Abstract
Non-invasive monitoring may be useful after kidney transplantation (KT), particularly for predicting acute rejection (AR). It is less clear whether chronic allograft nephropathy (CAN) is also associated with changes in urine cells. To identify non-invasive markers of allograft function in kidney transplant patients (KTP), mRNA levels of AGT, TGF-β1, EGFR, IFN-γ, TSP-1, and IL-10 in urine (Ur) samples were studied using QRT-PCR. Ninety-five KTP and 111 Ur samples were evaluated. Patients (Pts) were divided as, within six months (N = 31), and with more than six months post-KT (N = 64). KTP with more than six months post-KT were classified as KTP with stable kidney function (SKF) (N = 32), KTP with SKF (creatinine < 2 mg/dL) and proteinuria > 500 mg/24 h (N = 18), and KTP with biopsy proven CAN (N = 14). F-test was used to test for equality of variances between groups. IL-10 mRNA was decreased in Ur samples from KTP with less than six months post-KT (P = 0.005). For KTR groups with more than six months post-KT, AGT and EGFR mRNA were statistically different among KTP with SKF, KTP with SKF and proteinuria, and CAN Pts (P = 0.003, and P = 0.01), with KTP with SKF having higher mean expression. TSP-1 mRNA levels also were significantly different among these three groups (P = 0.04), with higher expression observed in CAN Pts. Using the random forest algorithm, AGT, EGFR, and TGF-β1 were identified as predictors of CAN, SKF, SKF with proteinuria. A characteristic pattern of mRNA levels in the different KTP groups was observed indicating that the mRNA levels in Ur cells might reflect allograft function.
INTRODUCTION
With improvements in immunosuppression, the incidence of acute rejection in renal transplant recipients has progressively decreased (1). Although long-term allograft survival is improving, late graft loss from chronic allograft nephropathy (CAN) remains a significant clinical problem. CAN remains the most common cause, after death, of late renal allograft loss (2). Thus, identification of markers associated with, or predictive of, CAN should prove clinically useful.
To determine the relative contributions of host and donor factors in the development of CAN, and the potential for their modification by therapeutic interventions, a prospective quantitative evaluation of renal allograft injury is required. Unfortunately, such studies have been limited by the lack of suitably sensitive assessments of early renal allograft injury.
This problem arises because renal injury is detected clinically as a rise in serum creatinine and the onset of proteinuria (3,4). However, a rise in serum creatinine is a late sign of renal injury because compensatory mechanisms within the kidney can maintain glomerular filtration rate (GFR) despite progressive structural injury (5–7). The ability to detect CAN is limited by the fact that biopsies usually are not performed unless there has been an elevation in serum creatinine (3,5–7). The ability to detect CAN is limited by the fact that biopsies usually are not performed unless there has been a reduction in creatinine clearance (3,5–7). Further, the diagnosis of CAN according to Banff criteria lacks the sensitivity to detect renal injury as it relies on a semiquantitative assessment of renal fibrosis. Therefore, there is a need to identify alternative methods to detect early renal injury that can be used to describe key determinants of long-term graft function.
There is accumulating evidence that non-invasive immunological monitoring may be useful in the early period after renal transplant (8–11), particularly with regard to predicting the presence of acute rejection. It is less clear whether CAN is also associated with consistent changes in peripheral blood or urine cells.
It is becoming increasingly clear that the response of the renal allograft to injury, namely glomerulosclerosis and interstitial fibrosis, is the same whatever the etiology of the renal injury (12). Further, cross sectional studies of renal tissue in patients with established CAN have suggested that the development of glomerulosclerosis and interstitial fibrosis is mediated by the enhanced expression of the profibrotic growth factor, transforming growth factor-β (TGF-β) (13–17). Primary regulation of TGF-β activity is controlled by cleavage of the latent TGF-β precursor. Thrombospondin, a disulfide linked trimer produced by connective tissue, is considered to be an important regulator of TGF-β activity in vivo (18).
We recently examined the global transcript profiles using kidney biopsies of transplant patients with CAN to elucidate the molecular signature present in CAN (19). TGF-β1, EGFR (epidermal growth factor receptor) and AGT (angiotensinogen) were found to be some of the differentially expressed genes in CAN (19).
In addition, recent studies have been advocated to dissect the role of cytokines in allograft rejection and long-term allograft outcomes. For example, there is evidence showing that the ratio of IFN-γ/IL-10 producing might be a tool to discriminate between non-rejecting patients and rejecting patients (20). On the other hand, Th2-derived cytokines for example, IL-10, TGF-β (involved in immunologic tolerance, either as growth factors for regulatory T-cell populations or as direct mediators of their regulatory functions), also are involved in chronic rejection process (20).
Although many cytokines can be measured in the urine at protein level, most of them are produced locally, exert effects within renal parenchyma, and are not excreted in the urine to any appreciable extent. It follows that the cytokine gene expression in urinary sediment at mRNA level would be a relevant marker of kidney diseases. In fact, many renal diseases, such as kidney allograft rejection, often are characterized by the infiltration of inflammatory cells, which produce a variety of cytokines, growth factors, chemokines, and other mediators of inflammation (8,10,11). We hypothesize that by measuring the gene expression of cytokines, growth factors, and growth factor receptors in urinary sediment, the extent of the renal damage can be evaluated and non-invasive surrogate markers of renal inflammation and fibrosis might be discovered.
In the present study, we evaluated the gene expression levels of a set of markers in urine samples of kidney transplant recipients with different post-transplantation times as a tool for non-invasive monitoring of graft function.
PATIENTS AND METHODS
Patients
Ninety-five kidney transplant patients (KTP) and 111 urine samples were evaluated. The KTP were divided as within six months (N = 31), and with more than six months post-KT (N = 64). In addition, KTP with more than six months post-KT were classified as KTP with stable kidney function (SKF) (defined by a creatinine level < 2 mg/dL) (N = 32), KTP with SKF (creatinine < 2 mg/dL) and proteinuria > 500 mg/24 h (N = 18), and KTP with biopsy proven CAN (N = 14). Gene expression levels of TGF-β1, EGFR, AGT, IFN-γ, thrombospondin-1 (TSP-1), and IL-10 were evaluated in urine samples of KTP using real time PCR Analysis (QRT-PCR). All biopsies were evaluated according to the Banff criteria (21). Variables included in the analysis, in addition to the gene expression levels, were patient age, acute rejection (AR), delayed graft function (DGF), cold ischemia time, warm ischemia time, cytomegalovirus (CMV), BK virus, and donor type (living or deceased donor).
Urine Samples
Urine (about 50 mL) from the KTP was allocated into 50 mL sterile conical tubes within two hours of post-collection. For patients with CAN, urine samples were collected at the time of biopsy. The tubes were centrifuged at 2,000g at room temperature for 20 min, and the supernatants were discarded. The pellets were re-suspended in 1 mL of phosphate buffered saline (PBS), transferred to cryotubes, then centrifuged at 16,000g for 5 min at room temperature. Supernatants were discarded. RNAlater (200 mL) (Ambion, Applied Biosystems, Foster City, California, USA) was added to the cryotube containing the urine cell pellet, then mixed and centrifuged at 16,000g for 30 s at room temperature prior to snap freezing and storage at −70° C. Isolation of RNA from urine pellets was performed using RNeasy kit (Qiagen, Valencia, California, USA) according to the manufacturer’s recommendations. The purity of the RNA preparations was tested by the 260/280 nm ratios.
Real Time PCR Analysis (QRT-PCR)
QRT-PCR was carried out for TGF-β1, EGFR, AGT, IFN-γ, TSP-1, and IL-10 genes in RNA from the urine samples, using pre-designed primer and probe sets from the Assays-on-Demand Genomic Assays (Applied Biosystems). Each assay consisted of two unlabeled PCR primers and an FAM dye-labeled TaqMan MGB probe. The endogenous control, β-2-microglobulin (B2M), was detected with a VIC dye-labeled TaqMan MGB probe (Human B2M (β-2-microglobulin) Endogenous Control, VIC/TAMRA Probe, Primer Limited, Applied Biosystems). Total RNA from each sample was subjected to reverse transcription using TaqMan Reverse Transcription Reagents (Applied Biosystems) according to the manufacturer’s protocol. Real-time PCR reactions were carried out in a 25μL reaction mixture using an ABI Prism 7700 sequence detection system (Applied Biosystems). All amplifications were carried out in duplicate and threshold cycle (Ct) scores were averaged for calculations of relative expression values. The Ct scores for genes of interest were normalized against Ct scores for the corresponding B2M control. Relative expression was determined by the following calculation where the amount of target is normalized to an endogenous reference (B2M RNA) and relative to an arbitrary calibrator (the reference class of patients used in the comparison)(22): Relative Expression = 2−ΔΔCt, where ΔΔCt = (ΔCt of experimental group) − (ΔCt of calibrator group).
Statistical Analysis
Descriptive statistics for continuous variables such as age and RT-PCR results (mean, median, standard deviation, minimum, and maximum) were reported, stratified by more than six months and less than six months. An F-test was used to test for equality of variances between the two groups. Variables with significantly different variances in the two groups were then compared using a two-sample t-test using Satterthwaite’s adjustment to the degrees; otherwise, a two-sample t-test was applied. Frequencies and percents were reported for categorical variables such as occurrence of acute rejection, presence of BK, and donor type (deceased donor/living donor). Examining the resulting P value from Fisher’s exact test assessed the significance of categorical variables.
The dataset was then restricted to patients more than six months. Continuous and categorical variables were compared among the following three groups: normal functioning kidney, normal functioning kidney with proteinuria, and CAN. For continuous variables, a one-way analysis of the variance model was fit and an F-test was used to determine whether there was a significant difference among the three groups. Fisher’s exact test was used in assessing the significance of the categorical variables.
RESULTS
Patients
This was a cross-sectional study of patients attending the Transplant Clinic at the Hume-Lee Transplant Center, Virginia Commonwealth University (VCU). Informed consent was obtained from all the subjects. The Western Institutional Review Board (WIRB) approved the study protocol.
Ninety-five patients were studied, 41 females/54 males, with a mean age of 43.0 ± 13.8 years old. The post-KT time was 38.2 ± 48.9 month, range = 1–280 months. Immunosuppression consisted of either a cyclosporine-based or tacrolimus-based regimen.
No significant differences were found in age, gender, post-transplantation time, and creatinine levels between KTP with stable kidney function, with and without proteinuria. The proteinuria levels were significantly different overall, with class comparisons revealing a significant difference between the KTP patients with stable kidney function with and without proteinuria [(means ± SD): 258.8 ± 158.9 compared with 2098 ± 670.7 mg/24 h, P < 0.0001; respectively]. The proteinuria level for the CAN group was 2712 ± 1632 mg/24 h. The creatinine levels were significantly different when comparing the three groups (P = 0.005). There were no significant differences between KTP with stable kidney function, with and without proteinuria, when considering cyclosporine levels (P = 0.7), mismatch HLA (P = 0.28), cold ischemia time (P = 0.32), and rejection episodes (P = 0.5).
CAN was defined as having plasma creatinine > 2.0 mg/dL, persistent proteinuria, and typical biopsy findings. Glomerular changes, interstitial fibrosis, tubular atrophy, and vascular changes were graded on a scale from 0 to 3. Among the 14 allograft biopsies from the patients of the CAN Group, three biopsies were graded as CAN grade 3; seven were graded as CAN grade 2, and four as CAN grade 1.
QRT-PCR Results
The 2−ΔΔCt method was used to calculate fold changes in the expression levels of the genes of interest compared with one of the normal urine samples. The 2−ΔΔCt method assumes that the efficiencies for the endogenous control amplicon (B2M) and the gene of interest amplicon are the same. Efficiencies were determined for the amplicons B2M, TGF-β1, EGFR, and TSP-1 on 1:5 dilution series. The slopes of Ct/log dilution plots for the reactions were −2.98, −3.02, −3.18, and −3.08, respectively; thus, all amplicons amplify with similar efficiencies.
TGF-β1, EGFR, AGT, IFN-γ, TSP-1, and IL-10 mRNA Levels in Urine Samples
mRNA levels comparisons between KTP with less and more than six months post-KT
IL-10 mRNA levels were decreased in KTP with less than six months post-KT (P = 0.005) (Table 1). From the comparison of the expression levels of the studied genes among KTP with less than six months post-KT that presented at least one acute rejection episode vs. KTP with less than six months post-KT without acute rejection episodes, we observed that IL-10 and IFN-γ were statistically significantly differentially expressed (2.92 ± 5.95 compared with 0.35 ± 0.45 mRNA IL-10/B2M fold change; P = 0.036; 23.39 ± 49.23 vs. 2.40 ± 3.59 mRNA IFN-γ/B2M fold change, P = 0.039, respectively) (Table 2).
Table 1.
Urinary levels of gene expression for the studied genes in kidney transplant patients.
| < 6 months post-transplantation | > 6 months post-transplantation | P value | |
|---|---|---|---|
| AGT | 0.08 | ||
| N | 31 | 64 | |
| Mean | 5.23 | 10.09 | |
| Median | 1 | 5.28 | |
| Std. Dev. | 11.15 | 13 | |
| (Minimum, Maximum) | (0.0005, 55.7) | (0.054, 64.0) | |
| EGFR | 0.16 | ||
| N | 31 | 64 | |
| Mean | 6.38 | 10.63 | |
| Median | 1.62 | 5.28 | |
| Std. Dev. | 14.19 | 13.58 | |
| (Minimum, Maximum) | (0.287, 68.594) | (0.054, 64.0) | |
| IFN-γ | 0.42a | ||
| N | 28 | 63 | |
| Mean | 6.15 | 11.49 | |
| Median | 1.16 | 1 | |
| Std. Dev. | 20.89 | 42.46 | |
| (Minimum, Maximum) | (0.011, 111.43) | (0.002, 315.17) | |
| IL10 | 0.005a | ||
| N | 30 | 64 | |
| Mean | 0.86 | 18.61 | |
| Median | 0.13 | 3.59 | |
| Std. Dev. | 2.71 | 48.83 | |
| (Minimum, Maximum) | (0.001, 14.93) | (0.041, 337.79) | |
| TGF-β1 | 0.26a | ||
| N | 31 | 64 | |
| Mean | 1.37 | 11.15 | |
| Median | 0.5 | 1.52 | |
| Std. Dev. | 2.73 | 68.37 | |
| (Minimum, Maximum) | (0.006, 14.93) | (0.134, 548.748) | |
| TSP-1 | 0.99a | ||
| N | 31 | 64 | |
| Mean | 5.48 | 5.52 | |
| Median | 0.47 | 1.036 | |
| Std. Dev. | 26.29 | 15.51 | |
| (Minimum, Maximum) | (0.033, 147.033) | (0.001, 103.968) |
Indicates a significant difference between variances noted via F-test; Satterthwaite’s approximation to the degrees of freedom used in t-test.
Table 2.
mRNA levels of IL-10 and IFN-γ between KTP with less than six months post-KT with and without acute rejection.
| Gene | KTP with < 6 months post-Tx with acute rejection | KTP with < 6 months post-Tx without acute rejection | P valuesa |
|---|---|---|---|
| IL-10 | 0.35 ± 0.45 | 2.92 ± 5.95 | 0.036 |
| IFN-γ | 23.39 ± 49.23 | 2.40 ± 3.59 | 0.039 |
Values expressed as mRNA specific gene/B2M fold change when compared with normal urine.
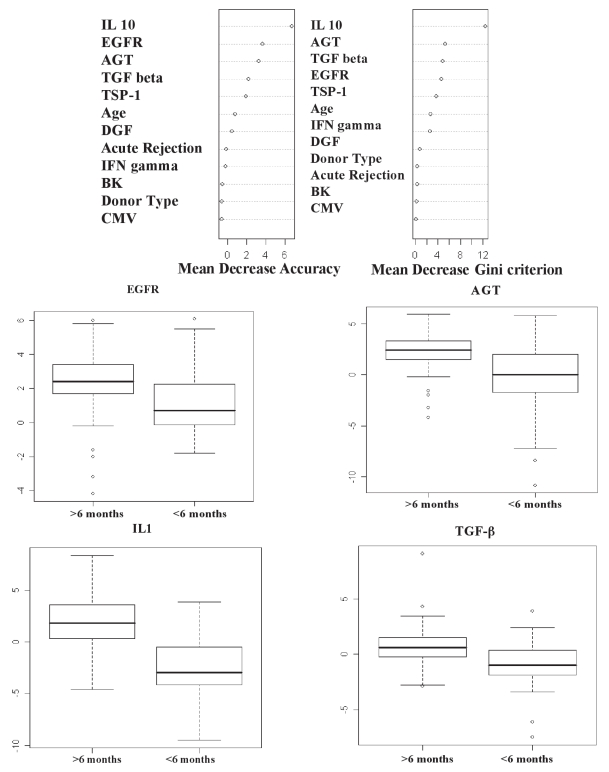
The random forest algorithm consisting of 2,500 classification trees was used to classify patients into one of two groups, more than six months post-KT vs. less than six months post-KT, using all variables. The unbiased estimate of generalization error, estimated using the out-of-bag observations, was 13.2%. Variable importance measures, assessed by mean decrease in accuracy and mean decrease in the Gini criterion, were subsequently clustered into K = two groups, based on the theoretical construct that one set of variables are important for discriminating the two groups, and the other set of variables are “noise” covariates. Variables found to be important markers of more than six months vs. less than six months were IL10, EGFR, AGT, and TGF-β1. Box plots of the log2 expression for these genes are displayed by group (Figure 1).
Figure 1.
A-Two variable importance measures for each predictor covariate from the random forest classification predicting < six months or ≥ six months. The mean decrease in accuracy (left panel) and mean decrease in Gini impurity (right panel). B-Box plots of the log2 RT-PCR expression for EGFR, AGT, IL10, and TGF-β1 displayed by < six months versus ≥ six months post-transplant.
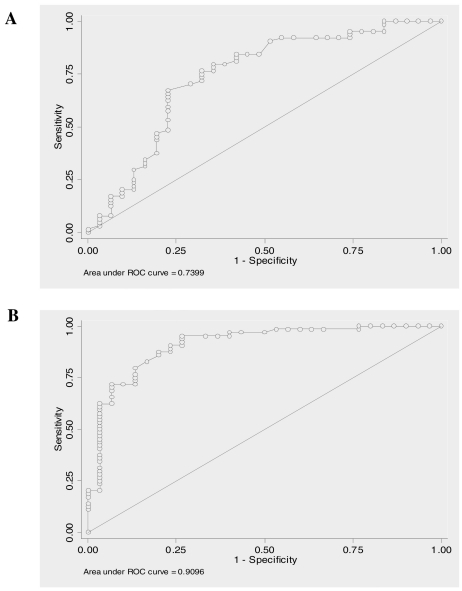
For each gene, a nonparametric receiver operator characteristic (ROC) analysis predicting less than six months vs. within six months was performed. For each gene, the area under the curve (AUC) and its associated 95% confidence interval (CI) were reported (Table 3). Also for the better predictor genes the ROC curve was graphically displayed (Figure 2).
Table 3.
Receiver operator characteristic (ROC) analysis predicting < six months versus ≥ six months post-KT.
| Gene | AUCa | 95% CI |
|---|---|---|
| AGT | 0.7399 | (0.62, 0.86) |
| EGFR | 0.7021 | (0.59, 0.82) |
| IFN-γ | 0.5522 | (0.42, 0.68) |
| IL10 | 0.9096 | (0.84, 0.98) |
| TGF-β | 0.7379 | (0.63, 0.85) |
| TSPD1 | 0.6983 | (0.59, 0.81) |
AUC: Area under the curve
Figure 2.
Receiver operating characteristic curves (ROC) for the better predictors. For each gene ROC analysis predicting < six months versus ≥ six months was performed. Afterward, the area under the ROC curve was estimated, A- AGT (AUC = 0.74); B- IL-10 (AUC = 0.91).
mRNA levels comparisons among subgroups of KTP with more than six months post-KT
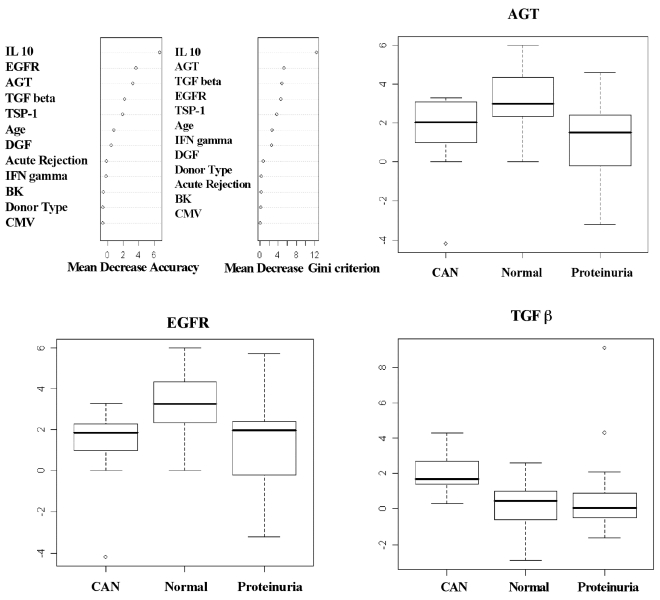
Among the different KTP groups with more than six months post-KT, mRNA levels of AGT EGFR, and TSP-1 were different among KTP with SKF, KTP with SKF and proteinuria, and CAN patients (P = 0.003, P = 0.01, and P = 0.04; respectively) with KTP with SKF having a higher mean (Table 4). Specifically, EGFR mRNA levels were lower in urine samples from CAN patients compared with urine samples from KTP with stable kidney function (P < 0.001). Moreover, EGFR levels in CAN samples were lower than in KTP with stable kidney function and proteinuria. Urinary TSP-1 mRNA levels were significantly higher in CAN patients than in the other two KTP groups (P < 0.001).
Table 4.
Levels of gene expression in KTP with more than six months post-transplantation.
| CAN | Normal functioning | Normal functioning with proteinuria | P value | |
|---|---|---|---|---|
| AGT | 0.003 | |||
| N | 14 | 32 | 18 | |
| Mean | 4.63 | 15.47 | 4.76 | |
| Median | 4.15 | 8.02 | 2.9 | |
| Std. Dev. | 3.44 | 16.02 | 6.3 | |
| (Minimum, Maximum) | (0.054, 9.849) | (1,64) | (0.109, 24.251) | |
| EGFR | 0.01 | |||
| N | 14 | 32 | 18 | |
| Mean | 4.23 | 15.41 | 7.12 | |
| Median | 3.65 | 9.71 | 4.01 | |
| Std. Dev. | 3.27 | 15.34 | 12.51 | |
| (Minimum, Maximum) | (0.054, 9.849) | (1, 64) | (0.109, 51.984) | |
| IFN-γ | 0.14 | |||
| N | 14 | 32 | 17 | |
| Mean | 9.59 | 3.25 | 28.57 | |
| Median | 1.19 | 0.85 | 1.62 | |
| Std. Dev. | 19.52 | 5.62 | 78.52 | |
| (Minimum, Maximum) | (0.117, 68.594) | (0.002, 25.992) | (0.125, 315.173) | |
| IL10 | 0.25 | |||
| N | 14 | 32 | 18 | |
| Mean | 22.6 | 8.99 | 32.6 | |
| Median | 7.32 | 2.1 | 6.37 | |
| Std. Dev. | 33.63 | 25.99 | 79.64 | |
| (Minimum, Maximum) | (1.231, 103.968) | (0.041, 147.033) | (0.985, 337.794) | |
| TGF-β1 | 0.29 | |||
| N | 14 | 32 | 18 | |
| Mean | 5.31 | 1.56 | 32.75 | |
| Median | 3.26 | 1.37 | 1.04 | |
| Std. Dev. | 5.06 | 1.22 | 128.85 | |
| (Minimum, Maximum) | (1.231, 19.698) | (0.134, 6.063) | (0.330, 548.748) | |
| TSP-1 | 0.04 | |||
| N | 14 | 32 | 18 | |
| Mean | 14.42 | 2.23 | 4.42 | |
| Median | 2.22 | 0.73 | 2.32 | |
| Std. Dev. | 30.63 | 4.32 | 7.27 | |
| (Minimum, Maximum) | (0.189, 103.968) | (0.001, 17.148) | (0.50, 32.00) |
TGF-β1 mRNA levels in urine samples from CAN patients were higher than in KTP with stable kidney function (P = 0.016). Interestingly, mRNA TGF-β1 levels were higher in KTP with stable kidney function and proteinuria (Table 4).
Acute rejection episodes were significantly different among the three groups (P = 0.02), with the highest rate observed in CAN patients. However, there were no statistically significant differences for BK, CMV, age, type of donor, or cold and warm ischemia time (Table 5). Similarly, using the same set of variables, the random forest algorithm consisting of 2,500 classification trees was used to classify patients in the more than six months group into one of three classes: CAN, normal functioning kidney, and normal functioning kidney with proteinuria. The unbiased estimate of generalization error, estimated using the out-of-bag observations, was 31.8%. Variable importance measures, assessed by mean decrease in accuracy and mean decrease in the Gini criterion, were subsequently clustered into K = 2 groups. This, based on the theoretical construct that one set of variables is important for discriminating between the two groups, and the other set of variables is composed of ‘noise’ covariates. Variables found to be important predictors of CAN, normal functioning kidney, and normal functioning kidney with proteinuria were AGT, EGFR, and TGF-β1. Box plots of the log2 expression for these genes are displayed by group (Figure 3).
Table 5.
Clinical characteristics among KTP groups with more than six months post-transplantation.
| CAN | Normal functioning | Normal functioning with proteinuria | P value | |
|---|---|---|---|---|
| Age (years) | 0.12 | |||
| N | 14 | 32 | 18 | |
| Mean | 36.7 | 45 | 38.7 | |
| Median | 39.5 | 46.5 | 38 | |
| Std. Dev. | 15.4 | 12.5 | 15.2 | |
| (Minimum, Maximum) | (14, 63) | (16, 70) | (13, 66) | |
| Cold Time (minutes) | 0.63 | |||
| N | 14 | 24 | 16 | |
| Mean | 632.6 | 459.8 | 591.3 | |
| Median | 652 | 41.5 | 330 | |
| Std. Dev. | 566.8 | 546.7 | 650.2 | |
| (Minimum, Maximum) | (18, 1800) | (16, 1470) | (16, 1698) | |
| Warm Time (minutes) | 0.69 | |||
| N | 14 | 25 | 15 | |
| Mean | 29.1 | 28 | 29.6 | |
| Median | 29 | 27 | 29 | |
| Std. Dev. | 4.1 | 7 | 6.3 | |
| (Minimum, Maximum) | (20, 35) | (2, 41) | (16, 46) | |
| Donor Type | 0.76 | |||
| Deceased | 9 (64%) | 17 (53%) | 10 (56%) | |
| Living | 5 (36%) | 15 (47%) | 8 (44%) | |
| Acute Rejection | 4 (29%) | 1 (3%) | 4 (22%) | 0.02 |
| BK | 3 (21%) | 7 (22%) | 4 (22%) | 1 |
| CMV | 2 (14%) | 4 (13%) | 2 (11%) | 1 |
| DGF | 2 (14%) | 3 (9%) | 5 (28%) | 0.23 |
Figure 3.
For ≥ six months samples, two variable importance measures for each predictor covariate from the random forest classification predicting CAN, normal, or proteinuria: the mean decrease in accuracy (left panel) and mean decrease in Gini impurity (right panel). For ≥ six-month samples, box plots of the log2 RT-PCR expression for EGFR, AGT, and TGF-β 1 displayed by CAN, stable kidney function, and stable kidney function with proteinuria. CAN: patients with biopsy proven CAN, Normal: kidney transplant patients with stable kidney function, Proteinuria: kidney transplant patients with stable kidney function (creatinine levels < 2 mg/dL) and proteinuria.
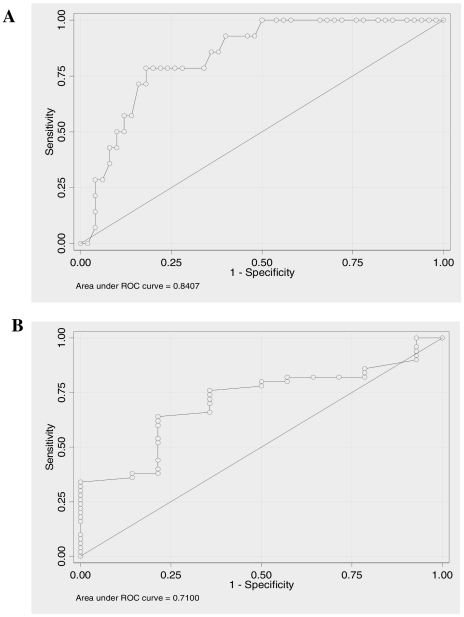
Lastly, for each gene, a nonparametric receiver operator characteristic (ROC) analysis predicting CAN versus SKF was performed. For each gene, the area under the curve (AUC) and its associated 95% confidence interval (CI) were reported (Table 6). Also for each gene the ROC curve was graphically displayed. AUC for TGF-β1 (AUC = 0.84) and EGFR (AUC = 0.71) are displayed in the Figure 4.
Table 6.
Receiver operator characteristic (ROC) analysis predicting CAN versus SKF in kidney transplant patients with more than six months post-kidney transplantation.
| Gene | AUC | 95% CI |
|---|---|---|
| AGT | 0.65 | (0.50, 0.80) |
| EGFR | 0.71 | (0.57, 0.85) |
| IFN-γ | 0.48 | (0.29, 0.66) |
| IL10 | 0.69 | (0.55, 0.83) |
| TGF-β | 0.84 | (0.74, 0.94) |
| TSPD1 | 0.66 | (0.50, 0.83) |
Figure 4.
Receiver operating characteristic curves (ROC) for the better predictors. For each studied gene a ROC analysis predicting CAN versus SKF was performed. Afterward, the area under the ROC curve was estimated, A- TGF-β1 (AUC = 0.84); B- EGFR (AUC = 0.71).
DISCUSSION
Kidney transplantation is the treatment of choice for patients with end-stage renal diseases. Acute rejection (AR), during the first month following kidney transplantation, is the major immunological event that may influence long-term graft outcomes (1–6). Chronic allograft nephropathy (CAN), leading to progressive allograft dysfunction, remains an important obstacle for long-term allograft survival (1–4). Therefore, kidney biopsy remains the major implement for the diagnosis and evaluation of graft function. An efficient and reliable test for continuous monitoring of the immune activation status of kidney transplant recipients is not available currently.
Although noninvasive and safe, peripheral blood evaluation of immune activation markers has yielded conflicting results for the prediction of AR of kidney allografts (23–24). More consistent results have been observed by examining the graft directly (25). Major contributions have been made recently in identifying the molecular signature within the allograft associated with both AR and chronic allograft dysfunction (19,26–29). In the context of early noninvasive diagnosis, the urine is attracting increased attention because of its potential diagnostic and pathophysiologic biomarker information (10,11).
In addition, urine represents an indirect way to visualize intragraft compartment. Recently Li et al (11) reported that mRNA levels of T cell-related cytotoxicity molecules perforin and granzyme B were enhanced in urine samples from AR patients. Other groups showed similar results (30–32). However, the role of the measurement of mRNA in urine samples as indicators of graft function at different times post-KT is less clear.
In the present study, we studied the level of expression of TGF-β1, EGFR, AGT, IFN-γ, TSP-1, and IL-10 genes in the urinary sediment of kidney transplant patients at different times post-KT. Gene expression levels were evaluated in urine samples from KTP with less and more than six months post-KT. In addition, KTP with more than six months post-KT were classified in subgroups for the analysis.
From the analysis of the markers in urine samples between KTP with less and more than six months post-transplantation, we observed differentially expressed genes. In KTP with less than six months post-KT IL-10 expression was significantly decreased, as was TGF-β1 though not significantly. TGF-β1 and IL-10 have been associated previously with CAN (33–35). It is well known that IL-10 promotes B cell proliferation and differentiation into plasma cells, increases antibody production as well as drives a shift from Th1 toward Th2-cell responses contributing to the pathogenesis of chronic allograft nephropathy.
Ordinarily, after six months post-transplantation, the number of nephrons is stable, resulting in an allograft with normal function (36). However, for an important number of grafts, the loss of nephrons continues, reflecting continued graft injury. This condition will end in fibrosis and atrophy, with the final event of loss of graft function (36). Proteinuria has been associated with faster progression of any renal disease and might be considered the first clinical manifestation of CAN (37,38). Patients with CAN have more than 0.5g proteinuria/24 h, 20–28%, compared with 6–8% of patients who do not have this condition (39). CAN is the result of cumulative damage to the kidney and is present in 30–50% of patients after six months post-transplantation (40). We sub-classified the KTP group with more than six months post-KT according to kidney function and proteinuria levels.
Different gene expression patterns were observed in the subgroups of KTP with more than six months post-KT. By studying the expression of our selected panel of markers in these groups of KTP we aimed to identify early markers of CAN progression. A group of KTP with normal allograft function was compared with KTP with histological diagnosis of CAN. Moreover, both previously described groups were compared with KTP with normal function but with proteinuria. As previously mentioned, proteinuria might be considered the first clinical manifestation of CAN (38–40).
AGT and EGFR mRNA levels of were statistically different among KTP with SKF, KTP with stable kidney function and proteinuria, and CAN patients. Moreover, TGF-β1 mRNA levels were significantly higher in CAN patients compared with KTP with SKF. However, TGF-β1 levels were higher in KTP with stable kidney function and proteinuria than in CAN patients. This finding might indicate that the activity of molecules involved in the development of fibrosis is higher in these patients. Moreover, AGT, EGFR, and TGF-β1 mRNA levels in urine were identified as good predictor markers of CAN with EGFR and TGF-β1 showing an AUC of 0.71 and 0.84 respectively.
EGF and EGFR have been implicated in expansion and deposition of extracellular matrix in renal injury (41). There is a growing evidence for the role of EGFR pathway in various types of renal lesions. However, it is still controversial whether the effect is beneficial or detrimental. In addition, while EGFR expression has been identified previously in normal and diseased native kidneys, its expression in renal allografts and its relation to allograft rejection or allograft survival remains unknown (42). In this study, EGFR was down expressed in CAN when compared with KTP with normal function. This finding is in concordance with our recent publication (19) where EGFR was down expressed in kidney biopsies with CAN when compared with normal allograft using microarrays. We also observed AGT down expressed in CAN biopsies (19). The AGT gene encodes the only glycoprotein known to be a precursor of the vasopresor angiotensin II (Ang II). Ang II is also a growth factor and a profibrogenic cytokine. It mediates the induction of TGF-β1 (43). Previous studies have shown that Ang II stimulation of mouse glomerular mesangial cells causes EGF receptor phosphorylation through the MMP-dependent generation of heparin-binding EGF (HB–EGF) and the EGF-receptor-dependent release of TGF-β (42,44). The elucidation of the interactions between these molecules and their role in the progression to chronic allograft dysfunction requires further evaluation and study.
In the present study we showed the potential utility of TGF-β1, AGT, and EGFR mRNA levels in urine samples of KTP as early markers of CAN progression. Prospective studies are needed for confirming these results. The evaluation of molecular markers in urine samples could represent an invaluable resource for monitoring KTP and predicting the development of CAN.
Footnotes
Online address: http://www.molmed.org
REFERENCES
- 1.Gjertson DW. Impact of delayed graft function and acute rejection on kidney graft survival. Clin Transpl. 2000;14:467. [PubMed] [Google Scholar]
- 2.Cecka JM. The UNOS scientific renal transplant registry. In: Cecka JM, Terasaki PI, editors. Clinical Transplants. UCLA Immunogenetics Center; 1999. 2000. p. 1. [PubMed] [Google Scholar]
- 3.Nankivell BJ, Chapman JR. Chronic allograft nephropathy: current concepts and future directions. Transplantation. 2006;81:643–54. doi: 10.1097/01.tp.0000190423.82154.01. [DOI] [PubMed] [Google Scholar]
- 4.Saurina A, et al. Conversion from calcineurin inhibitors to sirolimus in chronic allograft dysfunction: changes in glomerular haemodynamics and proteinuria. Nephrol Dial Transplant. 2006;21:488–93. doi: 10.1093/ndt/gfi266. [DOI] [PubMed] [Google Scholar]
- 5.Kasiske BL, et al. Long-term deterioration of kidney allograft function. Am J Transplant. 2005;5:1405–14. doi: 10.1111/j.1600-6143.2005.00853.x. [DOI] [PubMed] [Google Scholar]
- 6.Kasiske BL, et al. Comparing methods for monitoring serum creatinine to predict late renal allograft failure. Am J Kidney Dis. 2001;38:1065–73. doi: 10.1053/ajkd.2001.28605. [DOI] [PubMed] [Google Scholar]
- 7.Gill JS, Tonelli M, Mix CH, Pereira BJ. The change in allograft function among long-term kidney transplant recipients. J Am Soc Nephrol. 2003;14:1636–42. doi: 10.1097/01.asn.0000070621.06264.86. [DOI] [PubMed] [Google Scholar]
- 8.Tatapudi RR, et al. Noninvasive detection of renal allograft inflammation by measurements of mRNA for IP-10 and CXCR3 in urine. Kidney Int. 2004;65:2390–7. doi: 10.1111/j.1523-1755.2004.00663.x. [DOI] [PubMed] [Google Scholar]
- 9.Magee CC, Denton MD, Womer KL, Khoury SJ, Sayegh MH. Assessment by flow cytometry of intracellular cytokine production in the peripheral blood cells of renal transplant recipients. Clin Transplant. 2004;18:395–401. doi: 10.1111/j.1399-0012.2004.00179.x. [DOI] [PubMed] [Google Scholar]
- 10.Kotsch K, et al. Enhanced granulysin mRNA expression in urinary sediment in early and delayed acute renal allograft rejection. Transplantation. 2004;77:1866–75. doi: 10.1097/01.tp.0000131157.19937.3f. [DOI] [PubMed] [Google Scholar]
- 11.Li B, et al. Noninvasive diagnosis of renal-allograft rejection by measurement of messenger RNA for perforin and granzyme B in urine. NEngl J Med. 2001;344:947–54. doi: 10.1056/NEJM200103293441301. [DOI] [PubMed] [Google Scholar]
- 12.Mannon RB. Therapeutic targets in the treatment of allograft fibrosis. Am J Transplant. 2006;6:867–75. doi: 10.1111/j.1600-6143.2006.01261.x. [DOI] [PubMed] [Google Scholar]
- 13.Sharma VK, et al. Intragraft TGF-β1 mRNA: a correlate of interstitial fibrosis and chronic allograft nephropathy. Kidney Int. 1996;49:1297–303. doi: 10.1038/ki.1996.185. [DOI] [PubMed] [Google Scholar]
- 14.Horvath LZ, et al. Altered expression of transforming growth factor-beta S in chronic renal rejection. Kidney Int. 1996;50:489–98. doi: 10.1038/ki.1996.340. [DOI] [PubMed] [Google Scholar]
- 15.Lantz I, Dimeny E, Larsson E, Fellstrom B, Funa K. Increased immunoreactivity of transforming growth factor-beta in human kidney transplants. Transpl Immunol. 1996;4:209–14. doi: 10.1016/s0966-3274(96)80019-0. [DOI] [PubMed] [Google Scholar]
- 16.Mas V, Alvarellos T, Giraudo C, Massari P, De Boccardo G. Intragraft messenger RNA expression of angiotensinogen: relationship with transforming growth factor beta-1 and chronic allograft nephropathy in kidney transplant patients. Transplantation. 2002;74:718–21. doi: 10.1097/00007890-200209150-00022. [DOI] [PubMed] [Google Scholar]
- 17.Mas V, et al. Intragraft expression of transforming growth factor-beta 1 by a novel quantitative reverse transcription polymerase chain reaction ELISA in long lasting kidney recipients. Transplantation. 2000;70:612–6. doi: 10.1097/00007890-200008270-00014. [DOI] [PubMed] [Google Scholar]
- 18.Baboolal K, Jones GA, Janezic A, Griffiths DR, Jurewicz WA. Molecular and structural consequences of early renal allograft injury. Kidney Int. 2002;61:686–96. doi: 10.1046/j.1523-1755.2002.00149.x. [DOI] [PubMed] [Google Scholar]
- 19.Mas V, et al. Establishing the molecular pathways involved in chronic allograft nephropathy for testing new non-invasive diagnostic markers. Transplantation. 2007;83:448–57. doi: 10.1097/01.tp.0000251373.17997.9a. [DOI] [PubMed] [Google Scholar]
- 20.van den Boogaardt De, et al. The ratio of interferon-gamma and interleukin-10 producing donor-specific cells as an in vitro monitoring tool for renal transplant patients. Transplantation. 2006;82:844–8. doi: 10.1097/01.tp.0000229448.64363.18. [DOI] [PubMed] [Google Scholar]
- 21.Racusen LC, et al. The Banff 97 working classification of renal allograft pathology. Kidney Int. 1999;55:713–23. doi: 10.1046/j.1523-1755.1999.00299.x. [DOI] [PubMed] [Google Scholar]
- 22.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR the 2(−Delta Delta C (T)) Method. Methods. 2001;25:402–8. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 23.Dugré F, Gaudreau S, Belles-Isles M, Houde I, Roy R. Cytokine and cytotoxic molecule gene expression determined in peripheral blood mononuclear cells in the diagnosis of acute renal rejection. Transplantation. 2000;70:1074–80. doi: 10.1097/00007890-200010150-00014. [DOI] [PubMed] [Google Scholar]
- 24.Vasconcellos LM, et al. Cytotoxic lymphocyte gene expression in peripheral blood leukocytes correlates with rejecting renal allografts. Transplantation. 1998;66:562–6. doi: 10.1097/00007890-199809150-00002. [DOI] [PubMed] [Google Scholar]
- 25.Strehlau J, et al. Quantitative detection of immune activation transcripts as a diagnostic toll in kidney transplantation. Proc Natl Acad Sci. 1997;94:695–700. doi: 10.1073/pnas.94.2.695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Flechner SM, et al. Kidney transplant rejection and tissue injury by gene profiling of biopsies and peripheral blood lymphocytes. Am J Transplant. 2004;4:1475–85. doi: 10.1111/j.1600-6143.2004.00526.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sarwal M, et al. Molecular heterogeneity in acute renal allograft rejection identified by DNA microarray profiling. N Engl J Med. 2003;349:125–38. doi: 10.1056/NEJMoa035588. [DOI] [PubMed] [Google Scholar]
- 28.Scherer A, et al. Early prognosis of the development of renal chronic allograft rejection by gene expression profiling of human protocol biopsies. Transplantation. 2003;75:1323–30. doi: 10.1097/01.TP.0000068481.98801.10. [DOI] [PubMed] [Google Scholar]
- 29.Hotchkiss H, et al. Differential expression of profibrotic and growth factors in chronic allograft nephropathy. Transplantation. 2006;81:342–9. doi: 10.1097/01.tp.0000195773.24217.95. [DOI] [PubMed] [Google Scholar]
- 30.Yannaraki M, et al. Urinary cytotoxic molecular markers for a noninvasive diagnosis in acute renal transplant rejection. Transpl Int. 2006;19:759–68. doi: 10.1111/j.1432-2277.2006.00351.x. [DOI] [PubMed] [Google Scholar]
- 31.Matz M, et al. Early post-transplant urinary IP-10 expression after kidney transplantation is predictive of short- and long-term graft function. Kidney Int. 2006;69:1683–90. doi: 10.1038/sj.ki.5000343. [DOI] [PubMed] [Google Scholar]
- 32.Roelofs JJ, et al. Expression of urokinase plasminogen activator and its receptor during acute renal allograft rejection. Kidney Int. 2003;64:1845–53. doi: 10.1046/j.1523-1755.2003.00261.x. [DOI] [PubMed] [Google Scholar]
- 33.Nocera A, et al. Cytokine mRNA expression in chronically rejected human renal allografts. Clin Transplant. 2004;18:564–70. doi: 10.1111/j.1399-0012.2004.00227.x. [DOI] [PubMed] [Google Scholar]
- 34.Campistol JM, Inigo P, Larios S, Bescos M, Oppenheimer F. Role of transforming growth factor-beta1 in the progression of chronic allograft nephropathy. Nephrol Dial Transplant. 2001;16(Suppl 1):114–6. doi: 10.1093/ndt/16.suppl_1.114. [DOI] [PubMed] [Google Scholar]
- 35.Pribylova-Hribova P, et al. TGF-β1 mRNA upregulation influences chronic renal allograft dysfunction. Kidney Int. 2006;69:1872–9. doi: 10.1038/sj.ki.5000328. [DOI] [PubMed] [Google Scholar]
- 36.Halloran PF, Langone AJ, Helderman JH, Kaplan B. Assessing long-term nephron loss: is it time to kick the CAN grading system? Am J Transplant. 2004;11:1729. doi: 10.1111/j.1600-6143.2004.00662.x. [DOI] [PubMed] [Google Scholar]
- 37.Schuck O, et al. Predicting of histopathological grade of chronic allograft nephropathy from renal function and proteinuria. Ann Transplant. 2003;8:5–7. [PubMed] [Google Scholar]
- 38.Massy ZA, et al. Chronic renal allograft rejection: Immunologic and nonimmunologic risk factors. Kidney Int. 1996;49:518. doi: 10.1038/ki.1996.74. [DOI] [PubMed] [Google Scholar]
- 39.Solez K, Vincenti F, Filo RS. Histopathologic findings from 2-year protocol biopsies from a U.S. multicenter kidney transplant trial comparing tarolimus versus cyclosporine: a report of the FK506 Kidney Transplant Study Group. Transplantation. 1998;66:1736–40. doi: 10.1097/00007890-199812270-00029. [DOI] [PubMed] [Google Scholar]
- 40.Nankivell BJ, et al. The natural history of chronic allograft nephropathy. N Engl J Med. 2003;349:2326–33. doi: 10.1056/NEJMoa020009. [DOI] [PubMed] [Google Scholar]
- 41.Sis B, et al. Epidermal growth factor receptor expression in human renal allograft biopsies: an immunohistochemical study. Transpl Immunol. 2004;13:229–32. doi: 10.1016/j.trim.2004.05.001. [DOI] [PubMed] [Google Scholar]
- 42.Uchiyama-Tanaka Y, et al. Angiotensin II signaling and HB–EGF shedding via metalloproteinase in glomerular mesangial cells. Kidney Int. 2001;60:2153–63. doi: 10.1046/j.1523-1755.2001.00067.x. [DOI] [PubMed] [Google Scholar]
- 43.Wolf G. Renal injury due to renin-angiotensin-aldosterone system activation of the transforming growth factor-beta pathway. Kidney Int. 2006;70:1914–9. doi: 10.1038/sj.ki.5001846. [DOI] [PubMed] [Google Scholar]
- 44.Shah BH, Catt KJ. TACE-dependent EGF receptor activation in angiotensin-II-induced kidney disease. Trends Pharmacol Sci. 2006;27:235–7. doi: 10.1016/j.tips.2006.03.010. [DOI] [PubMed] [Google Scholar]