Abstract
It is often difficult to determine which of the sequence and structural differences between divergent members of multigene families are functionally important. Here we use a laboratory evolution approach to determine functionally important structural differences between two distantly related disulfide isomerases, DsbC and DsbG from Escherichia coli. Surprisingly, we found single amino acid substitutions in DsbG that were able to complement dsbC in vivo and have more DsbC-like isomerase activity in vitro. Crystal structures of the three strongest point mutants, DsbG K113E, DsbG V216M, and DsbG T200M, reveal changes in highly surface-exposed regions that cause DsbG to more closely resemble the distantly related DsbC. In this case, laboratory evolution appears to have taken a direct route to allow one protein family member to complement another, with single substitutions apparently bypassing much of the need for multiple changes that took place over ≈0.5 billion years of evolution. Our findings suggest that, for these two proteins at least, regions important in determining functional differences may represent only a tiny fraction of the overall protein structure.
Keywords: chaperone, protein folding, directed evolution
A number of models of the origin of life postulate that the primordial living cell contained only a small number of enzymes; years of gene duplication, divergence, and natural selection led to the current situation in which each organism possesses thousands of proteins with different functions (1). Identifying the mechanisms by which proteins can acquire new functions is important both in understanding this fundamental question of natural diversity and in elucidating the particular structural differences that allow related proteins to have distinct functions. Here we use a combination of directed evolution and structural biology to examine the functionally important structural differences of two distantly related Escherichia coli disulfide isomerases, DsbC and DsbG.
Directed evolution is an elegant method that is often used to alter enzymatic function (usually by broadening enzymatic specificity), but it also has been used to change the properties of one enzyme so that it has some of the functional properties of another related enzyme (2). One way to do this requires the presence of a unique and selectable phenotype for one family member, allowing the selection of gain-of-function mutations in a related protein. Analysis of those gain-of-function mutants can provide information about functional differences between the two family members. This technique has been used to alter the enzymatic activity of a number of proteins so that they now have common properties, including the E. coli paralogs aspartate aminotransferase (AATase) and tyrosine aminotransferase (3), the structurally similar HisA and TrpF (4), and the human θ class 1-1 and rat θ class 2-2 glutathione transferases (5). Directed evolution has also been used to explore the differences between various E. coli folding catalysts, such as DsbC and DsbA (6), Grx3 and Grx1 (7), and TrxA and DsbA (8). Here we use directed evolution to attempt to identify some of the fundamental structural differences between two distantly related disulfide isomerases, DsbC and DsbG.
In prokaryotes, correct disulfide bond formation in secreted proteins depends on both the DsbA/DsbB pathway, which catalyzes disulfide oxidation, and the DsbC/DsbG-DsbD pathway, which is thought to catalyze disulfide bond isomerization. DsbC and DsbG appear to perform distinct roles. DsbC directly interacts with substrate proteins containing nonnative disulfide bonds and appears to function by rearranging these nonnative disulfides to correct native disulfides (9). DsbC shares only 24% sequence identity with DsbG, and it is likely that DsbC and DsbG have unique in vivo functions: DsbC is required for the efficient expression of three periplasmic proteins, AppA, MepA, and RNase I, whereas DsbG does not appear to affect expression of these proteins (10, 11). Additionally, DsbG is active in some, but not all, in vivo and in vitro assays for DsbC activity (10). Overall, DsbG seems to have much weaker isomerase activity than DsbC in vivo. dsbG-null mutants have no defect in the folding of heterologous proteins containing multiple disulfide bonds, and no ability to catalyze disulfide bond rearrangement when the {30–51, 14–38} bovine pancreatic trypsin inhibitor intermediate is used as a substrate (12), but DsbG overexpression is able to restore the ability of dsbC mutants to express some of these proteins (10). This latter observation and DsbG's homology to DsbC have led to the conclusion that DsbG is a disulfide isomerase with restricted substrate specificity. dsbC strains are more sensitive to copper(II) than wild-type strains, whereas DsbG strains show wild-type sensitivity to copper(II). Copper(II) can catalyze the formation of incorrect disulfides, including disulfide-linked oligomers, which DsbC, but not DsbG, appears to be able to resolve in vivo (13).
Structural studies show that DsbC and DsbG have superficially similar structures: both are V-shaped homodimers consisting of two thioredoxin domains connected by a linker helix to an N-terminal dimerization domain (14–16). However, there are many structural differences between DsbC and DsbG. In particular, the putative substrate-binding cleft of DsbG is much larger (60 Å × 31 Å × 31 Å) than that of DsbC (40 Å × 25 Å × 25 Å), perhaps indicating that DsbG's substrates are larger than those of DsbC (14). Additionally, comparison of DsbG's surface with that of DsbC reveals many differences; specifically, DsbG contains a significant number of charged surface patches, in contrast to DsbC's largely hydrophobic and uncharged surface. These structural and surface charge differences support the hypothesis that DsbC and DsbG have differing substrate specificity. An important step in understanding the distinct roles of DsbC and DsbG is to define the sequence and structural features that distinguish them. However, even by close examination of the sequences and structures of DsbC and DsbG, it is very difficult to ascertain which differences contribute to differences in function and which are a result of genetic drift.
We decided to use this small multigene family as a model for studying the process of gene divergence. By attempting to bypass divergent evolution, we hoped to gain insight into the essential differences between two members of this gene family. We selected for gain-of-function point mutations in DsbG that were able to complement the dsbC copper sensitivity in vivo and show more DsbC-like isomerase activity in vitro. The crystal structures of the DsbC-like DsbG mutants reveal changes that make two regions of DsbG remarkably DsbC-like. This analysis suggests that only a tiny percentage of the structural features of DsbG need to be made more DsbC-like for DsbG to acquire at least some of DsbC's functional properties.
Results
Identification of dsbG Mutants That Complement dsbC Copper(II) Sensitivity.
We recently discovered that the DsbC disulfide isomerization pathway is required for in vivo resistance to the redox-active metal copper. Copper(II) catalyzes the formation of disulfide-bonded oligomers in vitro, and purified DsbC can resolve these incorrect oxidation states. Copper(II) is a powerful and nonspecific thiol oxidant. Exposure of cells to copper(II) is therefore likely to place an additional burden on the disulfide isomerization machinery of the cell. We proposed that the copper sensitivity of dsbC-null mutants is due to their lack of ability to isomerize proteins that have been misoxidized by copper(II) in vivo (13). Thus, copper(II) resistance appears to provide a powerful selection for disulfide isomerase activity in vivo, and it presents us with a unique opportunity to explore the in vivo requirements for effective disulfide isomerization.
Whole-Plasmid Mutagenesis for Gain-of-Function Testing.
We tested the ability of overexpressed wild-type DsbG to rescue the copper(II) sensitivity of dsbC-null strains. Wild-type DsbG, even when overexpressed ≈20-fold above the level expressed from the chromosome from the arabinose-inducible expression plasmid pBAD33a, was unable to complement the sensitivity of dsbC-null strains to 8 mM CuCl2. In contrast, wild-type DsbC expressed from pBAD33a fully restored copper(II) resistance to a dsbC-null strain. The inability of DsbG to complement the copper sensitivity of dsbC-null strains suggests that DsbG is unable to isomerize the incorrect disulfides formed by copper(II) in vivo. This suggestion is consistent with the lower isomerase activity exhibited by DsbG in vitro. We reasoned that by selecting for copper-resistant DsbG variants, we would find DsbG variants that are more DsbC-like in their isomerase activity.
To find copper-resistant dsbG variants, we created a plasmid library of DsbG mutants in pBAD33a and transformed them into the dsbC-null mutant AH131. Transformants were plated directly onto brain–heart infusion agar containing 8 mM CuCl2, chloramphenicol, and 0.1 mM arabinose to induce overexpression. Approximately 110,000 separate transformants were obtained after 18 different mutagenic PCRs. The transformants were replica plated onto media containing 8 mM CuCl2. Copper-resistant clones were retested on copper in the presence and absence of arabinose to confirm that copper resistance depended on induction of DsbG expression. Sequencing of either the complementing plasmids or randomly picked unselected clones revealed an average of 2.02 (range of 1–4) amino acid changes within the dsbG gene. Multiple clones often showed a mutation in one particular residue [see supporting information (SI) Table 1]. To determine whether these single point mutations in dsbG are, on their own, sufficient to complement copper resistance in a dsbC-null mutation, we introduced them individually into the dsbG gene by site-directed mutagenesis and tested their ability to restore growth of a dsbC-null strain on 8 mM CuCl2.
Four single point mutations (K113E, V216M, T200M, and N198I) strongly rescued copper(II) resistance, showing that these individual point mutants are independently sufficient to confer a dsbC-like phenotype on dsbG (Fig. 1). These four mutants form 102 to 104 times more viable colonies than wild-type dsbG on brain–heart infusion supplemented with 8 mM CuCl2. Three of these four residues (K113, V216, and N198) were found mutated to more than one amino acid: K113 was mutated to either Glu or Asn, V216 was mutated to either Met or Ala, and N198 was mutated to either Ile or Tyr. Additionally, the V216M and N198I mutations were obtained repeatedly (V216M three times, N198I twice) in completely separate PCRs (SI Table 1). This finding suggests that these particular residues play an important role in the dsbG mutants' ability to restore copper resistance. To help confirm this possibility, we reconstructed single amino acid changes in an otherwise unmutagenized dsbG background by mutagenesis with QuikChange (Stratagene, La Jolla, CA). These mutants are designated in SI Table 1 with a QC prefix to their number. A handful of other single point mutations were also sufficient to rescue copper(II) resistance, albeit to a lesser extent; these include A197, which lies adjacent to the strongly rescuing N198 and T200 mutations, as well as a number of other mutants where it seems likely that two or more point mutations together were required for complete rescue (SI Table 1 and data not shown). We cannot exclude the possibility that some of the second or third mutations found in our plasmids enhance rescue, but the observation that single point mutants in four separate positions complement well shows that multiple mutations are not always required for copper(II) resistance.
Fig. 1.
Copper-resistant phenotype of four DsbG mutants rescuing copper resistance of a dsbC-null strain (for complete list of mutants, see SI Table 1). Wt, wild-type.
DsbG is normally expressed from the chromosome at levels one-quarter that of DsbC, so one trivial mechanism by which these dsbG mutants can complement dsbC is that they do so by overexpression. This mechanism does not account for the effect of these mutations, however, because overexpression of wild-type DsbG could not complement the dsbC-null strain at this copper(II) concentration, and Western blot analysis revealed that the level of expression of DsbG was unchanged in the mutants relative to wild-type DsbG expressed from pBAD33a.
DsbG Mutants Show Enhanced Isomerase Activity in Vitro.
We purified the three mutants that best rescued dsbC-null copper(II) sensitivity (T200M, K113E, and V216M) and tested their ability to catalyze the isomerization of disulfide bonds in vitro. The standard measure of disulfide isomerase activity is to monitor the ability of an isomerase to unscramble incorrect disulfides present in a model protein substrate. The model protein we chose is hirudin, a small blood-clotting inhibitor that is commonly used in protein disulfide isomerization assays (17). Hirudin contains three disulfides in its native form. Addition of equimolar quantities of reduced DsbC to scrambled hirudin resulted in almost complete recovery of native hirudin (Fig. 2). Wild-type DsbG added in the same quantities was capable of generating only small quantities of native hirudin, consistent with previous reports that DsbG is a weaker isomerase in vivo than DsbC (10). Two of our DsbC-like DsbG mutants were much more active in unscrambling the disulfides than is wild-type DsbG. The T200M mutant was nearly as active as wild-type DsbC, and the K113E mutant was slightly less active than T200M. The V216M mutant was the most weakly active mutant in vitro, although it was still slightly more active than wild-type DsbG. Our results show that DsbG mutants isolated for their DsbC-like properties in vivo have enhanced DsbC-like isomerase activity in vitro.
Fig. 2.
Isomerase activity of DsbG mutants is enhanced. Disulfide-scrambled hirudin was incubated in the absence or in the presence of stoichiometric quantities of wild-type DsbC, DsbG, or the DsbG mutants and allowed to refold at 25°C for 18 h. Samples were acid trapped, and folding intermediates were separated by reversed-phase HPLC. The dotted lines labeled R and N denote the retention times of fully reduced and native hirudin, respectively.
DsbG Mutants Have More Reducing Redox Potentials.
We measured the redox potential of the T200M, K113E, and V216M DsbC-like mutants of DsbG proteins, as well as the wild-type DsbG and DsbC proteins, by equilibrium with glutathione (SI Fig. 5). Both K113 at −176 mV and T200M at −181 mV had much more reducing redox potentials than either wild-type DsbG at −125 mV or DsbC at −135 mV (12, 18). The redox potential of V216M was the same as that of wild-type DsbG (−131 mV).
Structures of DsbG Mutants Conferring Copper Resistance.
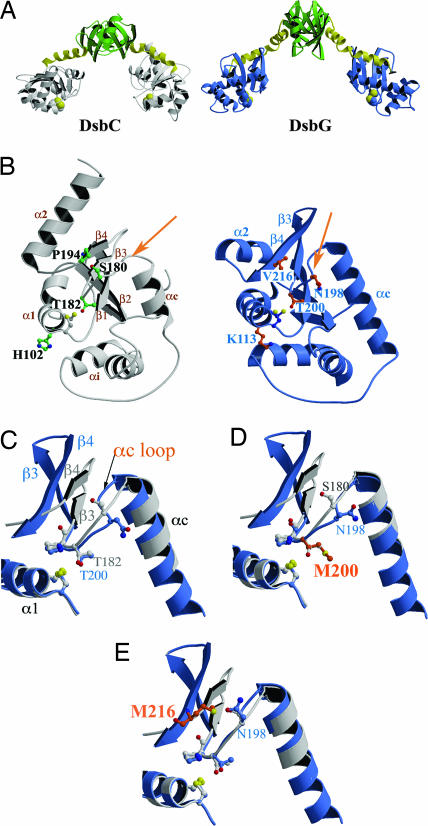
DsbC and DsbG are V-shaped homodimers. Each monomer contains an N-terminal dimerization domain and a catalytic thioredoxin domain connected by a linker helix (Fig. 3A). The four DsbG residues mutated in this work, K113, N198, T200, and V216, are surface-exposed and surround the redox active-site CXXC motif. The equivalent residues in DsbC, H102, S180, T182, and P194, are also exposed in DsbC (Fig. 3B) and cluster in two regions that interact with the partner protein DsbD (19). These regions are postulated to be involved in DsbC's interaction with misoxidized substrate proteins. This suggests that the residues mutated in DsbG form part of an interacting surface and that modifications alter the activity of the protein. To investigate this possibility, we crystallized three DsbG point mutants (K113E, T200M, and V216M) that best rescued dsbC-null copper sensitivity. These three mutants crystallized under conditions similar to wild-type DsbG and with the same C2 space group (14). Crystallographic statistics are given in SI Table 2. The root mean square deviations (rmsd) between the Cα atoms of wild-type DsbG and the mutants are 0.33 Å (K113E), 0.45 Å (T200M), and 1.15 Å (V216M), indicating no overall change in structure.
Fig. 3.
Comparison of the structures of DsbC, DsbG, and DsbC-like DsbG mutants identifies key regions functionally distinguishing the two proteins. Active-site cysteines are shown in yellow in all parts of the figure. (A) The structures of DsbC and DsbG show that they are both V-shaped homodimers with a thioredoxin-like domain (gray and blue, respectively) connected by a linker helix (yellow) to an N-terminal dimerization domain (green). (B) Comparison of the thioredoxin domains of DsbC (left, gray) and DsbG (right, blue). The αc loop (indicated by an arrow) links the connecting helix (αc) with strand β3. This loop has very different conformations in DsbC and DsbG. The residues mutated in DsbG that allowed it to gain DsbC-like properties are indicated (K113, N198, T200, and V216) and the corresponding residues in DsbC are also shown (H102, S180, T182, and P194, respectively). Two of these four residues are located on the αc loop (N198 and T200). (C) Superposition of wild-type DsbC (gray) and wild-type DsbG (blue), showing that they have very different αc loop conformations. (D) Superposition of wild-type DsbC (gray) and DsbG T200M (blue, mutated residue in orange; this work, Protein Data Bank ID code 2H0G), showing that the αc loop conformation of DsbG T200M is more like that of DsbC. (E) Superposition of wild-type DsbC (gray) and DsbG V216M (blue, mutated residue in orange; PDB ID code 2H0I), showing that the αc loop conformation of DsbG V216M is more like that of DsbC.
DsbG K113E Reverses Electrostatic Charge Near the Active Site.
K113 is located immediately after the redox active-site sequence, i.e., CPYCK, residues 109–113, of DsbG and contributes a positively charged patch next to the catalytic site; this positive charge is absent in DsbC (Fig. 4, compare DsbC wild type with DsbG wild type). In the DsbG structure, the side-chain amine of K113 interacts with the main-chain carbonyl oxygen of P108, fixing the K113 basic side chain close to the active site; the equivalent region in DsbC is more acidic than DsbG (Fig. 4). Upon mutation of DsbG K113 to E, the positive patch on the surface of DsbG is replaced with a negative patch, so that the active site is more acidic and therefore more DsbC-like. Our results thus suggest that mutation of K113 in DsbG destabilizes the thiolate form of C109, thereby making the protein more reducing.
Fig. 4.
Electrostatic surface of a portion of wild-type DsbC, wild-type DsbG, and variants of DsbG identifies a key difference between the two proteins. Positive and negative electrostatic potentials are shown in blue and red, respectively (saturation at 18 kT/e) for each of the proteins. Electrostatic surface representations were generated by using GRASP (28). These surfaces are derived from the crystal structures of wild-type DsbC (Protein Data Bank ID code 1EEJ), wild-type DsbG [PDB ID code 1V57 (8)], DsbG K113E (this work, PDB ID code 2H0H), and DsbG V216M (this work, PDB ID code 2H0I), and models of DsbG K113D, DsbG K113N, and DsbG K113W.
DsbG T200M Changes αc Loop Positioning.
The αc loop connects the linker helix αc to the β3 strand of the thioredoxin fold, and there is evidence that the equivalent loop in DsbA is involved in substrate binding (20). The T200M mutation affects the position of the αc loop, changing it to a conformation that is more like that of DsbC (Fig. 3 C–E). This change in conformation may open up access to the redox active-site and could affect the affinity of different substrate proteins for DsbG. The αc loop that is affected by the T200M mutant includes residue N198, which, when mutated to Ile, also allows DsbG to rescue ΔdsbC. We have been unable to solve the 3D structure of the weakly active N198I mutant, but suggest that it is also likely to modify the αc loop conformation.
DsbG V216M Affects Both Loop Positioning and Active-Site Charge.
The V216M mutation introduces a long hydrophobic side chain. Strikingly, mutation of this residue affects both the regions identified in the previously described two mutants, resulting in a shift of the αc loop to a conformation more like that of DsbC (Fig. 3 C–E) and an allosteric change in the K113 region adjacent to the active site. This latter change results in a rotation of the K113 side chain away from the active site, breaking the hydrogen bond with P108; this change unexpectedly reveals the acidic side chain of E163, thereby generating a negative patch on the surface, remarkably like that observed in DsbC (these allosteric changes are shown in SI Fig. 6). Thus, all of the DsbG mutants we have isolated that are dsbC-like in phenotype are also more DsbC-like in structure.
Analysis of our mutants' crystal structures suggested structural mechanisms by which these mutations might cause DsbG to behave more like DsbC. To assess whether our structural rationalizations were reasonable, we generated a complete spectrum of all possible amino acid substitutions at each of the four positions (K113, T200, V216, and N198) and determined which substitutions allowed DsbG to restore copper resistance. For the K113 position, we found that the amino acids that best rescue the dsbC-null phenotype are Asp, Asn, Glu, Gln, and surprisingly, Trp (SI Fig. 7). Models of the mutations (DsbG K113D, DsbG K113N, and DsbG K113W) suggest that they have the same effects as described for K113E; they all eliminate the positive charge close to the active site and abolish the hydrogen bond between residues 113 and 108. In the case of K113D, it is the acidic side chain that generates a negative patch, whereas for K113N and K113W, removal of the positively charged K113 residue reveals the side chain of E163, which maps to the same region of the protein and, in the absence of the K113 lysine side chain, contributes to a negative patch (Fig. 4 and SI Fig. 6).
Mutation of T200 to any of the hydrophobic amino acids makes DsbG DsbC-like. We suggest that these substitutions are likely to have effects similar to those that T200M has on the αc loop structure. The V216 position is best rescued by proline, which rescues copper resistance slightly better than the originally selected mutation, methionine. Interestingly, proline is the residue actually present at the corresponding position in wild-type DsbC. Finally, analysis of the N198 mutations shows that, generally, hydrophobic substitutions for asparagine allow some rescue of copper resistance.
Discussion
Multigene families are very common, and their functional analysis is a challenging problem. Often, individual members of a family have acquired distinct functions through sequence divergence. One particularly difficult problem is to determine which of the often numerous sequence and structural differences between family members are responsible for observed differences in function. Detailed examination of the sequence differences of two related proteins has, by itself, sometimes allowed identification of key residues distinguishing their functions (21). However, this method is often inadequate due to the multiple sequence differences present in most distantly related proteins. Without an intimate knowledge of the mechanism of action of the two proteins, it can be difficult to determine which sequence differences contribute to functional divergence and which simply represent genetic drift. In this paper, we have addressed this problem by using the approach of putting selective pressure to attempt to determine which of the structural differences between two distantly related protein folding catalysts are functionally important.
We examined the two E. coli disulfide isomerases, DsbC and DsbG. We recently found that dsbC-null strains are copper sensitive, whereas dsbG-null strains are not. We selected for gain-of-function mutants in dsbG that could rescue null mutations in dsbC. Surprisingly, point mutants in single nonconserved amino acids in DsbG resulted in a protein that could complement the copper sensitivity of null mutations in dsbC. Other workers have previously shown this type of outcome, in which directed evolution reveals changes in nonconserved residues that result in novel functions (21). However, few of such studies have examined the structural basis for the novel acquisition of function. We note that Jurgens et al. (4), using random mutagenesis and selection, managed to generate a single point mutant in HisA, an enzyme involved in an Amadori rearrangement in histidine biosynthesis that could at some level catalyze the same rearrangement in tryptophan biosynthesis. However, this HisA mutant enzyme was rather ineffective, with a kcat/Km value that was 10,000-fold lower than that of the analogous tryptophan enzyme.
In vitro analysis showed that our DsbG mutants have substantially increased disulfide isomerase activity; one mutant exhibited nearly as much disulfide isomerase activity as DsbC. Surprisingly, little is known about the in vivo requirements for disulfide isomerization. The presence of two active sites in the enzyme is important. Eukaryotic protein disulfide isomerase (PDI) normally has two thioredoxin-like catalytic domains, and DsbC is a dimer. A single catalytic domain of PDI in isolation has only 3–5% of the isomerase activity of wild-type PDI; there is also evidence that the dimerization of DsbC is required for its chaperone activity and that the two active sites need to be in a specific geometry relative to each other (22, 23). Our DsbG mutants migrate on gel filtration columns identically to wild-type DsbG and crystallize in the same space group, indicating that their association state is not profoundly altered. It is also likely that the redox potential of the isomerase is important for its function, because in general one of the most important parameters in determining the in vivo function of thiol disulfide exchange enzymes is their redox potential. There are several possible mechanisms of disulfide isomerization, but all require that the isomerase function as both an acceptor and a donor of disulfides. Thus, a priori, one would expect the redox potential of the isomerase to be finely balanced so that it can both accept and donate disulfides. Surprisingly, wild-type DsbC and DsbG both have very strongly oxidizing redox potentials (−135 mV and −125 mV, respectively), much more oxidizing than that of eukaryotic PDI (−188 mV for PDI's a domain and −152 mV for its a′ domain) (24) and nearly as oxidizing as DsbA (−122 mV), which is the most oxidizing thiol disulfide known. This observation implies that the ability of DsbC and DsbG to donate disulfides is vital for their action in vivo. Two of three of our most DsbC-like DsbG mutants have redox potentials shifted strongly in the reducing direction relative to wild-type DsbG and DsbC. Thus, one way of improving the ability of DsbG to resolve incorrect disulfides may be to enhance the reducing power of the isomerase. Raines and coworkers (25) selected thioredoxin variants that were capable of rescuing the lethal phenotype of null mutants in yeast PDI. These mutants made the protein more oxidizing. Recent evidence suggests that sulfhydryl oxidation, not disulfide isomerization, is the primary function of yeast PDI (26), so it appears that the mutants answered the selection for a sulfhydryl oxidase by mutating the active site so it is more oxidizing.
When we solved the crystal structures of three of the most DsbC-like DsbG mutants, we found that all mutations map to two small well defined surface-exposed regions. The crystal structures of three of these mutants that showed the largest effect on the complementation assays reveal changes in two areas of DsbG's thioredoxin domain that make it strikingly more similar to DsbC. These mutants (DsbG K113E, DsbG T200M, and DsbG V216M) affect a charged region and the αc loop, which lie directly adjacent to the active site. Both regions have been postulated to be involved in substrate binding, and both show distinct differences between DsbC and DsbG. In DsbC, the region adjacent to the active site is negatively charged; in DsbG, this region is positive. DsbG variants that rescue dsbC-null mutants cause this region to become less positively charged and therefore more like DsbC. Similarly, the αc loop shows distinct conformations in DsbC versus DsbG; DsbG mutants affecting the αc loop significantly change the structure of this loop, making it more similar to DsbC. Thus, the crystal structures of the DsbG mutants reveal changes in critical structural features, which probably would have remained unexplored by examination of sequence conservation alone. A standard way to analyze the essential differences between two gene families is to try to deduce the mutational path that evolution took after this gene duplication event; or, at a minimum, to try to establish residues that have clearly undergone positive selection. DsbC and DsbG share only 24% sequence identity, so the effort to retrace evolutionary changes was not successful. Even if this retracing were possible, it would require multiple amino acid substitutions. Because of the high level of divergence even within the DsbC and DsbG families, we were also unable to establish residues that are clearly under positive selection (see SI Fig. 8 and SI Table 3). In contrast, our technique of directly selecting for mutations that can functionally complement a dsbC growth phenotype provides a direct experimental route for determining the essential differences between DsbC and DsbG. Our work supports the conclusion that at least part of the functional differences between two distantly related proteins can be bridged by single point mutants that alter just the structural active-site region of one enzyme so that it more closely resembles that of the other.
Materials and Methods
Bacterial Strains and Plasmids.
AH131 (BL21 dsbC::kan) was used for expression studies (13). Plasmid pMB69 contains DsbC cloned under the arabinose promoter in pBAD33a (27) and was obtained from G. Georgiou (University of Texas, Austin). Plasmid pAH243 is also derived from pBAD33a and was constructed in this work. Briefly, His-tagged DsbG from pETDsbG2 (10) was PCR amplified; the PCR product was digested with XbaI and SalI and ligated into XbaI/SalI-digested pBAD33a. All plasmid construction was verified by DNA sequencing.
Whole-Plasmid Mutagenesis and Selection.
Primers used for whole-plasmid mutagenesis of pAH243 are 5′-CTCGGAGATCTTCCCCATCGGTGATGTCGG-3′ and 5′-TCACAAGATCTGGCTCGCCACTTCGGGCTCA-3′. These primers introduce a unique BglII restriction site into pAH243. To mutagenize pAH243, the Genemorph II random mutagenesis kit (Stratagene, La Jolla, CA) was used with minor modifications as follows. Briefly, denaturation temperature was increased to 98°C, the elongation time was increased to 2 min/kb of template, 4% DMSO was added to the PCR mixture, and a “hot-start” PCR was performed. PCR product was digested with BglII and DpnI and washed by using a Qiaquik kit (Qiagen, Valencia, CA). Linear mutagenized plasmid was ligated by using T4 DNA ligase, as directed by New England Biolabs (Beverly, MA), and precipitated by using the Pelletpaint protocol (Novagen, Madison, WI). Ligation product was resuspended in ≈3 ml of distilled deionized H2O and electroporated into electrocompetent AH131 cells. Transformants were plated directly onto copper selection plates containing various concentrations of copper, 34 mg/ml chloramphenicol, and 0.1 mM arabinose to induce expression of mutagenized DsbG. Possible copper-resistant colonies were streaked out on copper/chloramphenicol plates with and without arabinose to determine whether copper resistance was plasmid-dependent. DNA was extracted from positive clones and sequenced.
Bacterial Growth and Selection Conditions.
Initial selections were performed by using Bacto Brain Heart Infusion (BHI) medium (Difco, Sparks, MD), supplemented with 2% agar, 34 mg/ml chloramphenicol, and between 7 and 10 mM CuCl2. During the course of these experiments, the source used by Difco of certain components of this medium changed and the copper sensitivity of the dsbC-null strain relative to the wild-type strain decreased. Screening of all possible amino acid substitutions as shown in SI Fig. 7 was therefore performed on Terrific Broth (TB) (12 g/liter tryptone, 24 g/liter yeast extract, 4 ml/liter glycerol, 17 mM KH2PO4, and 72 mM K2HPO4) supplemented with 2% agar, 34 mg/ml chloramphenicol, and between 16 and 18 mM CuCl2, on which all strains showed similar phenotypes, as was seen on the original Bacto BHI.
Multisite-Directed Mutagenesis and Site-Directed Mutagenesis.
Multisite-directed mutagenesis was performed to mutagenize positions K113, N198, T200, and V216 of dsbG on pAH243 as indicated in the Stratagene multisite-directed mutagenesis kit. PCR was performed as directed, and the product was digested with DpnI, precipitated by using the Pelletpaint protocol (Novagen), and resuspended into ≈3 ml of distilled deionized H2O. Product was electroporated into AH573 or XL1-blue, DNA was extracted, and products were sequenced. DNA for each particular amino acid substitution was used to transform AH131. Particular point mutations were introduced into the wild-type dsbG gene on pAH243 by using a Stratagene Quikchange kit. All point mutations were verified by DNA sequencing.
Spot Titers.
Spot titrations for copper resistance were performed to quantify the relative copper resistance caused by each amino acid at a particular position. Briefly, strains were grown overnight in LB medium containing chloramphenicol (34 mg/ml) and diluted 1:100 into BHI containing chloramphenicol (34 mg/ml). Strains were grown to midlogarithmic phase at 37°C and serially diluted into 150 mM NaCl. Two milliliters of each dilution was plated onto TB plates containing 34 mg/ml chloramphenicol and between 17 and 18 mM CuCl2, with and without 0.1 mM arabinose. All spot titrations were performed in duplicate or triplicate.
Hirudin Refolding.
Scrambled hirudin was a kind gift from Shu Quan (University of Michigan), prepared according to Lu and Chang (17). Hirudin samples were diluted to 24 μM, incubated with or without 24 μM reduced DsbG or mutants in 100 mM sodium phosphate/1 mM EDTA, pH 7.0. Folding reactions were quenched by addition of 10% (vol/vol) formic acid after 18-h incubation at room temperature. The reaction products were separated by reversed-phase HPLC on a 218TP54 C18 column (Vydac, Hesperia, CA) at 55°C with an acetonitrile gradient (19–25%, vol/vol, 30 ml) in 0.1% trifluoroacetic acid. The eluted proteins were detected by their absorbance at 220 nm.
Protein Purification, Redox Potential Determination, and Crystallography.
Wild-type DsbC was purified by using a 6-His tag and nickel chromatography as described in ref. 11. All DsbG variants were purified from pBAD33a by using a 6-His tag and nickel chromatography. For crystallography, protein was further purified by using cation exchange (1-ml Hi-trap SP column; GE Healthcare Biosciences, Piscataway, NJ). Fractions containing DsbG were pooled and concentrated to at least 10 mg/ml in 25 mM Hepes buffer (pH 6.7)/50 mM NaCl. DsbC, DsbG, and mutants were reduced according to Hiniker and Bardwell (13). DsbC and DsbG concentrations were determined by absorbance at 280 nm, and molar concentration was calculated as protein monomers. The redox potentials of DsbG and mutants were determined as described by Bessette et al. (10). Crystals of DsbG mutants K113E, V216M, and T200M were grown by using hanging-drop and sitting-drop vapor diffusion methods at 277 K or 298 K, essentially as described for wild-type DsbG (14, 16), additional details are given in the legend to SI Fig. 6.
Supplementary Material
Acknowledgments
We thank Jianzhi Zhang and Ursula Jakob for their encouragement and their reading of this manuscript. This research was supported in part by a grant from the National Institutes of Health (to J.C.A.B.), Australian Research Council Grants (to B.H. and to J.L.M.), and a University of Queensland Early Career Research Award (to B.H.); J.C.A.B. is an Investigator of the Howard Hughes Medical Institute.
Footnotes
The authors declare no conflict of interest.
Data deposition: The atomic coordinates and structure factors for the three crystal structures have been deposited with the Protein Data Bank, www.pdb.org (PDB ID codes 2H0H, 2H0I, and 2H0G).
This article contains supporting information online at www.pnas.org/cgi/content/full/0704692104/DC1.
References
- 1.Bartel DP, Unrau PJ. Trends Cell Biol. 1999;9:M9–M13. [PubMed] [Google Scholar]
- 2.Yuan L, Kurek I, English J, Keenan R. Microbiol Mol Biol Rev. 2005;69:373–392. doi: 10.1128/MMBR.69.3.373-392.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Rothman SC, Kirsch JF. J Mol Biol. 2003;327:593–608. doi: 10.1016/s0022-2836(03)00095-0. [DOI] [PubMed] [Google Scholar]
- 4.Jurgens C, Strom A, Wegener D, Hettwer S, Wilmanns M, Sterner R. Proc Natl Acad Sci USA. 2000;97:9925–9930. doi: 10.1073/pnas.160255397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Griswold KE, Aiyappan NS, Iverson BL, Georgiou G. J Mol Biol. 2006;364:400–410. doi: 10.1016/j.jmb.2006.09.01. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bader MW, Hiniker A, Regeimbal J, Goldstone D, Haebel PW, Riemer J, Metcalf P, Bardwell JC. EMBO J. 2001;20:1555–1562. doi: 10.1093/emboj/20.7.1555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ortenberg R, Gon S, Porat A, Beckwith J. Proc Natl Acad Sci USA. 2004;101:7439–7444. doi: 10.1073/pnas.0401965101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Masip L, Pan JL, Haldar S, Penner-Hahn JE, DeLisa MP, Georgiou G, Bardwell JC, Collet JF. Science. 2004;303:1185–1189. doi: 10.1126/science.1092612. [DOI] [PubMed] [Google Scholar]
- 9.Rietsch A, Belin D, Martin N, Beckwith J. Proc Natl Acad Sci USA. 1996;93:13048–13053. doi: 10.1073/pnas.93.23.13048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bessette PH, Cotto JJ, Gilbert HF, Georgiou G. J Biol Chem. 1999;274:7784–7792. doi: 10.1074/jbc.274.12.7784. [DOI] [PubMed] [Google Scholar]
- 11.Hiniker A, Bardwell JC. J Biol Chem. 2004;279:12967–12973. doi: 10.1074/jbc.M311391200. [DOI] [PubMed] [Google Scholar]
- 12.van Straaten M, Missiakas D, Raina S, Darby NJ. FEBS Lett. 1998;428:255–258. doi: 10.1016/s0014-5793(98)00539-0. [DOI] [PubMed] [Google Scholar]
- 13.Hiniker A, Collet JF, Bardwell JC. J Biol Chem. 2005;280:33785–33791. doi: 10.1074/jbc.M505742200. [DOI] [PubMed] [Google Scholar]
- 14.Heras B, Edeling MA, Schirra HJ, Raina S, Martin JL. Proc Natl Acad Sci USA. 2004;101:8876–8881. doi: 10.1073/pnas.0402769101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Haebel PW, Goldstone D, Katzen F, Beckwith J, Metcalf P. EMBO J. 2002;21:4774–4784. doi: 10.1093/emboj/cdf489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Heras B, Edeling MA, Byriel KA, Jones A, Raina S, Martin JL. Structure (London) 2003;11:139–145. doi: 10.1016/s0969-2126(03)00005-4. [DOI] [PubMed] [Google Scholar]
- 17.Lu B. Y., Chang JY. Anal Biochem. 2005;339:94–103. doi: 10.1016/j.ab.2004.12.030. [DOI] [PubMed] [Google Scholar]
- 18.Zapun A, Missiakas D, Raina S, Creighton TE. Biochemistry. 1995;34:5075–5089. doi: 10.1021/bi00015a019. [DOI] [PubMed] [Google Scholar]
- 19.Rozhkova A, Stirnimann CU, Frei P, Grauschopf U, Brunisholz R, Grutter MG, Capitani G, Glockshuber R. EMBO J. 2004;23:1709–1719. doi: 10.1038/sj.emboj.7600178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kadokura H, Tian H, Zander T, Bardwell JC, Beckwith J. Science. 2004;303:534–537. doi: 10.1126/science.1091724. [DOI] [PubMed] [Google Scholar]
- 21.Joerger AC, Mayer S, Fersht AR. Proc Natl Acad Sci USA. 2003;100:5694–5699. doi: 10.1073/pnas.0531477100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zhao Z, Peng Y, Hao SF, Zeng ZH, Wang CC. J Biol Chem. 2003;278:43292–43298. doi: 10.1074/jbc.M306945200. [DOI] [PubMed] [Google Scholar]
- 23.Segatori L, Murphy L, Arredondo S, Kadokura H, Gilbert H, Beckwith J, Georgiou G. J Biol Chem. 2006;281:4911–4919. doi: 10.1074/jbc.M505453200. [DOI] [PubMed] [Google Scholar]
- 24.Wilkinson R, Xiao R, Gilbert HF. J Biol Chem. 2005;280:11483–11487. doi: 10.1074/jbc.M414203200. [DOI] [PubMed] [Google Scholar]
- 25.Chivers PT, Laboissiere MCA, Raines RT. EMBO J. 1996;15:2659–2667. [PMC free article] [PubMed] [Google Scholar]
- 26.Solovyov S, Xiao R, Gilbert HF. J Biol Chem. 2004;279:34095–34100. doi: 10.1074/jbc.M405640200. [DOI] [PubMed] [Google Scholar]
- 27.Guzman LM, Belin D, Carson MJ, Beckwith J. J Bacteriol. 1995;177:4121–4130. doi: 10.1128/jb.177.14.4121-4130.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Nicholls A, Sharp KA, Honig B. Proteins. 1991;11:281–296. doi: 10.1002/prot.340110407. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.