Abstract
A fundamental challenge in the redox biology of Mycobacterium tuberculosis (Mtb) is to understand the mechanisms involved in sensing redox signals such as oxygen (O2), nitric oxide (NO), and nutrient depletion, which are thought to play a crucial role in persistence. Here we show that Mtb WhiB3 responds to the dormancy signals NO and O2 through its iron-sulfur (Fe-S) cluster. To functionally assemble the WhiB3 Fe-S cluster, we identified and characterized the Mtb cysteine desulfurase (IscS; Rv3025c) and developed a native enzymatic reconstitution system for assembling Fe-S clusters in Mtb. EPR and UV-visible spectroscopy analysis of reduced WhiB3 is consistent with a one-electron reduction of EPR silent [4Fe-4S]2+ to EPR visible [4Fe-4S]+. Atmospheric O2 gradually degrades the WhiB3 [4Fe-4S]2+ cluster to generate a [3Fe-4S]+ intermediate. Furthermore, EPR analysis demonstrates that NO forms a protein-bound dinitrosyl–iron–dithiol complex with the Fe-S cluster, indicating that NO specifically targets the WhiB3 Fe-S cluster. Our data suggest that the mechanism of WhiB3 4Fe-4S cluster degradation is similar to that of fumarate nitrate regulator. Importantly, Mtb ΔwhiB3 shows enhanced growth on acetate medium, but a growth defect on media containing glucose, pyruvate, succinate, or fumarate as the sole carbon source. Our results implicate WhiB3 in metabolic switching and in sensing the physiologically relevant host signaling molecules NO and O2 through its [4Fe-4S] cluster. Taken together, our results suggest that WhiB3 is an intracellular redox sensor that integrates environmental redox signals with core intermediary metabolism.
Keywords: dormancy, redox, metabolism, iron–sulfur
The ability of Mycobacterium tuberculosis (Mtb) to maintain a state of latent tuberculosis infection is responsible for its remarkable success as a pathogen. Persistent infection is the result of complex interplay between the host immune system and bacterial survival mechanisms, and little is known about the specific bacterial and host components involved in this process. How Mtb can persist in human tissues for months to decades without replicating and then abruptly resume growth and cause active disease is a fundamental question in the tuberculosis field. Understanding the mechanistic basis of persistence has become the focus of much research, primarily because persistent bacilli are in a state of “drug unresponsiveness” wherein they are insensitive to antimycobacterial drugs. The role of oxygen tension in Mtb persistence has received wide attention because it has been demonstrated that although the tubercle bacilli require oxygen for replication, the bacilli can survive for years without oxygen in vitro (1). Accordingly, the mechanism of physiological adaptation in response to limiting O2 availability has been studied extensively (1). Studies have revealed that a rapid shift from aerobic to an O2-deficient environment results in bacterial death. However, the bacilli survive upon gradual depletion of O2, indicating that an ordered metabolic shutdown is necessary for adaptation to anoxia (1). Several groups have suggested the role of hypoxia-induced regulon (Dos) in the downshift of Mtb from the replicative to the persistent state. Of particular interest is the finding that a significant degree of overlap exists between Mtb gene expression patterns seen in response to hypoxia and those observed after exposure to NO. These studies revealed intriguing insights into the respiratory networks of Mtb that presumably lead to decreased energy production during in vivo adaptation (2). Long-term persistence followed by resumption of metabolic activity argues for the coordinated regulation of a subset of Mtb metabolic pathways during persistence (3). However, the molecular mechanism of how NO, hypoxia, and nutrient stress are sensed to induce metabolic adaptation is not known. Recent work suggested the involvement of a heme-containing cytochrome c oxidase and/or the presence of a fumarate nitrate regulator (FNR)-like sensor (2, 4, 5). As a result, it is postulated that Mtb possesses one or more sensors, which detect environmental redox signals such as O2, NO, and fluctuations in the intracellular redox state, to promote bacterial persistence.
Fe-S cluster proteins are known to play essential roles in sensing external signals as well as the intracellular redox state of microbial cells (6). Fe-S cluster transcription factors such as SoxR and FNR have been shown to be the primary sensors of NO and hypoxia and are responsible for the metabolic adaptation of Escherichia coli to environmental stresses (6). Mtb homologs of FNR and SoxR would be considered excellent candidates to sense and relay redox signals. However, a clear FNR-like protein is absent in Mtb. Therefore, one or more, yet-to-be-identified “redox sensors” likely play a crucial role in the downshift of Mtb to the dormant state. Of interest to us is the role of Mtb proteins in sensing environmental host signals. In an accompanying paper [Kumar et al. (28)], we demonstrated that Mtb DosS and DosT are heme proteins that directly sense O2, NO, and CO to modulate expression of the Mtb dormancy regulon. In addition, we have previously discovered a gene, whiB3, that plays a role in Mtb virulence (7). Therefore, of particular interest to us is the role of the Mtb WhiB family in sensing environmental host signals. In this work we hypothesize that Mtb WhiB3 is an Fe-S cluster protein capable of sensing redox signals through its Fe-S cluster. To test this hypothesis, we developed an endogenous reconstitution system to functionally assemble Mtb Fe-S clusters. We used EPR, UV-visible spectroscopy, and a sensitive radioactive assay to characterize how WhiB3 responds to O2 and NO, and we draw several important parallels with FNR. Finally, we analyzed the growth phenotype of MtbΔwhiB3 under nutritional stress. We anticipate that understanding the molecular and biochemical mechanism of this redox pathway will contribute toward understanding how Mtb maintains a state of latency.
Results
Mtb IscS (Rv3025c) Is a Functional Cysteine Desulfurase.
Bacteria have developed (at least) three distinct, highly conserved systems (Nif, Isc, and Suf) for the biogenesis of Fe-S clusters in vivo (8). In vitro, Fe-S clusters can be assembled anaerobically by using l-cysteine (sulfur donor), an l-cysteine desulfurase (e.g., NifS/IscS), an iron source, and a reducing agent (8). Most enzymatic Fe-S cluster assembly systems make use of NifS, the cysteine desulfurase of Azotobacter vinelandii. We were concerned that a non-native enzyme might not appropriately assemble Mtb Fe-S clusters. Therefore, we sought to identify the Mtb homolog of NifS and use it in an Mtb Fe-S assembly system. To date, a mycobacterial homolog of NifS/IscS remained uncharacterized, and no study has yet demonstrated the assembly of any Mtb Fe-S cluster protein using NifS/IscS. Rv3025c, an Mtb homolog of A. vinelandii NifS, shows the highest homology (37% identity, 55% homology), whereas three other Mtb homologs (Rv1464, Rv3778c, and Rv3700c) show relatively weak homology. Mtb IscS (Rv3025c) contains a conserved pyridoxal 5′-phosphate-binding site at Lys-205 and an active Cys-329 residue required for the formation of an enzyme-bound cysteinyl persulfide (8), findings consistent with Rv3025c encoding for an l-cysteine desulfurase. Overexpressed and purified IscS from E. coli exhibited a yellow color, and UV-visible spectroscopy of IscS showed a major absorbance peak at 420 nm, which are characteristic features of pyridoxal 5′-phosphate-dependent enzymes (9). The addition of l-cysteine, but not l-alanine or l-histidine, reproducibly increased absorbance at 315 nm and showed a concomitant decrease in absorbance at 420 nm [supporting information (SI) Fig. 6A]. These properties are highly suggestive of l-cysteine desulfurase activity (8, 9).
In the first step of Fe-S cluster assembly, NifS/IscS acquires sulfur by converting cysteine to alanine, resulting in the formation of cysteinyl persulfide. Under reducing conditions, persulfide-bound sulfur is released as free sulfide for the reconstitution of Fe-S cluster in the target protein (8). To evaluate the ability of Mtb IscS to use l-cysteine as a substrate to generate persulfide, we developed a sensitive 35S transfer assay. The results demonstrate the acquisition of 35S from 35S-labeled l-cysteine by IscS (see Experimental Procedures and SI Fig. 6B, lane 1). To assess the role of the cysteinyl residue in sulfur acquisition, IscS was alkylated by using N-ethylmaleimide and included in the 35S transfer assay. A drastic reduction in 35S incorporation from 35S-labeled cysteine in alkylated IscS was observed, thereby establishing a role for the highly reactive IscS cysteinyl thiolate in the formation of cysteinyl persulfide (SI Fig. 6B, lane 2). Finally, in the presence of the reducing agent DTT (SI Fig. 6B, lane 3), we demonstrated the partial loss of 35S from IscS, which is caused by the release of sulfide from persulfide-bound sulfur intermediate. Taken together, these data confirm that Mtb IscS has active cysteine desulfurase activity.
Mtb IscS Can Assemble an Fe-S Cluster in WhiB3.
Aerobically purified WhiB3 is reddish-brown in color, a characteristic feature of Fe-S cluster proteins. In addition, the primary sequence of WhiB3 contains a likely Fe-S cluster-binding motif consisting of four conserved cysteine residues C-X29-C-X2-C-X5-C (where X is any amino acid) (10). To demonstrate that Mtb IscS is capable of mobilizing S from l-cysteine to assemble the putative WhiB3 Fe-S cluster, we included 35S-labeled cysteine in the reaction mixture and monitored incorporation of 35S into WhiB3. We successfully demonstrated IscS-dependent transfer of 35S from l-cysteine to WhiB3 over time (see Fig. 1A). As expected, we identified multiple WhiB3 species on native polyacrylamide gels, which are attributable to homooligomerization and reconstitution intermediates (Fig. 1). Importantly, in our control reaction mixture we excluded IscS or cysteine and did not generate any signal (data not shown).
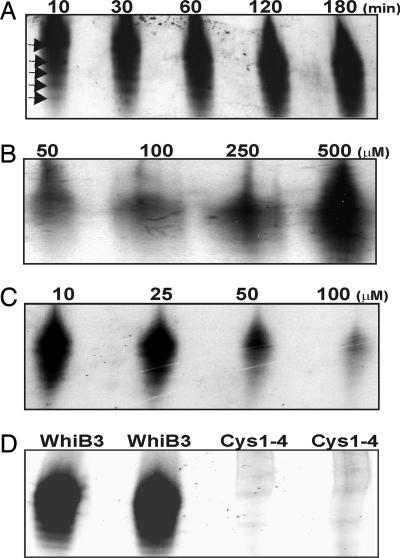
Fig. 1.
Mtb WhiB3 is an Fe-S cluster protein. (A) WhiB3 Fe-S cluster reconstitution catalyzed by IscS over time. At various time intervals (10, 30, 60, 120, and 180 min), aliquots from the reconstitution reactions were removed and analyzed by native PAGE followed by autoradiography. (B and C) Reconstitution of the WhiB3 Fe-S cluster was carried out for 1 h in the presence of 50, 100, 250, and 500 μM Fe2+ (B) or 10, 25, 50, and 100 μM Fe2+ chelator (2,2-dipyridyl) (C). (D) The four conserved cysteine residues in WhiB3 are essential for Fe-S cluster reconstitution. Fe-S cluster reconstitution was carried out with WhiB3 and WhiB3ΔCys-1–4 as described earlier and analyzed by native PAGE.
Next, we examined the requirement of Fe2+ for IscS-mediated Fe-S cluster assembly. Upon addition of increasing amounts of Fe2+, we observed a gradual increase in signal (Fig. 1B). In contrast, removal of Fe2+ with the iron chelator 2,2-dipyridyl caused a gradual decrease in signal (Fig. 1C). Lastly, we replaced all four cysteines in WhiB3 with alanine (WhiB3ΔCys-1–4). WhiB3ΔCys-1–4 was aerobically purified and used in the 35S transfer assay with IscS. The absence of 35S incorporation into WhiB3ΔCys-1–4 suggests that the four cysteine residues are ligands of the WhiB3 Fe-S cluster (Fig. 1D). Taken together, the above results show that WhiB3 is indeed an Fe-S cluster protein. The results also demonstrate that we have successfully developed an Mtb Fe-S cluster assembly system for WhiB3 and indicate a role for IscS in the biogenesis of Mtb Fe-S clusters.
Mtb WhiB3 Contains a 4Fe-4S Cluster.
Having established that Mtb WhiB3 is an Fe-S cluster protein, we now sought to determine the cluster type. We used UV-visible and EPR spectroscopy and thoroughly characterized the biophysical and biochemical properties of WhiB3. We found the UV-visible spectrum of aerobically purified WhiB3 (data not shown) to be indistinguishable from that of the bacterial type 2Fe-2S ferredoxins, exhibiting peaks at 413 nm and two broad shoulders near 458 and 330 nm (11). However, because WhiB3 was initially aerobically purified, we believe the 2Fe-2S cluster was most likely a degradation product. Therefore, to reconstitute the native WhiB3 Fe-S cluster, WhiB3 was then incubated under anaerobic conditions with freshly purified IscS, l-cysteine, DTT, and FeCl2 as described in Experimental Procedures. During anaerobic reconstitution, a greenish-brown color developed within 30 min (Fig. 2A, Inset). Upon further incubation, we observed a time-dependent increase in the intensity ≈413 nm with no other resolved features (Fig. 2A). Both the absorbance ≈413 nm and the color development are characteristic of FNR-type 4Fe-4S cluster formation (12). As shown in Fig. 2A, IscS completed assembly of the WhiB3 Fe-S cluster within 7 h, as is evidenced by the single broad shoulder at 413 nm and an A413/A280 ratio of 0.31. This ratio is consistent with the reconstitution of 4Fe-4S cluster in WhiB3. The UV-visible spectrum of reconstituted WhiB3 confers a molar absorption coefficient (ε) at 413 nm of 12,500 M−1 cm−1, which is similar to that of 4Fe-4S clusters (13). As additional verification, we performed a reconstitution reaction using WhiB3ΔCys-1–4 and observed no color development. As expected, we observed no peak at 413 nm (Fig. 2B), confirming the absence of a 4Fe-4S cluster in WhiB3ΔCys-1–4. To characterize further the redox behavior of the WhiB3 4Fe-4S cluster, anaerobically reconstituted WhiB3 was subjected to one-electron reduction with sodium dithionite (DTH). Reduction caused partial bleaching of the greenish-brown color with the concomitant loss of the peak at 413 nm (Fig. 2A). The spectral properties of DTH-reduced WhiB3 are characteristic of the reduction of a [4Fe-4S]2+ to a [4Fe-4S]+ state (10).
Fig. 2.
Spectroscopic characterization of WhiB3. UV-visible spectroscopy of WhiB3 Fe-S cluster reconstitution is shown. (A) Reconstitution was carried out inside an anaerobic glove box as described in Experimental Procedures. Time points at which samples were scanned by a UV-visible spectrophotometer are indicated. Note the time-dependent increase in the characteristic 4Fe-4S absorption peak at 413 nm. After completion of the Fe-S cluster reassembly, the Fe-S cluster was reduced by adding 5 mM DTH. (Inset) Development of a greenish-brown color during the WhiB3 Fe-S cluster reconstitution before (left cuvette) and after (right cuvette) the reconstitution reaction. (B) The four conserved cysteine residues (C23-C53-C56-C62) in WhiB3 are required for Fe-S cluster assembly. Fe-S cluster reconstitution in WhiB3 and WhiB3ΔCys-1–4 was carried out as described in Experimental Procedures and analyzed by UV-visible spectroscopy. (C and D) EDFS EPR spectra of IscS- (C) and NifS- (D) reconstituted WhiB3 after reduction with DTH. EPR spectra were obtained at 12 K, the EPR experimental conditions; π/2 and π pulses are 16 and 32 ns, τ = 180 ns. The spectra were acquired with 60 shots with a two-step phase cycle at a repetition rate of 1 kHz. Microwave frequency = 9.8046 GHz.
Finally, to identify conclusively the type of Fe-S cluster, we exploited echo-detected field sweep (EDFS) EPR and analyzed anaerobically reconstituted WhiB3. We observed no signal, suggestive of the presence of a nonparamagnetic species. However, upon reduction with DTH, WhiB3 exhibits a signal at g = 2.06 and 1.94 (Fig. 2C). This signal is consistent with a one-electron reduction of an EPR silent [4Fe-4S]2+ species to a paramagnetic [4Fe-4S]+ cluster with an electron spin S = 1/2 (10), and corroborated our UV-visible spectroscopic data. As an additional verification of the IscS-dependent reconstitution of the WhiB3 Fe-S cluster, we also tested the ability of the widely used A. vinelandii NifS to reassemble the WhiB3 Fe-S cluster. The A. vinelandii NifS-treated WhiB3 samples showed UV-visible and EPR characteristics virtually identical to those of the IscS-treated WhiB3 samples. However, the intensity of the absorbance at 413 nm and the EPR signals were much stronger compared with the IscS-treated WhiB3 sample (Fig. 2D). Altogether, the data generated by several independent approaches convincingly demonstrate that Mtb WhiB3 contains a redox-responsive 4Fe-4S cluster.
WhiB3 Responds to NO and O2.
It is widely believed that NO and O2 are key signaling molecules required for induction of the Mtb dormancy program. Therefore, determining the identity of Mtb proteins that react with these diatomic gases is important. In E. coli, the Fe-S cluster-containing transcription factors, FNR (4Fe-4S) and SoxR (2Fe-2S), have been shown to be direct sensors of NO and O2 (6). To establish whether WhiB3 is capable of reacting with either gas, in vitro reconstituted WhiB3 was independently exposed to O2 or NO. To assess sensitivity to atmospheric O2, air was bubbled through the sample for 2 min, and the oxygen-induced changes in the UV-visible spectrum were monitored over time. At an early time point (5 min), we observed a nominal but highly reproducible increase in absorbance at 413 nm followed by a progressive decrease over 60 min accompanied by an increase in absorbance in the 500- to 600-nm region (Fig. 3A). At subsequent time points (90–180 min), there was a significant reduction in the absorbance at 413 nm with the appearance of a new peak at 330 nm and a broad shoulder at 458 nm (Fig. 3A). These spectral features are similar to the UV-visible spectra of aerobically purified WhiB3 and are consistent with an oxygen-dependent conversion of a 4Fe-4S to a 2Fe-2S cluster. A plot of ΔA413 nm against time revealed the loss of ≈75% of the 4Fe-4S cluster within 180 min (Fig. 3A, Inset).
Fig. 3.
The Mtb WhiB3 Fe-S cluster responds to O2 and NO. (A) Anaerobically reconstituted WhiB3 was purified by size-exclusion column and exposed to O2 by bubbling air through the sample for 2 min. UV-visible spectra were acquired before and after air exposure at various time intervals as indicated. (Inset) Rate of the 4Fe-4S cluster loss was determined by calculating the percent loss of absorbance at 413 nm upon exposure to air at various time intervals. (B) The O2-treated sample of reconstituted WhiB3 was withdrawn immediately (5 min) after exposure to O2 (red line) and 60 min after exposure (blue line) and analyzed by EPR. EDFS EPR spectra were measured at 12 K; the π/2 and π pulses used were 16 and 32 ns with, τ = 180 ns. The spectrum was acquired with 60 shots with a two-step phase cycling at repetition rate of 1 kHz. Microwave frequency = 9.806 GHz. The appearance of a sharp signal at g = 2.01 indicates a [3Fe-4S]+ cluster. (C) Loss of the 4Fe-4S cluster in WhiB3 was also confirmed by exposing 35S-labeled WhiB3 to air as described before. At various time intervals (0, 30, 60, 90, and 120 min), samples were removed and analyzed by native PAGE followed by autoradiography (Upper) and SDS/PAGE followed by Coomassie blue staining to rule out degradation of WhiB3 upon air exposure (Lower). (D) Effect of NO on WhiB3. Reconstituted WhiB3 was exposed to various concentrations of proline NONOate (WhiB3:NO) as indicated and analyzed by EPR. (Inset) Aerobically purified WhiB3 was deoxygenated and exposed to a 10-fold molar excess of proline NONOate. EPR data were acquired by using a continuous-wave EPR spectrometer at a microwave frequency of 9.669 GHz operating at 100-kHz field modulation and 150 K. A modulation amplitude of 0.5 mT and microwave power of 2 mW were used. The peak at 2.036 g is consistent with the formation of monomeric DNIC (14).
More importantly, EDFS-EPR spectra of WhiB3 after air exposure (5 min) showed the appearance of a sharp signal centered at g = 2.01, which disappeared after prolonged exposure to air (60 min) (Fig. 3B). These observations indicate that the small increase in the absorbance at 413 nm at 5 min after air exposure was caused by the conversion of [4Fe-4S]2+ to the [3Fe-4S]+ intermediate. However, prolonged incubation (60 min) of air-exposed WhiB3 led to the conversion of the [3Fe-4S]+ intermediate to the EPR silent [2Fe-2S]2+ cluster followed by the loss of Fe-S cluster. Interestingly, a [3Fe-4S]+ intermediate was also observed during air exposure of the FNR Fe-S cluster (13).
Furthermore, to assess the effect of O2 on the WhiB3 Fe-S cluster in more detail, the anaerobically reconstituted WhiB3 Fe-S cluster was labeled with 35S, and the loss of incorporated 35S was measured upon exposure of WhiB3 to O2. As anticipated, we observed a gradual decline of radioactive signal with the complete loss of the signal at 120 min after air exposure (Fig. 3C). These results provide clear evidence that the Mtb WhiB3 4Fe-4S cluster is degraded by O2.
To investigate whether NO is a ligand of the WhiB3 Fe-S cluster and to assess the NO responsiveness of WhiB3, aerobically purified WhiB3 was treated with the fast NO-releasing compound proline NONOate and analyzed with continuous-wave EPR at liquid nitrogen temperatures. A weak EPR signal centered at g = 2.03 was observed, which is strongly indicative of a monomeric dinitrosyl–iron–dithiol (DNIC) complex (Fig. 3D, Inset) wherein the sulfide ligands are displaced by the formation of Fe-NO bonds and the iron atoms remain bound to the protein via cysteine ligands (14). In stark contrast to the aerobically purified WhiB3 sample, exposure of anaerobically reconstituted WhiB3 to proline NONOate showed a strong EPR signal at g = 2.036 (Fig. 3D). Furthermore, sequential addition of increasing amounts of proline NONOate significantly enhanced the EPR signal of monomeric DNIC in a concentration-dependent manner (Fig. 3D). In sum, these data conclusively show that the WhiB3 4Fe-4S cluster can react directly with NO and O2 and suggest that the WhiB3 Fe-S cluster functions as a built-in sensor of the proposed dormancy signals.
Role of Mtb WhiB3 in Metabolic Stress.
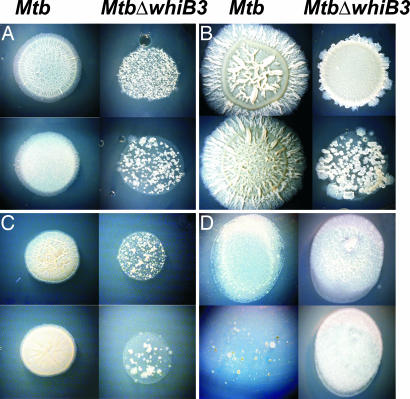
The results indicate that NO is a ligand of WhiB3 and that O2 reacts with WhiB3, both via its 4Fe-4S cluster. Nonetheless, MtbΔwhiB3 does not show a lethal phenotype upon exposure to bacteriostatic concentration of NO or in the Wayne model of gradual oxygen depletion (data not shown), suggesting that bacterial survival is not critically affected by these particular in vitro conditions. Importantly, recent studies have shown that nutrient depletion is a signal for mycobacterial persistence (15) and influences the expression of respiratory/metabolic enzymes responsible for modulating the intracellular redox state of Mtb (15). We tested the hypothesis that WhiB3 plays a role in the bacilli's response to nutritional stress. Growth of wild-type (WT) Mtb and MtbΔwhiB3 was compared by normalizing the cell density and spotting an equal volume (i.e., number) of cells onto solid Dubos medium (basal) or onto Dubos plates containing glucose, succinate, fumarate, pyruvate, citrate, or acetate as sole carbon sources. Dubos basal medium contains asparagine and casein hydrolysate as sole source of carbon and nitrogen. The spot colony phenotype indicates that MtbΔwhiB3 grew poorly on this medium as judged by the noticeably decreased colony diameter and fewer surviving colonies per spot, compared with WT Mtb (Fig. 4). More importantly, supplementation of Dubos basal medium with glucose, succinate, or fumarate did not rescue the MtbΔwhiB3 growth defect (Fig. 4). Also, MtbΔwhiB3 showed a slight but reproducible growth defect on pyruvate but not on citrate media (data not shown). In stark contrast, MtbΔwhiB3 grows significantly better than WT Mtb on basal medium supplemented with the short-chain fatty acid (C2) acetate. Furthermore, MtbΔwhiB3 showed a striking difference in spot colony rugosity compared with WT Mtb. For example, on glucose plates, WT Mtb exhibited a cord-like structure with an organized inner ring, well defined edges, and an overall rugose appearance, whereas the MtbΔwhiB3 colony displayed less cording with irregular edges and structure (Fig. 4). Stable expression of whiB3 in MtbΔwhiB3 resulted in complementation of the growth defect on Dubos basal medium (SI Fig. 7).
Fig. 4.
Growth phenotype of MtbΔwhiB3. Identical volumes (30 μl) and equal number of cells (3 × 105, 3 × 104) of WT Mtb and MtbΔwhiB3 were spotted on Dubos–agar medium either in the absence [basal (A)] or in the presence of 50 mM glucose (B), 50 mM succinate (C) or 25 mM acetate (D) as sole carbon substrate. Growth was analyzed every week after inoculation, and photographs were taken at 4–6 weeks after inoculation at ×7 magnification by using a Zeiss stereo microscope. (Left) WT Mtb. (Right) MtbΔwhiB3.
Furthermore, liquid growth assays confirmed that MtbΔwhiB3 was compromised for growth in 7H9 basal media supplemented with either glucose or succinate as sole carbon sources (SI Fig. 8). Finally, growth was restored by complementing MtbΔwhiB3 with WT WhiB3 but not WhiB3ΔCys-1–4 (SI Fig. 8). These results demonstrate that WhiB3 plays an important role in regulating the core intermediary metabolism of Mtb. Because the intracellular redox state is dramatically influenced by nutrients (16), these results serve to implicate further the WhiB3 Fe-S cluster as a critical sensory component of the intracellular redox state of Mtb.
Discussion
Little is known regarding the biochemical and genetic mechanisms of redox sensing in Mtb. Here we show that Mtb WhiB3 is a redox-responsive protein that through its Fe-S cluster responds to physiologically relevant host signals thought to be involved in persistence. We provide biochemical evidence that Mtb WhiB3 contains a redox-responsive 4Fe-4S cluster that specifically reacts with O2 and NO. We also provide evidence for a link among environmental redox signals, nutritional stress, and WhiB3. These findings will provide mechanistic insight into how Mtb senses and responds to protective host molecules and nutritional stress, to favor bacilli persistence.
Our first challenge was to accurately assemble the WhiB3 Fe-S cluster. Rather than exploiting the widely used A. vinelandii NifS for Fe-S cluster assembly, we chose to identify and use the endogenous Mtb cysteine desulfurase. Through a series of spectroscopic and sensitive labeling experiments, we demonstrated that IscS is a pyridoxal 5′-phosphate-containing enzyme capable of assembling the WhiB3 Fe-S cluster. Intriguingly, Mtb does not contain other Isc components involved in Fe-S cluster assembly (e.g., IscA and IscU) but does possesses the complete repertoire of Suf machinery. The characterization of the Mtb Fe-S cluster assembly components is particularly interesting because in other systems NifS/IscS has been implicated in the repair of oxygen and NO-damaged Fe-S cluster proteins (8). We used the 35S transfer assay and UV-visible and EPR spectroscopy to characterize the WhiB3 Fe-S cluster thoroughly. Upon treatment with DTH, the EPR silent [4Fe-4S]2+ cluster undergoes a one-electron reduction to generate a paramagnetic state, which results in an EPR signal distinctive of a [4Fe-4S]+ cluster (10). The complete absence of 4Fe-4S cluster in WhiB3ΔCys-1–4 confirms the role of four conserved residues (C23-C53-C57-C62) in coordinating an Fe-S cluster. Because the E. coli Fe-S cluster-containing transcriptional regulators, FNR and SoxR, are sensors of O2 and NO, respectively (6), we studied the effect of exposure to O2 and NO on the WhiB3 Fe-S cluster and were able to draw several parallels between the mechanism of cluster degradation of WhiB3 and FNR. For example, in response to O2, the transformation of the WhiB3 [4Fe-4S]2+ cluster via [3Fe-4S]+ and [2Fe-2S] cluster intermediates is mechanistically similar to the O2-induced modification of the FNR [4Fe-4S]2+ cluster, which involves two steps (13). First, oxidation of the WhiB3 cubane [4Fe-4S]2+ cluster by O2 rapidly generates the [3Fe-4S]+ intermediate and releases one electron and Fe2+ for the two-electron reduction of O2 to produce H2O2. Second, in a nonredox step, H2O2 converts the [3Fe-4S]+ intermediate to a planar [2Fe-2S]2+ cluster (Fig. 5). Finally, H2O2 and/or superoxide destroys the 2Fe-2S cluster (17). These spectroscopic data are further supported by the sensitive Fe-S cluster 35S-labeling assay demonstrating that Fe2+ and S are essential for WhiB3 Fe-S cluster configuration. Importantly, when anaerobically reconstituted WhiB3 was exposed to O2, we demonstrated an associated loss of the WhiB3 Fe-S cluster, suggesting that O2 gradually decays the Fe-S cluster. These data strongly suggest that similar to FNR, WhiB3 activity is regulated via its Fe-S cluster by O2 and thus functions as a redox sensor. Notably, the O2 exposure properties of the FNR and WhiB3 Fe-S clusters may reflect important biological differences between E. coli that is capable of a rapid transition from aerobic to anaerobic environment and Mtb that requires a gradual transition. Crucial differences between WhiB3 and FNR are also apparent. For example, when exposed to O2, loss of the FNR [4Fe-4S]2+ cluster is much more rapid. In the case of FNR, 10-min air exposure caused a ≈50% decrease in the 4Fe-4S cluster absorbance (12), whereas 120 min was required to cause a similar decrease for WhiB3. Finally, as is the case for FNR and SoxR Fe-S clusters, we provide clear evidence that NO also targets the WhiB3 Fe-S cluster to generate a characteristic DNIC complex. The interaction of NO with the WhiB3 4Fe-4S cluster could modulate its activity in vivo by direct nitrosylation of the Fe-S cluster as described for SoxR (6) and FNR (14).
Fig. 5.
Hypothetical model depicting WhiB3 as an intracellular redox sensor. In an oxygen-rich environment, the WhiB3 [4Fe-4S]+ cluster is oxidized to [4Fe-4S]2+ and undergoes sequential conversion, yielding [3Fe-4S]+ with the concomitant production of H2O2 and [2Fe-2S]2+ intermediates and eventually complete loss of the cluster. Mtb IscS can activate apo-WhiB3 in vivo to generate WhiB3 [4Fe-4S]2+. The [4Fe-4S]2+ cluster can undergo reduction by thioredoxin and mycothiols to generate [4Fe-4S]+. In vivo growth of Mtb is accompanied by carbon source and O2 depletion thereby causing fluctuations in the intracellular redox state. WhiB3 senses these changes in the redox state through its [4Fe-4S] cluster to regulate catabolic [glycolysis, tricarboxylic acid (TCA) cycle, glyoxylate cycle] and anabolic (polyketides biosynthesis) metabolism for maintaining redox balance and persistence. The O2- and NO-sensing properties of WhiB3 and the growth defect exhibited by MtbΔwhiB3 on various carbon sources suggest a role as redox sensor responsible for integrating environmental signals to core intermediary metabolism.
The fact that WhiB3 contains a redox-sensitive 4Fe-4S cluster and is cytoplasmically located and constitutively expressed (18) suggests a role for WhiB3 in sensing physiological changes associated with cellular metabolism. Because the intracellular redox environment is directly coupled to central metabolism, we assessed the role of WhiB3 in carbon source utilization. The severe growth impairment of MtbΔwhiB3 observed on various carbohydrates implicates WhiB3 in sensing and responding to the changes in the cellular redox state (energy levels), thereby enabling Mtb to alleviate the potential harmful effect of redox imbalance. Interestingly, MtbΔwhiB3 grew better on acetate as the sole carbon source, pointing toward WhiB3 as a potential regulator of fatty acid metabolism in Mtb. Several lines of evidence have demonstrated that Mtb prefers fatty acids rather than carbohydrates as carbon substrates during infection (19, 20). These studies suggest that Mtb senses through a yet-to-be identified mechanism, low concentrations of glycolytic substrate and/or oxygen, and initiates a metabolic switchover to the preferred in vivo carbon source, fatty acids (19, 21). The redox-sensing properties of WhiB3, together with the growth properties of MtbΔwhiB3 on carbon-limiting media, in particular acetate, implicate WhiB3 in the regulation of this metabolic switchover event. Lastly, the dramatic effect of whiB3 on colony rugosity (Fig. 4) suggests alterations in cell wall lipid composition and may well be in support of our earlier hypothesis that whiB3 regulates the production of polyketides to modulate the host immune system (7).
Nutritional stress is known to influence the electron transport chain, proton-motive force, nucleotide pools, and redox potential (16). Previous studies have demonstrated a link between nutrient depletion and Mtb persistence (15) and between aerobic carbon starvation and anaerobiosis that tightly regulate the intracellular redox balance of bacteria (22). Interestingly, induction of the Mtb Dos dormancy pathway by hypoxia, NO (2, 23), and palmitate (a reduced carbon source) (5) suggests an overlap between the Mtb adaptive response to extracellular redox signals and carbon source utilization, which cause changes in the intracellular redox state.
The observation that MtbΔwhiB3 showed a growth defect on different carbon sources, but not in the Wayne model (in which there is a sufficient carbon source, but oxygen is gradually depleted), and none upon exposure to bacteriostatic concentrations of NO (which inhibits aerobic respiration), strongly supports our model (Fig. 5) and current literature that aerobic respiration and carbon source utilization are intricately linked to fluctuations in the intracellular redox environment. The latter is supported by elegant studies showing that in response to carbon source starvation, E. coli FNR and ArcA (24, 25) regulate the synthesis of key enzymes involved in glycolysis and tricarboxylic acid cycle. Finally, the severe growth impairment of MtbΔwhiB3 on various carbon sources as well as the induction of WhiB2 (15), WhiB1 (26), and WhiB3 (27) upon nutrient limitation, by exogenous cAMP, and during infection, respectively, strongly suggests that some Mtb WhiB family members play an important role in sensing fluctuations in the intracellular redox state.
In summary, we have identified and characterized a native enzymatic reconstitution system enabling the in vitro assembly of Fe-S clusters in Mtb. We have shown that Mtb WhiB3 contains a redox-responsive [4Fe-4S]2+ cluster that upon oxygen exposure generates a [3Fe-4S]+ species and is eventually destroyed, suggesting that WhiB3 is a sensor of O2. We have also shown that NO is a ligand of WhiB3. Finally, we have shown that WhiB3 is essential for optimal growth on a number of carbon sources, including tricarboxylic acid cycle intermediates. Based on this cumulative evidence, we propose that Mtb WhiB3 is a redox-dependent transcription factor that senses intracellular redox changes through its Fe-S cluster and integrates environmental signals with core intermediary metabolism essential for survival. The ability of WhiB3 to react with diverse redox signals has broad implications for understanding the mechanism of Mtb persistence.
Experimental Procedures
Culture Conditions.
Mtb H37Rv and MtbΔwhiB3 mutant were cultured in Middlebrook 7H9/7H10 medium as described previously (7). E. coli cultures were grown in LB medium. Whenever required, antibiotics were added to the culture medium (for E. coli, 100 μg/ml carbenicillin and 150 μg/ml hygromycin; for Mtb, 50 μg/ml hygromycin).
Overexpression and Purification of WhiB3, WhiB3ΔCys-1–4, and Mtb IscS.
WhiB3, WhiB3ΔCys-1–4, and Mtb IscS were purified as described (see SI Experimental Procedures).
Iron-Sulfur Cluster Assembly.
The Fe-S cluster reconstitution process was performed under anoxic conditions in an anaerobic glove box. Aerobically purified WhiB3 and WhiB3ΔCys-1–4 (30 μM each) were completely degassed by using compressed argon gas and anaerobically equilibrated with buffer containing 50 mM sodium phosphate, pH 7.5, and 150 mM NaCl. This was followed by the addition of a 10-fold molar excess Fe(II) chloride, 5 mM DTT, and 2.5 μg of IscS. The reaction was initiated by adding 2 mM l-cysteine and subsequently monitored over time by UV-visible spectroscopy for Fe-S cluster assembly. The reaction mixture was terminated by size-exclusion chromatography to remove low molecular polymers. For reduction, a sample of fully reconstituted WhiB3, after passing through a size-exclusion column, was treated with 5 mM DTH and analyzed by UV-visible spectroscopy. For EPR spectroscopy, reconstituted WhiB3 before (oxidized) and after (reduced) DTH treatment was transferred to EPR tubes and immediately frozen in liquid nitrogen. To study the effect of O2 on Fe-S cluster stability, a sample of fully reconstituted WhiB3 was purified by size-exclusion chromatography, transferred to an anaerobic cuvette, and air bubbled through the mixture for 2 min. The cuvette was then sealed and monitored by UV-visible spectroscopy (DU800; Beckman Coulter, Fullerton, CA). For EPR analysis at liquid helium temperatures, reconstituted WhiB3 was transferred to an EPR tube 3 and 60 min after air exposure and immediately frozen in liquid nitrogen. For NO exposure, purified WhiB3 and anaerobically reconstituted WhiB3 were treated inside the anaerobic glove box with aliquots of freshly prepared proline NONOate (Cayman Chemicals, Ann Arbor, MI). Aliquots of WhiB3 were placed in an anaerobic cuvette and titrated by injection with aliquots of a 2.5 mM stock solution of proline NONOate. Samples were then transferred to EPR tubes and immediately frozen in liquid nitrogen.
EPR Spectroscopy.
EPR measurements were performed as described (see SI Experimental Procedures).
Supplementary Material
Acknowledgments
We thank Anton Poliakov for electrospray ionization MS analysis and members of the A.J.C.S. laboratory and Mary Hondalus for critical review of this manuscript. We also thank Denis Dean (Virginia Tech, Blacksburg, VA) for providing the A. vinelandii nifS plasmid. This work was supported by National Institute of Allergy and Infectious Diseases, National Institutes of Health Grant R01AI058131 (to A.J.C.S.). A.S. is the recipient of a postdoctoral fellowship from the Heiser Foundation. L.G is the recipient of National Institutes of Health Cell and Molecular Biology Training Grant NIH T32 GM008111-19, and J.T is the recipient of Agent of International Health and Biodefense Concern Training Grant 1T32AI55438.
Abbreviations
- DNIC
dinitrosyl–iron–dithiol
- DTH
sodium dithionite
- EDFS
echo-detected field sweep
- FNR
fumarate nitrate regulator
- Mtb
Mycobacterium tuberculosis.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/cgi/content/full/0700490104/DC1.
References
- 1.Wayne LG, Sohaskey CD. Annu Rev Microbiol. 2001;55:139–163. doi: 10.1146/annurev.micro.55.1.139. [DOI] [PubMed] [Google Scholar]
- 2.Voskuil MI, Schnappinger D, Visconti KC, Harrell MI, Dolganov GM, Sherman DR, Schoolnik GK. J Exp Med. 2003;198:705–713. doi: 10.1084/jem.20030205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Loebel RO, Shorr E, Richardson HB. J Bacteriol. 1933;26:167–200. doi: 10.1128/jb.26.2.167-200.1933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ohno H, Zhu G, Mohan VP, Chu D, Kohno S, Jacobs WR, Jr, Chan J. Cell Microbiol. 2003;5:637–648. doi: 10.1046/j.1462-5822.2003.00307.x. [DOI] [PubMed] [Google Scholar]
- 5.Boshoff HI, Myers TG, Copp BR, McNeil MR, Wilson MA, Barry CE., III J Biol Chem. 2004;279:40174–40184. doi: 10.1074/jbc.M406796200. [DOI] [PubMed] [Google Scholar]
- 6.Green J, Paget MS. Nat Rev Microbiol. 2004;2:954–966. doi: 10.1038/nrmicro1022. [DOI] [PubMed] [Google Scholar]
- 7.Steyn AJ, Collins DM, Hondalus MK, Jacobs WR, Jr, Kawakami RP, Bloom BR. Proc Natl Acad Sci USA. 2002;99:3147–3152. doi: 10.1073/pnas.052705399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Johnson DC, Dean DR, Smith AD, Johnson MK. Annu Rev Biochem. 2005;74:247–281. doi: 10.1146/annurev.biochem.74.082803.133518. [DOI] [PubMed] [Google Scholar]
- 9.Heidenreich T, Wollers S, Mendel RR, Bittner F. J Biol Chem. 2005;280:4213–4218. doi: 10.1074/jbc.M411195200. [DOI] [PubMed] [Google Scholar]
- 10.Jakimowicz P, Cheesman MR, Bishai WR, Chater KF, Thomson AJ, Buttner MJ. J Biol Chem. 2005;280:8309–8315. doi: 10.1074/jbc.M412622200. [DOI] [PubMed] [Google Scholar]
- 11.Ta DT, Vickery LE. J Biol Chem. 1992;267:11120–11125. [PubMed] [Google Scholar]
- 12.Khoroshilova N, Popescu C, Munck E, Beinert H, Kiley PJ. Proc Natl Acad Sci USA. 1997;94:6087–6092. doi: 10.1073/pnas.94.12.6087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Crack J, Green J, Thomson AJ. J Biol Chem. 2004;279:9278–9286. doi: 10.1074/jbc.M309878200. [DOI] [PubMed] [Google Scholar]
- 14.Cruz-Ramos H, Crack J, Wu G, Hughes MN, Scott C, Thomson AJ, Green J, Poole RK. EMBO J. 2002;21:3235–3244. doi: 10.1093/emboj/cdf339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Betts JC, Lukey PT, Robb LC, McAdam RA, Duncan K. Mol Microbiol. 2002;43:717–731. doi: 10.1046/j.1365-2958.2002.02779.x. [DOI] [PubMed] [Google Scholar]
- 16.Taylor BL, Zhulin IB. Microbiol Mol Biol Rev. 1999;63:479–506. doi: 10.1128/mmbr.63.2.479-506.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sutton VR, Stubna A, Patschkowski T, Munck E, Beinert H, Kiley PJ. Biochemistry. 2004;43:791–798. doi: 10.1021/bi0357053. [DOI] [PubMed] [Google Scholar]
- 18.Hutter B, Dick T. Res Microbiol. 1999;150:295–301. doi: 10.1016/s0923-2508(99)80055-2. [DOI] [PubMed] [Google Scholar]
- 19.Munoz-Elias EJ, McKinney JD. Nat Med. 2005;11:638–644. doi: 10.1038/nm1252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Graham JE, Clark-Curtiss JE. Proc Natl Acad Sci USA. 1999;96:11554–11559. doi: 10.1073/pnas.96.20.11554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bloch H, Segal W. J Bacteriol. 1956;72:132–141. doi: 10.1128/jb.72.2.132-141.1956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Nystrom T. Mol Microbiol. 1994;12:833–843. doi: 10.1111/j.1365-2958.1994.tb01069.x. [DOI] [PubMed] [Google Scholar]
- 23.Park HD, Guinn KM, Harrell MI, Liao R, Voskuil MI, Tompa M, Schoolnik GK, Sherman DR. Mol Microbiol. 2003;48:833–843. doi: 10.1046/j.1365-2958.2003.03474.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Shalel-Levanon S, San KY, Bennett GN. Biotechnol Bioeng. 2005;92:147–159. doi: 10.1002/bit.20583. [DOI] [PubMed] [Google Scholar]
- 25.Levanon SS, San KY, Bennett GN. Biotechnol Bioeng. 2005;89:556–564. doi: 10.1002/bit.20381. [DOI] [PubMed] [Google Scholar]
- 26.Agarwal N, Raghunand TR, Bishai WR. Microbiology. 2006;152:2749–2756. doi: 10.1099/mic.0.28924-0. [DOI] [PubMed] [Google Scholar]
- 27.Banaiee N, Jacobs WR, Jr, Ernst JD. Infect Immun. 2006;74:6449–6457. doi: 10.1128/IAI.00190-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kumar A, Toledo JC, Patel RP, Lancaster JR, Jr, Steyn AJC. Proc Natl Acad Sci USA. 2007;104:11568–11573. doi: 10.1073/pnas.0705054104. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.