| 0·2% Triton X-100 | 0·561 | 0·553 |
GAD was captured on the solid-phase via the N-MoAb used at a dilution of 1/1000.
Epitope mapping the N-MoAb using random peptide phage-display libraries
To understand the different reactivities of N- and C-terminus-specific antibodies with native and denatured GAD65, the amino acid residues that contribute to the epitopes of the antibodies were determined by selecting phages that expressed peptides, which were bound by the antibodies, from random peptide libraries of T7 and M13 phages.
An M13 phage library expressing linear 12-mer peptide inserts on the gene III protein was screened by three rounds of immunopanning with the N-MoAb. A T7 phage peptide library expressing linear 9-mer peptide inserts was also screened with the N-MoAb in four rounds of immunopanning. In each case, the specific plaques from the final round were further selected for binding of N-MoAb in an immunoblotting assay. The DNA inserts of selected phage clones were sequenced, and the corresponding amino acid sequences were aligned with each other to highlight common motifs; they were then aligned with the relevant portion of GAD-65 (amino acids 4–11) to show the sequence homologies (Fig. 2).
Fig. 2.

Sequences of peptides selected by N-MoAb from an M13 pIII linear 12-mer phage peptide library (a), or a T7 linear 9-mer phage peptide library (b). The number assigned to each clone is indicated on the left. Amino acid homologies (including conservative substitutions) between peptides are indicated by vertical lines; amino acid homologies with the relevant section of GAD65 sequence (shown above the peptide sequences) are indicated in bold. *Indicates the position of a stop codon in the DNA insert encoding the peptide.
The peptides selected by the N-MoAb from the M13 library showed homologies with each other and with the sequence of GAD65 between amino acids 4–11 (Fig. 2a), as did the peptides selected from the T7 library (Fig. 2b). Indeed, the consensus from the two sets of sequences indicates that all these amino acids contribute to the epitope except for Phe8, which was not represented in any of the peptides. Thus, the consensus epitope for the N-MoAb is PGSG-WSF, spanning residues 4–11 of GAD-65. Stop codons were found at the amino acid position 9 of all the inserts selected from the T7 library. This could be because the N-MoAb binds only when the insert is at the extreme C-terminus of the surface coat protein, or because the insert is unstable within the surface coat protein, and is therefore not expressed except at the extreme C-terminus.
Epitope mapping the C-MoAb using a random peptide phage-display library
The M13 phage pIII linear 12-mer library was screened with the C-MoAb as described above for the N-MoAb. The DNA inserts of 15 selected clones were sequenced and the corresponding peptides aligned with each other and with the relevant portion of GAD65 (amino acids 572–582) (Fig. 3). The consensus epitope for the C-MoAb thus defined was FLI-EIE-L spanning amino acids 573–581 of GAD65.
Fig. 3.

Sequences of peptides selected by C-MoAb from an M13 pIII linear 12-mer phage peptide library. The number assigned to each clone is indicated on the left. Amino acid homologies (including conservative substitutions) between peptides are indicated by vertical lines; amino acid homologueies with the relevant section of GAD65 sequence (shown above the peptide sequences) are indicated in bold.
Epitope mapping the C-pc antibody using random peptide phage-display libraries
The rabbit C-pc antibody antiserum was used to screen the M13 phage pIII linear 12-mer random peptide library, and a T7 9-mer library that was designed to have peptides constrained by a disulphide bridge formed by cysteines at the ends of the inserts.
The DNA inserts of four selected M13 clones were sequenced and the corresponding peptides aligned with each other and with the relevant portion of GAD65 (amino acids 572–582) (Fig. 4a). These alignments suggest a concensus epitope of DFLIEEIE spanning amino acids 572–579 of GAD65.
Fig. 4.

Sequences of peptides selected by C-pc antibody from an M13 pIII linear 12-mer phage peptide library (a), or a T7 constrained 9-mer phage peptide library (b). The number assigned to each clone is indicated on the left. Amino acid homologueies (including conservative substitutions) between peptides are indicated by vertical lines; amino acid homologies with the relevant section of GAD65 sequence (shown above the peptide sequences) are indicated in bold. *Indicates the position of a stop codon in the DNA insert encoding the peptide.
The DNA inserts of six of the selected T7 clones aligned with the GAD65 sequence (Fig. 4b) and suggested a consensus epitope of GQDL, spanning amino acids 582–585 of GAD65. This is different from the epitope indicated by the peptides selected from the M13 library discussed above (amino acids 572–579). However, this is consistent with the polyclonal nature of the antibodies in the C-pc antibody rabbit antiserum that was raised against the peptide representing amino acids 572–585 of GAD65, with different antibodies recognizing distinct epitopes within the peptide.
DISCUSSION
A recent comprehensive study demonstrated convincingly that the major linear (as opposed to conformational) epitopes of GAD recognized by autoantibodies in the sera of APS-II, SMS and type 1 Dm patients occur in the N-terminal and C-terminal regions, and that the antibodies to these epitopes arise more frequently in SMS and APS-II than in type 1 Dm[7]. This is consistent with previous studies [11,13–15], and also with the findings of the present study. Thus, we found that antibodies specific to linear epitopes within peptides representing amino acids at the N-terminus or C-terminus of GAD65 can be detected in a minority of patients with SMS or APS-II. We reasoned that the heterogeneity between patients in the occurrence of these antibodies may be because it is only in a minority of patients that the processes required for exposure of the linear epitopes in GAD65 occur to a sufficient extent to facilitate recognition by B cells specific for the epitopes.
It is widely recognized that immunoprecipitation assays with radio-labelled GAD are the most sensitive and specific for detecting high-affinity serum antibodies to GAD [29]. However, the most reliable of these employ recombinant in vitro translated GAD, whereas we used natural GAD that should avoid any possible epitope modifications associated with the way GAD folds during in vitro translation. It was therefore appropriate for us to use ELISAs in this context. We were surprised that the antibodies specific to the C-terminus of GAD (C-MoAb and C-pc antibody) did not bind GAD efficiently in the capture ELISA as well as the direct ELISA. This raised the possibility that the conformation of the C-terminus of GAD65 differs in the fluid phase (or when bound by the N-MoAb as a capture antibody) compared with when it is bound to plastic. Presumably, in the former situations GAD is in its native conformation, whereas it may undergo partial denaturation as a result of adherence to plastic. Consistent with this, we observed that deliberate mild denaturation of GAD with 0·2% Triton X-100 greatly enhanced reactivity with C-terminus-specific antibodies. However, a number of other possible explanations should also be considered. First, it is possible that the C-terminus-specific antibodies bind to a fragment of GAD that contains no other epitopes, and that this fragment is not formed in the presence of Triton. This seems unlikely because much of the reactivity of SMS sera is directed at the C-terminal region of GAD65, and this is not affected by the presence or absence of Triton. A second possibility is that the GAD in the rat brain preparation forms complexes (with GAD or other molecules) and that this masks the epitope(s) for the C-terminus-specific antibodies. Western blotting experiments have shown that the rbGAD preparation does contain high molecular weight forms of GAD, but the C-pc antibody recognizes these in addition to the monomeric GAD in the preparation [30]. Indeed, GAD65 is thought to occur mainly as dimers [2], but the proposed site of dimerization involves the middle region of GAD (amino acids 201–461) [19] which would not interfere with the binding of C-terminus-specific antibodies. Furthermore, it also seems unlikely that other molecules, either proteins or lipids, could interact with GAD in such a way as to interfere with the binding of C-terminus-specific antibodies without affecting the interactions with GAD-6 or SMS antibodies. A third possibility is that the C-terminus is normally buried within the native structure of GAD65, and is exposed by mild denaturation of the molecule. However, this is inconsistent with the model of the C-terminal region of GAD65 proposed by Schwartz et al. on the basis of homology with dialkylglycine decarboxylase [19].
In the GAD65 model by Schwartz et al., amino acids 568–582 form an α-helix with a hydrophilic face exposed on the surface of GAD, and an opposite hydrophobic face that interacts with internal β-pleated sheets [19] (Fig. 5b). We believe that this α-helical secondary structure of the C-terminus of GAD65 may explain the lack of reactivity of the C-MoAb and C-pc antibody with GAD in its native conformation. Both of these C-terminus-specific reagents react with the linear peptide representing residues 572–585 of GAD65, and the peptides selected from the phage display libraries indicated the probable contact residues of these antibodies (Figs 3 and 4). However, as shown in Fig. 5a, in an α-helical conformation these amino acids are distributed all the way around the helix and so could not interact simultaneously with an antibody combining site. In the three-dimensional model of the C-terminal region of GAD65, the only contact residues for C-MoAb and/or C-pc antibody that are surface exposed and reasonably accessible for antibody binding are Asp572, Phe573, Ile575, Glu576 and Glu579 (Fig. 5b). These amino acids have surface accessibilities ranging from about 5% to 30% (indicated in Fig. 5a), as calculated by the Swiss-PdbViewer programme version 3·7b2. By contrast, residues Leu574, Glu577, Ile578 and Leu581 have accessibilities of <4% (Fig. 5a), and are relatively buried in the C-terminal model shown in Fig. 5b. Furthermore, the rigid α-helical native structure may not allow appropriate orientation of the amino acid side chains for interaction with C-MoAb and C-pc antibody. Thus, these antibodies may not interact with GAD unless the helix is denatured. In addition, as indicated in Fig. 4b, some antibodies in the C-pc antibody preparation recognize the C-terminal four amino acids Gly582, Gln583, Asp584 and Leu585 that have accessibilities of about 4%, 35%, 10% and 40%, respectively, in the model of the C-terminal region of GAD65 (Fig. 5b). The Gly582 appears to be an important contact residue, despite its poor accessibility in the model of the native structure, as it is represented in all the selected peptides (Fig. 4b). Once again, therefore, the native structure may need to be denatured to expose this amino acid. Thus, the C-MoAb and C-pc antibody may not bind effectively to the native form of the C-terminus of GAD65, but do bind when this is partially denatured (linearized) when GAD is bound directly to plastic or exposed to 0·2% Triton X-100. Furthermore, it seems likely that the C-terminus of GAD-65 must undergo denaturation in vivo in order to facilitate recognition by B cells and the generation of antibodies specific for linear epitopes in the C-terminal sequence, as found in a minority of patients (Table 1 and Fig. 1c).
Fig. 5.
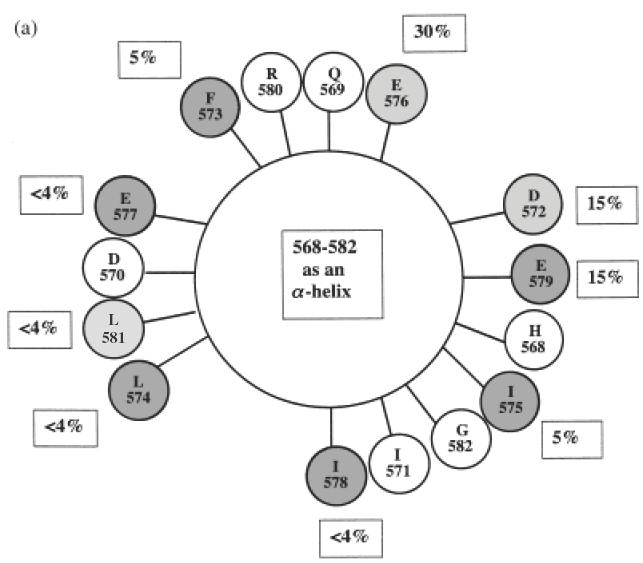
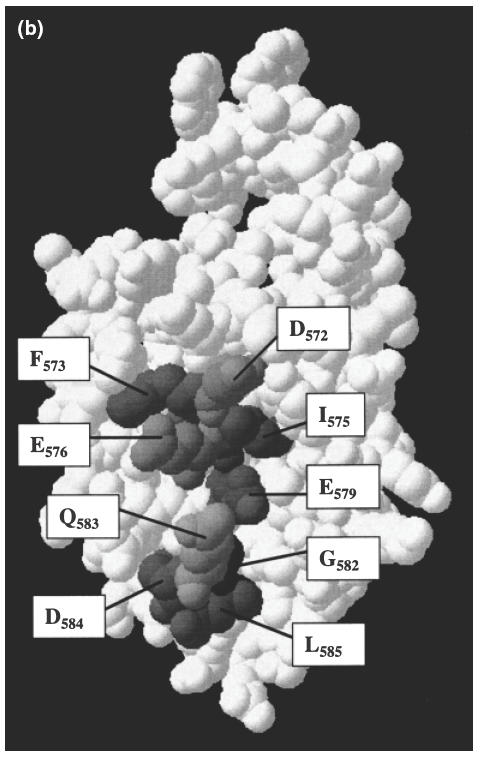
(a) Helix wheel projection of residues 568–582 of GAD65, predicted by Schwartz et al. [19]. The shading of the residues indicates the involvement of each amino acid in the epitopes of C-MoAb and/or C-pc antibody as predicted by the selection of phage-displayed peptides by these antibodies (see Figs 3 and 4), as follows: no shading, no involvement; light shading, C-MoAb epitope only; medium shading, C-pc antibody epitope only; dark shading, both C-MoAb and C-pc antibody epitopes. Percentages indicate residue accessibility within the three-dimensional model of this region of GAD65 (see Fig. 5b) as predicted by the Swiss-PdbViewer version 3·7b2. (b) Three-dimensional model of the C-terminal region of GAD65 [19] shaded to indicate accessible amino acids recognized by C-MoAb and C-pc antibody.
The N-MoAb recognizes a linear epitope in a peptide representing residues 4–17 of GAD65, and our screening of the phage peptide display libraries localized this epitope more precisely to the sequence PGSG-WSF spanning amino acids 4–11 of GAD65 (Fig. 2). In contrast to the C-MoAb and C-pc antibody, the N-MoAb did interact with its epitope in native GAD65 in the capture ELISA, as well as binding mildly denatured GAD in the direct ELISA or following Triton treatment. This suggests that the N-terminal sequence is linear and surface exposed in the native structure of GAD-65. Indeed, it is unlikely that this sequence forms an α-helical secondary structure because this would place the proposed contact residues for the N-MoAb all the way round the α-helix, so that they could not interact simultaneously with the N-MoAb's combining site (Fig. 6). Consistent with this, Schwartz et al. predict that the first α-helix in GAD65 spans amino acids 26–48, i.e. further from the N-terminus than the epitope recognized by the N-MoAb [19]. Furthermore, although it has not yet been possible to model the three-dimensional structure of the N-terminal region of GAD65, our studies provide evidence for the surface exposure of amino acids in the region of residues 4–11.
Fig. 6.
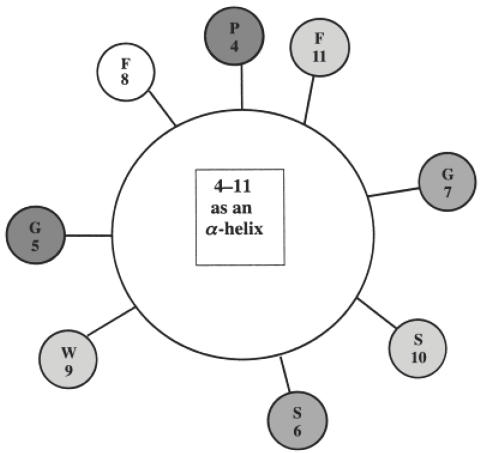
Helix wheel projection of residues 4–11 of GAD65. The shading of the residues indicates the involvement of each amino acid in the epitope of N-MoAb as predicted by the selection of phage-displayed peptides by N-MoAb, as follows: no shading, no involvement; light shading, predicted by selected M13 pIII linear 12-mer peptides; medium shading, predicted by selected T7 linear 9-mer peptides; dark shading, predicted by both selected M13 linear 12-mer peptides and T7 linear 9-mer peptides.
This therefore raises the question of why antibodies specific to N-terminal linear epitopes arise in only a minority of SMS and APS patients if the epitopes are naturally surface exposed and denaturation of the GAD65 structure is not necessary for their generation. It may be relevant that, in vivo, GAD65 is bound reversibly to membranes via sites in the N-terminal region [2]. We suggest that this would mask N-terminal epitopes from B cell recognition unless the GAD was released from the membranes.
We thus propose that different mechanisms may be involved in exposing linear epitopes at the N-terminus and C-terminus of GAD65 for recognition by autoreactive B cells and the generation of autoantibodies. The C-terminus is surface exposed in native GAD65, but its α-helical secondary structure must be denatured to generate linear epitopes. The C-terminus may be particularly susceptible to such denaturation compared with other, more constrained internal regions of GAD65. In contrast, the N-terminus is both surface exposed and linear in the native structure, but may be masked by membrane interactions, which must be broken to facilitate recognition by B cells. In both cases, however, it may be only in a minority of patients that the pathophysiological processes that denature GAD65 and/or release it from its membrane interactions, occur to a sufficient extent for B cell recognition and antibody generation to the linear epitopes thus exposed.
Acknowledgments
This work was supported by grants from Diabetes UK and the Special Trustees for Nottingham University Hospitals. T.A.M.A. Al-bukhari was supported by a scholarship from the Saudi Arabian Cultural Bureau. We are very grateful to Professor S. Baekkeskov (University of California, San Francisco) for kindly providing the pdf files containing the coordinates of the three-dimensional model of GAD65, and to Dr M. R. Christie (King's College London) for his help. We thank Ms S. Hyde for technical assistance in the preparation of rbGAD. We are grateful to the following for supplying sera from SMS patients: Dr G. Lennox and Dr A. P. Nisbett (University Hospital Nottingham), Dr H. Willison (Southern General Hospital, Glasgow) and Dr B. R. F. Lecky (The Walton Centre, Liverpool).
REFERENCES
- 1.Leslie RDG, Atkinson MA, Notkins AL. Autoantigens IA-2 and GAD in type 1 (insulin-dependent) diabetes. Diabetologia. 1999;42:3–14. doi: 10.1007/s001250051105. [DOI] [PubMed] [Google Scholar]
- 2.Kanaani J, Lissin D, Kash SF, Baekkeskov S. The hydrophilic isoform of glutamate decarboxylase, GAD67, is targeted to membranes and nerve terminals independent of dimerization with the hydrophilic membrane-anchored isoform, GAD65. J Biol Chem. 1999;274:37200–9. doi: 10.1074/jbc.274.52.37200. [DOI] [PubMed] [Google Scholar]
- 3.Bonifacio E, Lampasona V, Bernasconi L, Ziegler A-G. Maturation of the humoral autoimmune response to epitopes of GAD in preclinical childhood type 1 diabetes. Diabetes. 2000;49:202–8. doi: 10.2337/diabetes.49.2.202. [DOI] [PubMed] [Google Scholar]
- 4.Solimena M, Folli F, Denis-Donini S. Autoantibodies to glutamate decarboxylase in a patient with stiff man syndrome, epilepsy and type 1 diabetes mellitus. N Engl J Med. 1988;318:1012–20. doi: 10.1056/NEJM198804213181602. [DOI] [PubMed] [Google Scholar]
- 5.Baekkeskov S, Aanstoot HJ, Christgau S, et al. Identification of the 64k autoantigen in insulin-dependent diabetes as the GABA synthesizing enzyme glutamic acid decarboxylase. Nature. 1990;347:151–6. doi: 10.1038/347151a0. [DOI] [PubMed] [Google Scholar]
- 6.Christie MR, Genovese S, Cassidy D, et al. Antibodies to islet 37k antigen, but not to glutamate decarboxylase, discriminate rapid progression to IDDM in endocrine autoimmunity. Diabetes. 1994;43:1254–9. doi: 10.2337/diab.43.10.1254. [DOI] [PubMed] [Google Scholar]
- 7.Sohnlein P, Muller M, Syren K, The Childhood Diabetes in Finland Study Group, Richter W. Epitope spreading and a varying but not disease-specific GAD65 antibody response in type 1 diabetes. Diabetologia. 2000;43:210–7. doi: 10.1007/s001250050031. [DOI] [PubMed] [Google Scholar]
- 8.Falorni A, Ackefors M, Carlberg C, et al. Diagnostic sensitivity of immunodominant epitopes of glutamic acid decarboxylase (GAD65) autoantibodies in childhood IDDM. Diabetologia. 1996;39:10911098. doi: 10.1007/BF00400659. [DOI] [PubMed] [Google Scholar]
- 9.Daw K, Powers AC. Two distinct glutamic acid decarboxylase autoantibody specificities in IDDM target different epitopes. Diabetes. 1995;44:216–20. doi: 10.2337/diab.44.2.216. [DOI] [PubMed] [Google Scholar]
- 10.Butler MH, Solimena M, Dirkx R, Jr, Hayday A, De Camilli P. Identification of a dominant epitope of glutamic acid decarboxylase (GAD-65) recognized by autoantibodies in stiff-man syndrome. J Exp Med. 1993;178:2097–106. doi: 10.1084/jem.178.6.2097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kim J, Namchuk M, Bugawan T, et al. Higher autoantibody levels and recognition of a linear NH2-terminal epitope in the autoantigen GAD65, distinguish stiff-man syndrome from insulin-dependent diabetes mellitus. J Exp Med. 1994;180:595–606. doi: 10.1084/jem.180.2.595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Li L, Hagopian WA, Brashear HR, Daniels T, Lernmark A. Identification of autoantibody epitopes of glutamic acid decarboxylase in stiff-man syndrome patients. J Immunol. 1994;152:930–4. [PubMed] [Google Scholar]
- 13.Daw K, Ujihara N, Atkinson M, Powers AC. Glutamic acid decarboxylase autoantibodies in stiff-man syndrome and insulin-dependent diabetes mellitus exhibit similarities and differences in epitope recognition. J Immunol. 1996;156:818–25. [PubMed] [Google Scholar]
- 14.Mauch L, Abney CC, Berg H, Scherbaum WA, Liedvogel B, Northemann W. Characterization of a linear epitopes within the human pancreatic 64-kDa glutamic acid deacarboxylase and its autoimmune recognition by sera from insulin-dependent diabetes mellitus patients. Eur J Biochem. 1993;212:597–603. doi: 10.1111/j.1432-1033.1993.tb17698.x. [DOI] [PubMed] [Google Scholar]
- 15.Rharbaoui F, Granier C, Kellou M, et al. Peptide specificity of high-titer anti-glutamic acid decarboxylase (GAD) 65 autoantibodies. Immunol Lett. 1998;62:123–30. doi: 10.1016/s0165-2478(98)00036-4. [DOI] [PubMed] [Google Scholar]
- 16.Ziegler B, Strebelow M, Rjasanowski I, Schlosser M, Ziegler M. A monoclonal antibody-based characterization of autoantibodies against glutamic acid decarboxylase in adults with latent autoimmune diabetes. Autoimmunity. 1998;28:61–8. doi: 10.3109/08916939809003868. [DOI] [PubMed] [Google Scholar]
- 17.Davenport C, Radford PM, Al-bukhari TAMA, Lai M, Bottazzo G-F, Todd I. Heterogeneity in the occurrence of a subset of autoantibodies to glutamic acid decarboxylase in autoimmune polyendocrine patients with islet cell antibodies. Clin Exp Immunol. 1998;111:497–505. doi: 10.1046/j.1365-2249.1998.00526.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bjork E, Velloso LA, Kampe O, Karlsson FA. GAD-autoantibodies in insulin-dependent diabetes mellitus, stiff-man syndrome and autoimmune polyendocrine syndrome type I recognise different epitopes. Diabetes. 1994;43:161–5. doi: 10.2337/diab.43.1.161. [DOI] [PubMed] [Google Scholar]
- 19.Schwartz HL, Chandonia J-M, Kash SF, et al. High-resolution autoreactive epitope mapping and structural modelling of the 65 kDa form of human glutamic acid decarboxylase. J Mol Biol. 1999;287:983–99. doi: 10.1006/jmbi.1999.2655. [DOI] [PubMed] [Google Scholar]
- 20.Richter W, Shi Y, Baekkeskov S. Autoreactive epitopes defined by diabetes-associated human monoclonal antibodies are localized in the middle and C-terminal domains of the smaller form of glutamate decarboxylase. Proc Natl Acad Sci USA. 1993;90:2832–6. doi: 10.1073/pnas.90.7.2832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Syren K, Lindsay L, Stoehrer B, et al. Immune reactivitiy of diabetes-associated human monoclonal autoantibodies defines multiple epitopes and detects two domain boundaries in glutamate decarboxylase. J Immunol. 1996;157:5208–14. [PubMed] [Google Scholar]
- 22.Powers AC, Bavik K, Tremble J, Daw K, Scherbaum WA, Banga JP. Comparative analysis of epitope recognition of glutamic acid decarboxylase (GAD) by autoantibodies from different autoimmune disorders. Clin Exp Immunol. 1999;118:349–56. doi: 10.1046/j.1365-2249.1999.01030.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Myers MA, Davies JM, Tong JC, et al. Conformational epitopes on the diabetes autoantigen GAD65 identified by peptide phage display and molecular modelling. J Immunol. 2000;165:3830–8. doi: 10.4049/jimmunol.165.7.3830. [DOI] [PubMed] [Google Scholar]
- 24.Chang Y, Gottlieb D. Characterization of the proteins purified with monoclonal antibodies to glutamic acid decarboxylase. J Neurosci. 1988;8:2123–30. doi: 10.1523/JNEUROSCI.08-06-02123.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ujihara N, Daw K, Gianani R, Boel E, Yu L, Powers AC. Identification of glutamic acid decarboxylase autoantibody heterogeneity and epitope regions in type 1 diabetes. Diabetes. 1994;43:968–75. doi: 10.2337/diab.43.8.968. [DOI] [PubMed] [Google Scholar]
- 26.Ziegler B, Augstein P, Luhder F, et al. Monoclonal antibodies specific to the glutamic acid decarboxylase 65 kDa isoform derived from a non-obese diabetic (NOD) mouse. Diabetes Res. 1994;25:47–64. [PubMed] [Google Scholar]
- 27.Ziegler B, Augstein P, Schroder D, et al. Glutamate decarboxylase (GAD) is not detectable on the surface of rat islet cells examined by cytofluorometry and complement-dependent antibody-mediated cytotoxicity of monoclonal GAD antibodies. Horm Metab Res. 1996;28:11–5. doi: 10.1055/s-2007-979121. [DOI] [PubMed] [Google Scholar]
- 28.Davenport C, Lovell H, James RFL, Todd I. Brain-reactive autoantibodies in BB/d rats do not recognize glutamic acid decarboxylase. Clin Exp Immunol. 1995;101:127–35. doi: 10.1111/j.1365-2249.1995.tb02288.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Schmidli RS, Colman PG, Bonifacio E. Disease sensitivity and specificity of 52 assays for glutamic acid decarboxylase antibodies. The Second International GADAB Workshop. Diabetes. 1995;44:636–40. doi: 10.2337/diab.44.6.636. [DOI] [PubMed] [Google Scholar]
- 30.Trigwell SM, Radford PM, Page SR, et al. Islet glutamic acid decarboxylase modified by reactive oxygen species is recognized by antibodies from patients with type 1 diabetes mellitus. Clin Exp Immunol. 2001;126:242–9. doi: 10.1046/j.1365-2249.2001.01653.x. [DOI] [PMC free article] [PubMed] [Google Scholar]


