Abstract
It is still unclear why some patients with HIV progress more slowly than others to developing full blown AIDS. In this study using flow cytometry we have investigated the TCRBV repertoire of peripheral blood T lymphocytes in 17 long-term non-progressing HIV patients (LTNP) to determine if there is a biased usage of T cell receptor V gene products. Patients were identified from hospital records and entered into the study. Three colour flow cytometry was used to determine the expression of the TCRBV3S5, BV5S1, BV5S2, BV5S3, BV6S1, BV7S1, BV9, BV11, BV12, BV13, BV14, BV16, BV17, BV18, BV20, BV21S3, BV22, and BV23 by CD8 and CD4 positive cells isolated from the peripheral blood of patients and controls. Increases in the absolute numbers of CD8+ T cells expressing TCRBV2 and 8 were observed in the HIV-LTNP population (P < 0·05 in both cases). No differences were seen in numbers of CD8+ T cells expressing other TCRBV or in any TCRBV within the CD4+ T cell popu-lation. At follow up (1–2 years later), those patients in which CD4 levels were below 500 × 106/l were those initially found to have lower levels of TCRBV8 +ve CD8 cells. A significant increase in the absolute numbers of T cells coexpressing the gamma delta (γδ) T cell receptor and CD8 were also seen in the HIV-LTNP patients compared with controls (P = 0·002). The increase in CD8+ T cells in the HIV-LTNP patients may be interpreted as either an antigen specific, or group of antigen specific responses to viral antigen, or less likely a viral superantigen. A low level of TCRBV8, CD8+ T cells might be predictive of a more rapid disease progression and might indicate a protective role for this population in HIV infected patients. The increase in γδT cells bearing the CD8 coreceptor suggests a role for this cell type in the response to HIV infection.
Keywords: TCR, CD8, cellular immunity, HIV progression
INTRODUCTION
A proportion of HIV infected individuals remain asymptomatic for a long period post infection. These patients have a strong antiviral immune response and appear to have low levels of plasma viral RNA [1]. A skewing of T cell receptor usage in HIV has previously been reported by a number of investigators although many such studies have not reported the relationship of the changes to CD4+ or CD8+ populations. The possibility of a superantigen effect of HIV has been investigated by a number of groups and could explain increases, or decreases of TCRBV fami-lies of T cells found in patients with the disease. Superantigen effects of crude HIV, gp160 and gp120 have also been observed on normal lymphocytes [2,3].
There are conflicting reports regarding the changes in parti-cular TCRBV expressing T cells in HIV infected individuals. For example, a significant reduction in the numbers of lymphocytes expressing TCRBV gene products 14, 15, 16 and 17, in patients with HIV compared with controls was observed by some investigators using PCR analysis [4]. In addition, significant changes in both CD4 and CD8 positive TCRBV populations of T cells from HIV infected patients have also been shown to occur [5,6]. It seems likely that the stage of disease is of importance in determining the altered percentages of T cells bearing certain TCRBV gene products. For example, expansion of certain TCRBV families has been shown in the CD8+ population in the primary T cell response to virus [7] and in infants infected with HIV in utero [8].
In this study we have investigated the T cell receptor usage of both CD4 and CD8 bearing T cells in the peripheral blood of asymptomatic HIV infected persons who have been known to be seropositive for the virus for more than eight years.
PATIENTS AND METHODS
Patients
Patients and controls were recruited from a central London genito-urinary medicine clinic. Patients attending routine HIV clinics were identified as long-term non-progressing (LTNP) from clinical records and were defined as individuals known to have been seropositive for HIV for at least 8 years and having absolute CD4 T cells counts above 500 × 106 cells/l (n = 15)or a CD4% greater than 30% of total lymphocytes (n = 2) within the previous six months. Controls were males attending the routine genito-urinary medicine clinic for an HIV antibody test but were subsequently shown to be antibody negative for the virus.
All of the samples were from homosexual male patients (n = 17) and controls (n = 12). The median age of the patient group was 40·5 years and the range 28–48 years. The median age of the control group was 32 years and the range 26–45 years. At study entry in the patient group the median CD4 count was 530 × 106/l, range 360–1300 (CD4 percentage median 33, range 14–52) and median time since first positive HIV antibody test 9·2 years, range 8 years to 11·5 years. All patients were categorized as asymptomatic. The study was reviewed and approved by the University College London Hospitals ethics committee. All study participants gave written informed consent.
Isolation of peripheral blood mononuclear cells
All peripheral blood samples were received blind and the code remained unbroken until all of the results were determined. Peripheral blood was obtained by venepuncture, collected into mucous heparin and centrifuged over Ficoll hypaque (1·077 g/ml, Nycomed, Oslo, Norway) to isolate the mononuclear cell fraction. The mononuclear cells were incubated for 40 minutes at room temperature with Permeafix (Ortho). Preliminary studies showed that the fixation procedure did not affect the subsequent staining but inactivated the virus.
Staining procedure: three colour flow cytometry
Cells were suspended in 0·01 m phosphate buffered saline (PBS – Sigma, Poole, UK) containing 1% bovine serum albumin and 0·05% sodium azide (staining buffer) and dispensed into 96 U bottomed microtitre plates (Nunc, Merck Ltd, Lutterworth, UK) at 1 × 105 cells per well. The plates were then centrifuged and the supernatant aspirated. The cells were gently resuspended and 20 μl of the appropriate anti TCRBV monoclonal antibody added to the well. All antibodies were pretitrated to give optimum staining. The plate was then incubated for 40 min on ice. The cells were washed twice in staining buffer and then 20 μl of a 1 in 20 dilution of rabbit antimouse immunoglobulin conjugated to fluorescein isothiocyanate (FITC, Dakopatts, Ely, UK) added to each well. After a further incubation for 40 min on ice the cells were again washed twice and 20 μl of a 1 in 10 dilution of normal mouse serum added to each well to block nonspecific binding of the second antibody to unbound sites on the rabbit antimouse FITC antibody. After 40 min on ice the cells were washed again and 10 μl of a 1 in 5 dilution of anti CD4 conjugated to phycoerythrin (PE, Dakopatts) and 5 μl of anti CD8 conjugated to cychrome C (Pharmingen, San Diego, CA, USA) were added to each well. The cells were again washed and resuspended in 0·5% paraformaldehyde in PBS and left in the dark for at least one hour before being analysed by flow cytometry. Monoclonal antibodies used to stain T cell populations were CD3-FITC, CD4-PE (Both Dako) and CD8-Cy (Pharmingen). The monoclonal antibodies used to identify different TCRBV expressed by CD4 and CD8 ± cells were TCRBV2(MPB2/D5), BV3(Jovi 3), BV5S1(Immunol 57), BV5S2 (36213), BV5S3(2D2), BV6S1(CRI 304·4), BV7S1(3GSD5), BV8(JR2), BV9(FIN9), BV11(C21), BV12(VER2·32·1), BV13 (BAM13), BV14(CAS1·1·3), BV16(Tamaya 1·2), BV17(E17.SF3), BV18(BA62·6), BV20(ELL1·4), BV21S3(IG125), BV22 (Immu546), BV23(HUT78·1) – clonal origins are shown in parentheses. Gamma delta T cells were stained with TCRδ1 monoclonal antibodies.
Flow cytometric analysis
All samples were read on a FACScan flow cytometer equipped with a 488-nm argon laser. Compensation of the three fluorochromes was determined for each patient sample and the background fluorescence by appropriate isotype control antibodies IgG1-PE (Dako), IgG1-Cy Pharmingen), IgM (Dako), IgG1 (Dako), IgG2a (Dako) and IgG2b (Dako). At least 7000 events per sample were analysed using WinMDI software.
Statistical analyses
All statistical analyses were performed using the Instat statistical package software. All comparisons were made using the Mann–Whitney-U-test and the resulting P-value adjusted using the Bonferoni correction.
RESULTS
Although there was a significant increase in the CD8+ cells in the HIV-LTNP patients – median, 935 × 106 cells/l, range, 741·4 × 106–1483 × 106 compared to median, 340 × 106 cells/l range, 271·1 × 106–567·6 × 106 for the HIV sero-negative samples P < 0·0006), only absolute numbers of CD8+ T cells using TCRBV2 and 8, but no other tested TCRBV families, were significantly elevated in the HIV-LTNP group compared with controls (Table 1 and Fig. 1). Both the total lymphocyte numbers and CD4+ T cell numbers were comparable between the two groups. When absolute numbers of T cells were determined, no differences in expression of TCRBV by the CD4+ population were seen (data not shown). No differences in the percentages of CD8 positive cells expressing any TCRBV gene product were seen. Longitudinal studies of three control individuals over a period of 5 years showed stability of TCRBV2 expression (7·7 ± 0·2%; 12·1 ± 1·1%; 8·9 ± 1·5%) and TCRBV8 expression (4·9 ± 0·9%; 4·3 ± 0·4%; 2·8 ± 0·5%).
Table 1.
Significant differences in Vβ2 and Vbβ CD8+ cells and γδT cells between controls and HIV-LT patients
| TCR used | CD8 Control median | CD8 Control 95% CL | CD8 HIV-LT median | CD8 HIV-LT 95% CL* | P-value** |
|---|---|---|---|---|---|
| Vβ2 | 13·5+ | 10·63–27·01 | 36·0 | 31·2–58·87 | 0·021 |
| Vβ8 | 14·0 | 11·56–29·14 | 41·0 | 36·73–65·74 | 0·021 |
| γδ | 17·0 | 10·10–26·37 | 80·0 | 55·8–111·19 | 0·002 |
| All CD8 | 340 | 271–567 | 935·0 | 741–1483 | 0·0006 |
confidence limits
P-values from Mann–Whitney test, Bonferoni corrected.
data are expressed as absolute counts × 10−6/l.
Fig. 1.
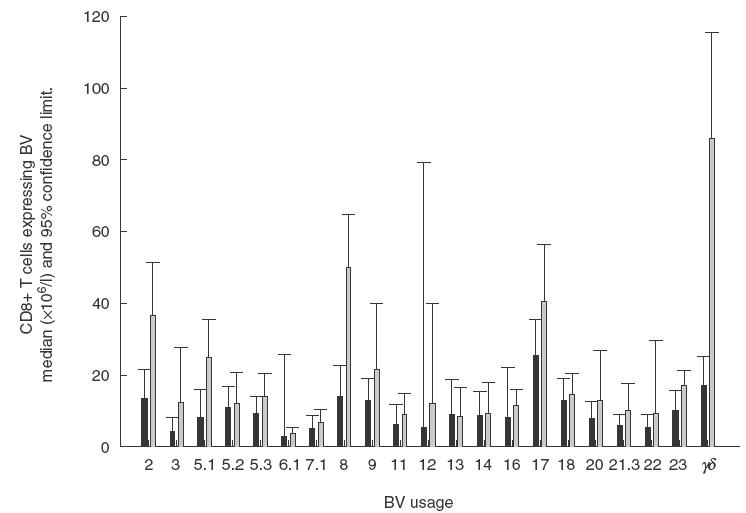
Expression of TCR by CD8 ± T cells from controls (▪) and HIV-LT patients ( ). Data are expressed as absolute median counts with 95% confidence limits (× 10−6/l).
). Data are expressed as absolute median counts with 95% confidence limits (× 10−6/l).
CD4 counts were available in 16 patients on clinical follow up 1–2 years later. The patients were grouped into those whose CD4 counts had remained or dropped below 500 × 106l−1 (n = 5) and those whose counts remained above 500 × 106l−1 (n = 11). Table 2 shows the median absolute counts of CD8+ T cells expressing different Vβ families at initial analysis in relation to CD4 counts at follow up. It is noteworthy that the group with <500 × 106l−1 CD4 counts at follow up had lower levels of CD8+ TCRBV8+ cells at initial analysis than those with >500 × 106l−1 CD4 counts compared with the other TCRBV families. Although it was difficult to evaluate the statistical significance due to the smaller sample size (N = 5) a value of P = 0·07 was obtained indicating a value close to significance at the 5% level.
Table 2.
Relationship between CD8 T cells expressing particular TCRBV families in HIV-LTNP patients with CD4 T cell counts measured at follow up after 2 years
| Initial TCRBV family counts | ||||
|---|---|---|---|---|
| CD4 counts at follow up | Vβ 5·1 | Vβ 7·1 | Vβ 2 | Vβ 8 |
| >500 × 106/l (n = 11) | 3·42* | 0·8 | 6·7 | 6·2 |
| (2·11) | (0·46) | (6·3) | (2·9) | |
| <500 × 106l−1 (n = 5) | 2·46 | 1·03 | 4·08 | 2·4 |
| (1·8) | (1·01) | (2·07) | (2·9) | |
Median absolute CD8 counts are expressed × 10−7/l (± SD).
The gamma delta (γδ) subset of T cells showed a significant difference in the expression of CD8 between the controls and HIV-LTNP patients. There was a 4–5 fold increase in the absolute numbers of γδ T cells coexpressing CD8 in the HIV-LTNP (median,80·0 × 106 cells/l; range, 55·8 × 106–111·2 × 106) compared to 17·0 × 106 cells/l (range, 10·1 × 106–26·3 × 106, P = 0·002; see Table 1).
Plasma HIV RNA levels were not measured at time of study entry but were measured in 16 patients as part of their routine clinical follow up, 14 within 18 months of their study sampling date and 2 within 2–4 years. The median level was 5460 copies/ml, range ≤1000–32 800 (Chiron branch DNA assay version 2), 15 had levels ≤15 000 copies/ml. This level of viral load is supportive of the definition of the patient group as long-term non-progressors.
DISCUSSION
Our data indicate that there are increases in certain TCRBV families in the CD8+ population of T cells, but not CD4+ T cells, in long-term non-progressing HIV patients. This is shown as an increase in absolute numbers of cells expressing certain T cell receptor elements. Increased numbers of CD8+ T cells were seen to bear TCRBV2 and 8. Other reports have noted a decrease in the proportion of cells expressing certain TCRBV families (in this case BV5S1, 12 and 2 [6]). From our study we suggest that this apparent decrease may be due to an increase in the absolute numbers of one of the other families not examined.
It has been reported that there is no change in the expression of T cell receptor usage in patients with HIV infection. For example, Boyer et al. [9] using PCR techniques, showed no differences in the usage of T cell receptor TCRBV products when comparing HIV antibody positive and HIV antibody negative individuals. However, the majority of studies now suggest that there are expansions in the T cell population bearing one or more TCRBV element, and that the expansion is linked to certain HIV proteins [2,3,10], although as yet there is no specific TCRBV family associated with HIV infection [11–13].
The expansion or deletion of particular TCRBV bearing T cells might be related to the stage of disease. In a study of primary HIV infection, Pantaleo et al. [7] showed expansions of CD8+ cells in which the TCRBV usage was restricted in each of six individuals but varied between each subject. Analysis of junctional sequences in the expanded population showed considerable oligoclonality. Similar findings have been observed with infants vertically infected with HIV-1 [8]. The variation of expanded TCRBV populations between patients might be the result of an early response to the virus. It is possible that this early TCRBV expansion, especially of CD4+ T cells, could be advantageous to the virus in that certain TCRBV populations might be more susceptible to viral entry. In fact, it has been documented that HIV replicates more efficiently in ‘normal’ T cells expressing TCRBV12 [14,] than in CD4T cells using other TCRBV families and a decrease in TCRBV12 cells has been observed in HIV patients in some studies [5,6]. In our HIV-LTNP patients we were unable to demonstrate an alteration of this TCRBV segment (data not shown), but in our experiments we were looking much later in the disease process.
A role for HIV superantigens has been supported by a number of studies. In vitro, experiments by Akolar and colleagues suggested that viral antigens, particularly gp160 and gp120, may be capable of acting as superantigens in that these antigens specifically activate T cells bearing certain TCRBV gene products [2,3]. In these experiments, the TCRBV elements shown to be consistently affected were TCRBV2 and 3, although other expansions were seen in individual samples. These proteins (gp160 and gp120 antigens) have more recently been associated with TCRBV ex-pansions in the CD4 population [10]. Gp120 has been reported to have a superantigen activity for a subset of IgVH3 expressing B cells [15] and certain synthetic HIV peptides for both CD4+and CD8+ T cells [16]. However, others have shown no superantigenic activity with HIV proteins [17] and it is now the general consensus that superantigens in HIV infection play little role in T cell expansions in vivo.
Longitudinal studies by others have shown expansions in the CD8 positive population at all stages of the disease [18] but unlike our studies no particular TCRBV was increased.
In our studies, the increase in the absolute numbers of CD8+ T cells bearing particular, common, TCRBV gene products in the peripheral blood of HIV-LTNP patients is not surprising since there was an increase in absolute numbers of CD8 positive T cells. However, what was particularly noteworthy was that only TCRBV 2 and 8 were selectively expanded in the CD8+ T cells, whilst others were decreased, although not significantly. Since LTNP patients have a relatively stable HIV specific effector repertoire of CD8+ T cells which correlates with relatively stable viraemia and CD4+ T cell counts [19], it is possible that the effective cytolytic T cells are present within the TCRBV8 family. Lower levels of these CD8+ TCRBV8+ T cells could therefore be a contributory factor associated with progression. This is consistent with our findings that at follow up 1–2 years later those patients whose CD4 T cell levels had remained or fallen below 500 × 106/l, had lower numbers of circulating CD8, TCRBV8 T cells at initial analysis, compared with those CD4 levels remained above 500 × 106/l. It therefore seems likely that CD8 T cells play a role in controlling progression in both in early and later stages of the disease. Oligoclonal expansions of CD8 positive cells in children vertically infected with HIV have been documented [8,20], although these cells had varied TCRBV usage between individuals. In addition, reported expansions in the peripheral blood of HIV infected children born to HIV positive mothers [9] lasted for approximately 3 months after birth and were postulated to be an early response to the virus. Interestingly, these expansions were also seen in HIV uninfected children born to infected mothers suggesting that the infant has responded to, and cleared the virus. It will be important to determine the levels of specific TCRBV segments bearing CD8 T cells in individual patients, throughout the course of their disease. A recent report by Wilson et al. [21] has shown, using tetrameric complexes, that the expansions of CD8 families seen are antigen specific. It will now be important to study the functional activity of the CD8 TCRBV2 and especially TCRBV8 T cells in the HIV-LTNP patients to determine the possible mechanism(s) by which they could control disease progression.
The influence of HLA on TCRBV usage by the peripheral T cell population is unclear. On the one hand it has been suggested that HLA class I and II does influence the TCRBV repertoire [22] whilst other studies have failed to show a link with either of the HLA genes [23,24]. In this study we did not analyse HLA haplotypes since there were too few patients for statistical analysis.
Our data show a significant increase in the absolute numbers of γδ T cells expressing low levels of CD8, in the peripheral blood of HIV-LTNP patients. Two subsets of circulating γδ T cells, those bearing TCRDV1 and those bearing TCRDV2/GV9, make up over 95% of the γδT cells in the peripheral blood [25]. Both of these γδ cell subsets have been implicated in the immune response against HIV. The TCRDV2 subset has been shown to have a strong lytic and proliferative in vitro response to HIV infected cells [26] with a concommitant production of CC chemokines. The same authors, however, show that this subset is decreased or functionally disabled in patients with HIV [27]. This might represent a common antiviral response since in other viral infections, such as herpes virus, the TCRDV2 population seems to be the responding subset [for review see 28]. Other investigators have shown that the TCRDV1 subset of cells is responsive to HIV [29,30]. Moreover, this expansion has been suggested to be specific for HIV since it is not seen in other viral infections [31]. Another report showed a marked increase in the percentage of γδ T cells and these expressed low levels of CD8 [32]. This is consistent with our data where we found that the level of CD8 expressed by the γδ T cells was lower than the major CD8 population (data not shown). It has been shown that the γδ T cell population in patients with HIV infection has an increase in the expression of activation markers such as HLA-DR and CD38 when compared to control individuals [33]. Wallace et al. show that γδ T cell clones using either TCRDV1 or DV2 isolated from HIV antibody negative individuals have the ability to lyse HIV infected target cells [34] suggesting that both of these populations of cells may be able to respond to virally infected cells without prior exposure to the virus. However, to further confuse the role of γδ T cells, an increase in a new population of γδ T cells bearing both TCRDV2 and DV3 chains has recently been reported in a patient with HIV. Interestingly this new population also bore CD8 [35].
The information above suggests a role for the γδ T cell in combating viral infection. This might be a general antiviral res-ponse, as in the TCRDV2 cell population, or a more specific HIV response by TCRDV1 bearing cells. Our study did not subdivide the γδT cell population of the LTNP patients into its component parts but this might be a worthwhile subject of study. The role of γδ T cells in the HIV-LTNP patients several years after infection with HIV is unclear. It is possible that these CD8+ γδ T cells are helping to maintain the status quo of the chronically infected HIV-LTNP patients. A functional study of γδ T cells in these patients might also shed light on their possible protective role.
Acknowledgments
This work was supported by the Special Trustees of the Middlesex Hospital, London.
REFERENCES
- 1.Cao Y, Qin L, Zhang L, Safrit J, Ho HH. Virologic and immunologic characterization of long-term survivors of human immunodeficiency virus type 1 infection. New Eng J Med. 1995;332:201–8. doi: 10.1056/NEJM199501263320401. [DOI] [PubMed] [Google Scholar]
- 2.Akolar PN, Chirmule N, Gulwani-Akolar B, et al. Vβ-specific activation of T cells by the HIV glycoprotein gp160. Scand J Immunol. 1995;41:487–98. doi: 10.1111/j.1365-3083.1995.tb03597.x. [DOI] [PubMed] [Google Scholar]
- 3.Akolar PN, Gulwani-Akolar B, Silver J. Differential patterns of T-cell receptor BV-specific activation of T cells by gp120 from different HIV strains. Scand J Immunol. 1995;42:598–606. doi: 10.1111/j.1365-3083.1995.tb03702.x. [DOI] [PubMed] [Google Scholar]
- 4.Imberti L, Sottini A, Bettinardi A, Puoti M, Primi D. Selective depletion in HIV infection of T cells that bear specific T cell receptor Vβ sequences. Science. 1991;254:860–2. doi: 10.1126/science.1948066. [DOI] [PubMed] [Google Scholar]
- 5.Hodara VL, Jeddi-Tehrani M, Grunewald J, et al. HIV infection leads to differential expression of T-cell receptor Vβ genes in CD4+ and CD8+ T cells. AIDS. 1993;7:633–8. doi: 10.1097/00002030-199305000-00004. [DOI] [PubMed] [Google Scholar]
- 6.Bansal AS, Green LM, Khoo SH, et al. HIV induces deletion of T cell receptor variable gene product-specific T cells. Clin Exp Immunol. 1993;94:17–20. doi: 10.1111/j.1365-2249.1993.tb05970.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Pantaleo G, Demarest JF, Soudeyns H, et al. Major expansion of CD8+ T cells with a predominant Vβ usage during the primary immune response to HIV. Nature. 1994;370:463–7. doi: 10.1038/370463a0. [DOI] [PubMed] [Google Scholar]
- 8.Halapi E, Gigliotti O, Hodara V, et al. Detection of CD8 T-cell expansions with restricted T-cells receptor V gene usage in infants vertically infected with HIV-1. AIDS. 1996;10:1621–6. doi: 10.1097/00002030-199612000-00005. [DOI] [PubMed] [Google Scholar]
- 9.Boyer V, Smith LR, Ferre F, et al. T cell receptor Vβ repertoire in HIV-infected individuals. lack of evidence for selective Vβ deletion. Clin Exp Immunol. 1993;92:437–41. doi: 10.1111/j.1365-2249.1993.tb03417.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Li Pira G, Oppezzi L, Seri M, et al. Repertoire breadth of human CD4+ T cells specific for HIV gp120 and p66 (primary antigens) or for PPD and tetanus toxoid (secondary antigens) Human Immunol. 1998;59:137–48. doi: 10.1016/s0198-8859(98)00004-4. [DOI] [PubMed] [Google Scholar]
- 11.Cheynier R, Henrichwark S, Wain-Hobson S. Somatic hypermutation of the T cell receptor V beta gene in microdissected splenic white pulps from HIV-1-positive patients. Eur J Immunol. 1998;28:1604–10. doi: 10.1002/(SICI)1521-4141(199805)28:05<1604::AID-IMMU1604>3.0.CO;2-R. [DOI] [PubMed] [Google Scholar]
- 12.King DJ, Amjadi P, Tilling R, et al. ‘Naïve’ and ‘memory’ CD4+ T-cells and T-cell receptor (TCR) V beta repertoire dynamics are independent of the levels of viremia following HIV seroconversion. Immunol Lett. 1999;66:199–206. doi: 10.1016/s0165-2478(98)00158-8. [DOI] [PubMed] [Google Scholar]
- 13.Mion M, Indraccolo S, Feroli F, et al. TCR expression and clonality analysis in peripheral blood and lymph nodes of HIV infected patients. Human Immunol. 1997;57:93–103. doi: 10.1016/s0198-8859(97)00205-x. [DOI] [PubMed] [Google Scholar]
- 14.Dobrescu K, Kabak S, Mehta K, et al. Human immunodeficiency virus 1 reservoir in CD4+ T cells is restricted to certain Vβ subsets. Proc Natl Acad Sci USA. 1995;92:5563–7. doi: 10.1073/pnas.92.12.5563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Neshat MN, Goodglick L, Lim K, Braun J. Mapping the B cell superantigen binding site for HIV-1 gp120 on a V(H)3 Ig. Int Immunol. 2000;12:305–12. doi: 10.1093/intimm/12.3.305. [DOI] [PubMed] [Google Scholar]
- 16.Eick A, Larned J, Jason J. Effects of HIV-1 peptides on T-cell receptor variable beta chain families. Hum Immunol. 2000;61:993–1000. doi: 10.1016/s0198-8859(00)00176-2. [DOI] [PubMed] [Google Scholar]
- 17.Lapatschek MS, Durr S, Sutter G, Wagner H, Miethke T. Functional evaluation of HIV/SIV Nef as superantigen. Virology. 2001;282:329–37. doi: 10.1006/viro.2001.0844. [DOI] [PubMed] [Google Scholar]
- 18.Gorochov G, Neumann AU, Kereveur A, et al. Perturbation of CD8± and CD8+ T cell repertoires during progression to AIDS and regulation of the CD4+ repertoire during antiviral therapy. Nat Med. 1998;4:145–6. doi: 10.1038/nm0298-215. [DOI] [PubMed] [Google Scholar]
- 19.Propato A, Schiaffella E, Vicenzi E, et al. Spreading of HIV-specific CD8 (+) T-cell repertoire in long-term nonprogressors and its role in the control of viral load and disease activity. Hum Immunol. 2001;62:561–76. doi: 10.1016/s0198-8859(01)00245-2. [DOI] [PubMed] [Google Scholar]
- 20.Than S, Kharbanda M, Chitnis V, et al. Clonal dominance patterns of CD8 T cells in relation to disease progression in HIV-infected children. J Immunol. 1999;162:3680–6. [PubMed] [Google Scholar]
- 21.Wilson JDK, Ogg GS, Allen RL, et al. Oligoclonal expansions of CD8+ T cells in chronic HIV infection are antigen specific. J Exp Med. 1998;188:785–90. doi: 10.1084/jem.188.4.785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Reed EF, Tugulea SL, Suciu-Foca N. Influence of HLA class I and class II antigens on the peripheral T-cell repertoire. Hum Immunol. 1994;40:111–22. doi: 10.1016/0198-8859(94)90055-8. [DOI] [PubMed] [Google Scholar]
- 23.Brinchmann JE, Janson CH, Klem LB, Haaheim LL, Spurkland A. Strict adherence to a common rank order of T-cell receptor Vβ usage in human leucocyte antigen disparate individuals. Scand J Immunol. 1996;44:179–84. doi: 10.1046/j.1365-3083.1996.d01-294.x. [DOI] [PubMed] [Google Scholar]
- 24.Rieux-Laucat F, Le Deist F, Selz F, Fischer A, de Villartay JP. Normal T cell receptor V beta usage in the primary immunodeficiency associated with HLA class II deficiency. Eur J Immunol. 1993;23:928–34. doi: 10.1002/eji.1830230425. [DOI] [PubMed] [Google Scholar]
- 25.Moretta L, Ciccone E, Mingari MC, et al. Human T lymphocytes expressing gamma/delta T cell antigen receptor. Clin Immunol Immunopathol. 1989;50:S117–23. doi: 10.1016/0090-1229(89)90118-9. [DOI] [PubMed] [Google Scholar]
- 26.Poccia F, Battistini L, Cipriani B, et al. Phosphoantigen-reactive Vgamma 9 Vdelta 2 T lymphocytes suppress in vitro human immunodeficiency virus type 1 replication by cell-released antiviral factors including CC chemokines. J Infect Dis. 1999;180:858–61. doi: 10.1086/314925. [DOI] [PubMed] [Google Scholar]
- 27.Poccia F, Wallace M, Colizzi V, et al. Possible protective and pathogenic roles of gamma delta T lymphocytes in HIV-infections. Int J Mol Med. 1998;1:409–13. doi: 10.3892/ijmm.1.2.409. [DOI] [PubMed] [Google Scholar]
- 28.Sciammas R, Bluestone JA. TCR gamma delta cells and viruses. Microbes Infection. 1999;1:203–12. doi: 10.1016/s1286-4579(99)80035-5. [DOI] [PubMed] [Google Scholar]
- 29.Autran B, Triebel F, Katlama C, Rozenbaum W, Hercend T, Debre P. T cell receptor γ/δ+ lymphocyte subsets during HIV infection. Clin Exp Immunol. 1989;75:206–10. [PMC free article] [PubMed] [Google Scholar]
- 30.Boullier S, Dadaglio G, Lafeuillade A, et al. V delta 1 T cells are expanded in the blood throughout HIV infection display a cytotoxic activity and are primed for TNF-alpha and IFN-gamma production but are not selected in lymph nodes. J Immunol. 1997;159:3629–37. [PubMed] [Google Scholar]
- 31.Rossol R, Dobmeyer JM, Dobmeyer TS, et al. Increase in Vdelta1+ gamma delta T cells in the peripheral blood and bone marrow as a selective feature of HIV-1 but not other virus infections. Br J Haematol. 1998;100:728–34. doi: 10.1046/j.1365-2141.1998.00630.x. [DOI] [PubMed] [Google Scholar]
- 32.De Paoli P, Gennari D, Martelli P, Basaglia G, Crovatto M, Battistin S. A subset of γδ T lymphocytes is increased during HIV-1 infection. Clin Exp Immunol. 1991;83:187–91. doi: 10.1111/j.1365-2249.1991.tb05612.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Norazmi MN, Arifin H, Jamaruddin MA. Increased level of activated γδ lymphocytes correlates with disease severity in HIV infection. Immunol Cell Biol. 1995;73:245–8. doi: 10.1038/icb.1995.40. [DOI] [PubMed] [Google Scholar]
- 34.Wallace M, Bartz SR, Chang WL, MacKenzie DA, Pauza CD, Malkovsky M. γδ T lymphocyte responses to HIV. Clin Exp Immunol. 1996;103:177–84. doi: 10.1046/j.1365-2249.1996.d01-625.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Taupin JL, Halary F, Dechanet J, et al. An enlarged subpopulation of T lymphocytes bearing two distinct gammadelta TCR in an HIV-positive patient. Int Immunol. 1999;11:545–52. doi: 10.1093/intimm/11.4.545. [DOI] [PubMed] [Google Scholar]


