Abstract
Estrogen drives both transcriptional activation and proteolysis of estrogen receptor α (ERα; encoded by ESR1). Here we observed variable and overlapping ESR1 mRNA levels in 200 ERα-negative and 50 ERα-positive primary breast cancers examined, which suggests important posttranscriptional ERα regulation. Our results indicate that Src cooperates with estrogen to activate ERα proteolysis. Inducible Src stimulated ligand-activated ERα transcriptional activity and reduced ERα t1/2. Src and ERα levels were inversely correlated in primary breast cancers. ERα-negative primary breast cancers and cell lines showed increased Src levels and/or activity compared with ERα-positive cancers and cells. ERα t1/2 was reduced in ERα-negative cell lines. In both ERα-positive and -negative cell lines, both proteasome and Src inhibitors increased ERα levels. Src inhibition impaired ligand-activated ERα ubiquitylation and increased ERα levels. Src siRNA impaired ligand-activated ERα loss in BT-20 cells. Pretreatment with Src increased ERα ubiquitylation and degradation in vitro. These findings provide what we believe to be a novel link between Src activation and ERα proteolysis and support a model whereby crosstalk between liganded ERα and Src drives ERα transcriptional activity and targets ERα for ubiquitin-dependent proteolysis. Oncogenic Src activation may promote not only proliferation, but also estrogen-activated ERα loss in a subset of ERα-negative breast cancers, altering prognosis and response to therapy.
Introduction
Estrogen regulates the proliferation and development of tissues expressing estrogen receptors (ERs) and is a risk factor for breast cancer development. One-third of new breast cancers lack detectable ERα protein; these ERα– cancers have a worse prognosis than do ERα+ breast cancers (1). ERα– breast cancers do not respond to hormone response modifiers like tamoxifen (2) and often show de novo or acquired resistance to chemotherapy (1). While there are 2 forms of ERs, ERα and ERβ (3–5), considerably more is known about the role of ERα in human breast cancer, and in this study we investigated ERα exclusively. While estrogen is mitogenic for cultured ERα+ breast cancer lines, ERα– breast cancer lines proliferate in the absence of estrogen, and ERα– breast cancers are generally believed to be estrogen independent.
Factors responsible for the ERα– status of breast cancers remain largely unknown. Deletions, rearrangements, and point mutations in the ESR1 gene, which encodes ERα, are too uncommon to account for the ERα– phenotype (6, 7). ERα promoter hypermethylation has been observed in a minority (up to 25%) of ERα– breast carcinomas (6). In 3 early nonquantitative studies, ESR1 mRNA was detected in a majority (67%–71%) of 64 primary ERα– cancers (8–10), indicating posttranscriptional or posttranslational control of ERα levels in human breast cancers. Transcriptional profiling has demonstrated that ESR1 mRNA is detected but variably reduced in ERα– compared with ERα+ cancers (11–13). The distinct gene expression profiles of ERα+ and ERα– cancers have led to the hypothesis that these 2 tumor groups arise from different cellular origins: ERα–/Her2– tumors are derived from the basal epithelium, while ERα+ cancers have a luminal epithelial origin (14, 15). The results of our present study shed further light on mechanisms regulating ERα levels.
ERα is a 66-kDa nuclear hormone receptor (HR) transcription factor (16). Upon ligand binding, ERα dimerizes and associates with coactivators and chromatin remodeling factors to activate transcription of genes containing estrogen response elements (EREs) (17). ERα contains 2 transcription activation functions, AF-1 and AF-2. AF-1 can be phosphorylated and activated in a ligand-independent manner following growth factor stimulation, while AF-2 is activated by ligand-stimulated changes in ERα conformation (18, 19). The ERα phosphorylation state affects coactivator binding and ERα-DNA binding affinity.
In addition to transcriptional activation, ligand-ERα binding rapidly activates crosstalk with mitogenic signaling kinases (reviewed in refs. 20, 21). Estrogen-ERα binding promotes a rapid and transient interaction of ERα with cellular Src (cSrc), binding to Shc and Ras-MAPK activation (22–25). In some cell types, estrogen stimulates tripartite ERα, cSrc, and PI3K complex formation, leading to PKB/AKT and MAPK activation (26). Signaling kinases activated by liganded ERα not only activate mitogenic cascades, but also phosphorylate the ERα and its coactivators, generating a feed-forward loop that augments ERα transcriptional activity (20, 21, 27).
The ERα can also be phosphorylated and activated in a ligand-independent manner in response to peptide growth factors including IGF-I (28), TGF-α (29), and EGF (30, 31) that activate PKB and MAPK signaling pathways and cause ERα phosphorylation and ERα-dependent gene transcription. Phosphorylation of aminoterminal (30, 32, 33) and carboxyterminal (34–36) sites on the ERα increase ERα transcriptional activity.
Estrogen binding to the ERα rapidly stimulates ERα ubiquitylation and proteolysis (37–39). Unliganded ERα is very stable, with a t1/2 of up to 5 days (37). Upon ligand binding, the ERα t1/2 drops dramatically, to 3–5 hours (37, 39). The detection of ubiquitinated ERα in vivo in uterine tissue (37) and the finding that proteasome inhibition abrogates estrogen-stimulated ERα loss confirmed an in vivo role for proteasome-mediated ERα degradation in regulating ERα levels (38, 39).
ERα ubiquitination and proteasome activity are intimately linked to ERα-dependent transcriptional activation (40, 41). Ligand binding activates both ERα-dependent transcription and ERα ubiquitination (40). Proteasome inhibitors and mutations that inhibit coactivator binding both abrogate ligand-mediated ERα proteolysis and ERE transcriptional activity (41). Different ligands stimulate ERα proteolysis to different degrees (42), and ubiquitin ligases BRCA1 (43), MDM2 (44), and E6AP (45) can all stimulate estrogen-induced transcriptional activity.
The 60-kDa cSrc tyrosine kinase regulates cellular proliferation and motility as well as tumor metastasis (46). Increased levels and/or activity of cSrc have been observed in primary breast cancers (47), but to our knowledge, an association with ERα levels has not previously been reported. Here we demonstrate that both ERα+ and ERα– primary human breast carcinomas expressed ESR1 mRNA. Crosstalk between liganded ERα and cSrc appeared to promote proteasomal degradation of the ERα. cSrc inhibition impaired ligand-activated ERα ubiquitylation and ERα proteolysis, while Src induction shortened the ERα t1/2. Src induction also increased ERα-driven transcription. ERα– breast cancer specimens and cell lines showed elevated cSrc levels and/or activity compared with ERα+ tumors and cell lines, and ERα proteolysis was increased in ERα– cell lines. Src stimulated both ERα ubiquitylation and proteasome-dependent ERα degradation in vitro. These observations and the inverse correlation between cSrc and ERα levels in primary breast cancers suggest that Src may promote cell proliferation by stimulating transcription-coupled ERα degradation in human breast cancers.
Results
ERα– breast cancers express ESR1 mRNA.
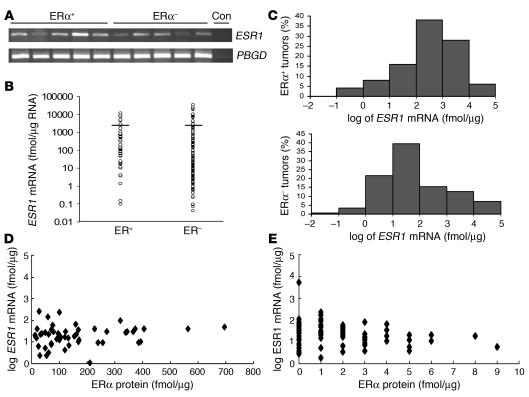
ESR1 mRNA expression was quantitated in 200 ERα– and 50 ERα+ primary human breast cancers. ERα protein levels were determined in a single clinical reference laboratory by ligand binding assay (LBA). Tumor ESR1 mRNA was quantitated by quantitative real-time RT-PCR (QPCR). Crossing-point values for each tumor sample were compared to a standard curve generated from serial dilutions of ERα cDNA plasmid (data not shown). Quantitation of housekeeping gene human porphobilinogen deaminase (PBGD) expression demonstrated similar mRNA quality in both ERα+ and ERα– breast cancers. ESR1 mRNA was detected in all of 50 ERα+ and 200 ERα– cancers (Figure 1A). The ESR1 mRNA values (Figure 1B) and distribution (Figure 1C) showed considerable variability and overlap in ESR1 mRNA concentrations in ERα+ and ERα– tumors. The mean ESR1 mRNA concentration was 1.14 × 103 fmol/μg RNA in ERα+ cancers (range, 1.02 × 10–1 to 1.19 × 104 fmol/μg RNA) and 1.27 × 103 fmol/μg RNA in ERα– cancers (range, 4.55 × 10–2 to 3.56 × 104 fmol/μg RNA). While the lowest and highest ESR1 mRNA concentrations were similar and the mean ESR1 mRNA values did not differ significantly between the ERα+ and ERα– cancers (P > 0.50), the modal ESR1 mRNA value in the ERα– tumors was approximately 1 log lower than in the ERα+ tumors. When ERα protein concentrations were graphed versus ESR1 mRNA values, there was no clear relationship between ERα protein and ESR1 mRNA levels for either ERα+ or ERα– cancers (Figure 1, D and E).
Figure 1. ERα– and ERα+ human breast cancers express ESR1 mRNA.
(A) PCR of ESR1 and human PBGD from breast tumors. Blots are representative of 50 ERα+ and 200 ERα– cancers. Con, control. (B) ESR1 mRNA concentrations. The horizontal line denotes the mean. (C) Frequency of ESR1 mRNA concentrations rounded to the nearest logarithm value. (D and E) ESR1 mRNA (fmol/μg total RNA) plotted versus ERα protein by LBA (fmol/μg cytosolic protein) in the same human breast cancers.
Serum growth factors cooperate with estrogen to activate ERα proteolysis.
As a baseline for further study, we showed that addition of β-estradiol, the primary estrogen type in humans, to estrogen-deprived MCF-7 cells stimulated a rapid reduction of ERα protein that was impaired by proteasome inhibition with n-acetyl-Leu-Leu-norleucinal (Figure 2A). The ERα t1/2 was greater than 24 hours in estrogen-depleted MCF-7 cells. Within 6 hours of estradiol addition, the ERα t1/2 fell to 5 hours (Figure 2B). A significant reduction in ERα t1/2 was also noted within 1 hour of ligand addition (data not shown).
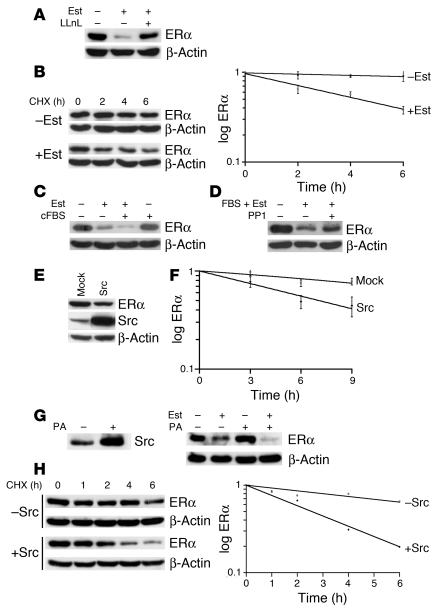
Figure 2. Src promotes estrogen-dependent ERα degradation.
(A) ERα before and 6 hours after addition of estradiol (Est) with or without the proteasome inhibitor n-acetyl-Leu-Leu-norleucinal (LLnL) to estrogen-depleted MCF-7 cells. Equal loading was confirmed by β-actin. (B) ERα t1/2 was assayed by CHX chase in estrogen-depleted cells and at 2, 4, and 6 hours after addition of estradiol. Graph shows results of densitometric analysis of 3 CHX chase experiments (mean ± SEM). (C) Cells were grown in 0.1% cFBS for 48 hours and then treated with estradiol alone, 5% cFBS plus estradiol, or 5% cFBS alone. ERα and β-actin were assayed 6 hours later. (D) Serum- and estrogen-deprived MCF-7 cells were transferred to 5% FBS plus estradiol with or without added Src inhibitor PP1, and ERα was assayed 6 hours later. (E and F) MCF-7 cells were transfected with PCI-Src Y530F (Src) or empty vector (Mock). After 24 hours, (E) ERα and Src levels were assayed and (F) ERα t1/2 was assayed by CHX chase (mean ± SEM). (G and H) The MCFpINDSrc2 line was estrogen depleted for 72 hours, and Src was induced or not for 24 hours with PA prior to addition of estradiol. (G) Src at time of estradiol addition (left), and ERα and β-actin before and 6 hours after estradiol (Est + or –) was added (right). (H) CHX pulse chase, starting 6 hours after estradiol addition with (+Src) or without (–Src) prior induction of Src by PA.
Crosstalk between cSrc, PI3K, and receptor tyrosine kinases and liganded ERα leads to ERα phosphorylation and activation of ERα transcriptional activity (21, 28). To determine whether crosstalk between ERα and signaling pathways may also modulate ligand-activated ERα proteolysis, we tested whether addition of growth factors would affect estrogen-stimulated ERα loss. MCF-7 cells were growth factor and estrogen deprived in 0.1% charcoal-stripped FBS (cFBS) for 48 hours. Estradiol together with 5% cFBS reduced ERα levels more rapidly than did estradiol alone (Figure 2C). Growth factor stimulation with 5% cFBS without added estradiol was not sufficient to trigger ERα proteolysis. Thus, serum growth factors may activate signaling kinases to promote estrogen-activated ERα proteolysis.
Src promotes estrogen-stimulated ERα degradation.
Liganded ERα binds cSrc leading to cSrc activation (22). Treatment of MCF-7 cells with the Src inhibitor PP1 caused a dose-dependent accumulation of ERα over 48 hours (data not shown). PP1 also impaired the fall in ERα levels observed when estrogen- and growth factor–starved cells were transferred to serum together with estradiol (Figure 2D). Thus, cSrc may promote ligand-activated ERα proteolysis. Transfection of activated Src (PCI-Src Y530F) reduced ERα levels (Figure 2E). The ERα t1/2 fell from 14 hours in asynchronously proliferating MCF-7 cells to 9 hours at 24 hours after Src transfection (Figure 2F).
To assay further the effect of Src on ERα stability, 2 different MCF-7 derivatives were generated to inducibly express activated Src-Y350F. Results of experiments using 1 of these Src inducible MCF-7 lines, designated MCFpINDSrc2, are described below. MCFpINDSrc2 cells were deprived of estradiol and growth factors for 72 hours. Src was induced by treatment with ponasterone A (PA) for 24 hours prior to addition of estradiol (Figure 2G, left). Induction of activated Src did not reduce ERα levels in the absence of estradiol. However, within 6 hours of estradiol addition, ERα levels were markedly lower in Src-induced cells (Figure 2G, right), and ERα t1/2 was reduced to 2.6 hours in cells stimulated by estradiol together with Src induction, compared with an ERα t1/2 of 8.1 hours in cells treated with estradiol alone (Figure 2H). Treatment with PA alone did not reduce ERα t1/2 (data not shown). At 24 hours after Src induction, QPCR showed a modest increase in ESR1 mRNA expression compared with uninduced cells (data not shown); thus, the more rapid ERα protein loss in estradiol-treated, Src-induced cells was not due to reduced ESR1 mRNA expression. Data in a second Src-inducible cell line also confirmed these findings (data not shown). Thus, Src appears to cooperate with estrogen to stimulate ERα proteolysis. Proteasome inhibition reduced the effect of expression of activated Src (data not shown).
Src promotes ligand-activated ERα transcriptional activity.
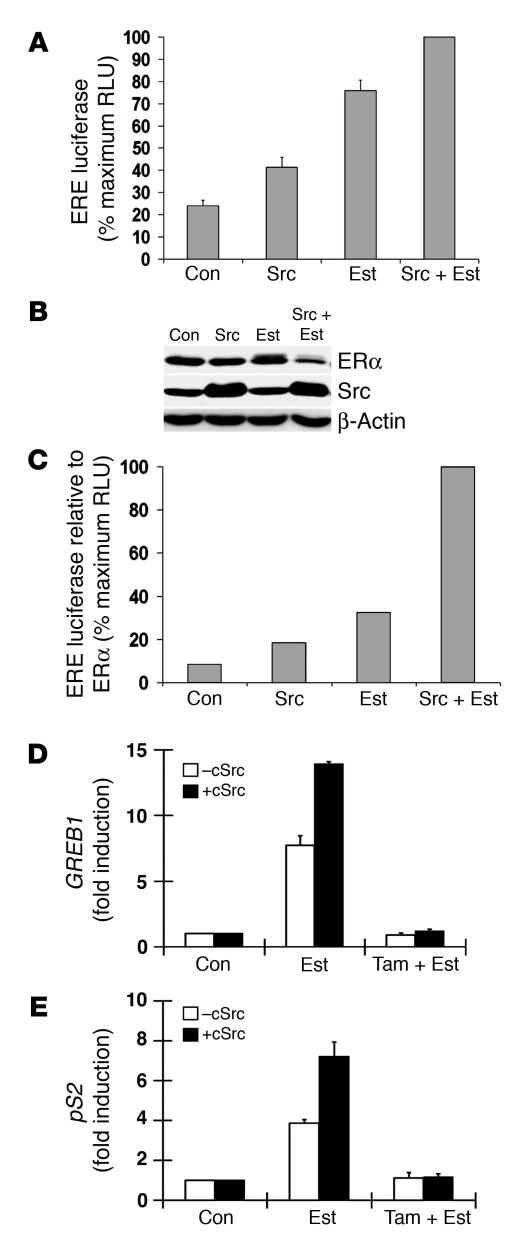
Activation of many transcriptional factors is linked to factor proteolysis (48). Because our data suggested that Src promotes ligand activated ERα proteolysis, we assayed effects of Src on ERα transcriptional activity. In cells grown in the presence of full serum and estradiol, addition of 10–8 M estradiol and Src transfection each reproducibly increased ERE luciferase activity within 4 hours, albeit less notably with Src alone (Figure 3A). Estradiol addition together with Src transfection increased ERE luciferase activity beyond that induced by estradiol alone. Src transfection and estradiol stimulation also decreased ERα levels beyond that seen with estradiol alone (Figure 3B). For ERE luciferase activity relative to available ERα (i.e., correcting for the reduced ERα level at 4 hours), Src transfection and estradiol stimulation had more than additive effects (Figure 3C).
Figure 3. Src promotes estrogen-dependent ERα transcriptional activity.
(A–C) MCF-7 was transfected with ERE luciferase reporter and either PCI-Src Y530F or empty vector control and then stimulated with estradiol. Shown are (A) ERE luciferase activity as well as ERα and Src levels (B) before and (C) 4 hours after Src transfection, estradiol addition, or both. (C) Relative ERE luciferase activity corrected for ERα level. (D and E) MCFpINDSrc2 cells were estrogen depleted for 72 hours, and cSrc was induced or not for 24 hours prior to the addition of estradiol (Est) or estradiol plus tamoxifen (Est + Tam). (D and E) QPCR of cellular (D) GREB1 and (E) pS2.
To investigate further if Src affects estrogen-mediated transcription activity of ERα, we used QPCR to quantitate estrogen stimulated expression of cellular ERα target genes, pS2 and GREB1 with and without prior Src induction in the MCFpINDSrc2 line. MCFpINDSrc2 cells were estrogen deprived for 3 days, and Src was induced or not within the last 24 hours of starvation. Within 3 hours of estradiol addition, GREB1 and pS2 mRNA levels increased by 8- and 4-fold, respectively, compared with 14- and 7-fold higher than baseline when Src was inducted prior to estradiol addition. Src induction alone did not activate GREB1 or pS2 (data not shown). Neither gene was activated when cells were treated with estradiol together with tamoxifen, with or without Src induction. Thus the effect of Src on these genes was ERα mediated, and Src increased the transcriptional potency of ERα on these ERα target genes (Figure 3, D and E).
MEK and PI3K are not sufficient to promote ligand-mediated ERα proteolysis.
MEK inhibition of asynchronous MCF-7 cells with U0126 for 48 hours reduced ERα levels (Supplemental Figure 1A; supplemental material available online with this article; doi:10.1172/JCI21739DS1). In estrogen-deprived MCF-7 cells, MEK inhibition prior to estradiol addition led to a greater loss of ERα (Supplemental Figure 1B) and a shorter ERα t1/2 (data not shown) than with estradiol alone. Thus, in these assay conditions, MEK effectors appear to oppose ligand-stimulated ERα proteolysis.
Treatment with the PI3K inhibitor LY294002 did not affect ERα levels in asynchronous MCF-7 cells (Supplemental Figure 1C). In estrogen-deprived MCF-7 cells, PI3K inhibition prior to estrogen repletion inhibited PKB phosphorylation and cell cycle progression, but did not affect estrogen-mediated ERα loss (Supplemental Figure 1, D and E). Thus, estrogen-stimulated ERα proteolysis does not require PI3K/PKB action or cell cycle entry.
ERα protein levels and stability are reduced in breast cancer lines with activated cSrc.
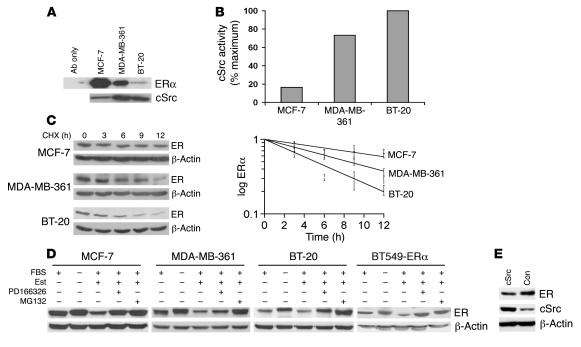
The BT-20 breast cancer line shows both cSrc and EGFR activation, while Her2 and cSrc are activated in MDA-MB-361 (49). ESR1 mRNA was detected in MCF-7, BT-20, and MDA-MB-361 by nonquantitative RT-PCR (data not shown). Although the BT-20 cell line has been characterized as ERα–, low but detectable ERα protein was present on ERα immunoprecipitation from 1 mg cell lysate (Figure 4A). cSrc kinase activities were increased (Figure 4B), while the level and t1/2 of ERα were reduced in BT-20 and MDA-MB-361 compared with MCF-7 cells (Figure 4C). The calculated ERα t1/2 was 14 hours in asynchronous MCF-7, 9 hours in MDA-MB-361, and 5 hours in BT-20 cells.
Figure 4. Estrogen regulation of ERα levels in ERα+ and ERα– breast cancer lines.
(A) ERα was detected by immunoblotting ERα precipitates from 1 mg cell lysate of asynchronous ERα+ MCF-7, MDA-MB-361, and ERα–BT-20 cells. (B) cSrc activity in asynchronous MCF-7, MDA-MB-361, and BT-20 cells. (C) ERα t1/2 in asynchronous cells as assayed by CHX chase, calculated from 3 independent assays (mean ± SEM). (D) After 48 hours serum and estrogen deprivation in 0.1% cFBS, MCF-7, MDA-MB-361, BT-20, and BT549-ERα cells were stimulated with estradiol plus 5% FBS with or without prior addition of MG132 or Src inhibitor PD166326, and ERα was assayed 6 hours later. (E) BT-20 cells were transfected with siRNA to cSrc or nonspecific control siRNA (Con) and deprived of estrogen for 48 hours. Cells were then treated with estradiol for 4 hours prior to Western blot analysis of cSrc and ERα levels.
To further assay effects of Src and proteasome function on ERα levels in ERα– breast cancer lines, the BT549 line was transfected with ERα to generate the stable line BT549-ERα. Asynchronous BT549-ERα cells had elevated Src activity and the ERα t1/2 was 3.9 hours (data not shown). In both ERα+ (MCF-7 and MDA-MB-361) and ERα– (BT-20 and BT549-ERα) lines, ERα levels increased with estrogen deprivation. Estradiol-stimulated ERα loss was impaired by proteasome inhibition with MG132 and also by Src inhibition with PD166326 (Figure 4D). For BT-20 cells, blots were exposed longer and more protein was loaded than for ERα+ lines.
Because PD166326 inhibited Src and Src family kinases, we further tested the specific role of Src in ERα regulation by transfecting BT-20 cells with either siRNA to Src or control siRNA and then depriving them of estrogen for 48 hours. Reduction of Src expression by Src siRNA impaired estrogen-stimulated ERα loss in BT-20 cells (Figure 4E). Thus Src appears to activate estrogen-stimulated ERα proteolysis in both ERα+ and ERα– breast lines.
Src inhibition impairs estrogen-stimulated ERα ubiquitylation in vivo.
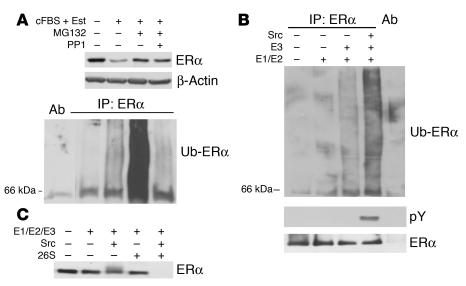
To test the effect of Src inhibition on ligand-driven ERα ubiquitylation, MCF-7 cells were estrogen deprived and then stimulated with estradiol with or without prior addition of the proteasome inhibitor MG132 or the Src inhibitor PP1. The ERα was immunoprecipitated from equal amounts of protein lysate, and complexes were resolved, immunoblotted with anti-ubiquitin antibody, and stripped and reprobed for total ERα. ERα levels were maximal in estrogen-deprived cells. Although ERα levels were reduced 6 hours after estradiol stimulation, detection of ERα ubiquitylation was modestly increased. When estrogen-deprived cells were treated with estradiol and MG132, the ERα protein level remained elevated, and ubiquitylated ERα was readily detected (Figure 5A). In contrast, while Src inhibition with PP1 prevented estrogen-stimulated loss of the ERα protein and maintained high ERα protein levels, ERα ubiquitylation was minimal (Figure 5A). Thus, Src inhibition impaired ligand-activated ERα ubiquitylation and prevented ligand-mediated loss of ERα.
Figure 5. Src stimulates ERα ubiquitylation and degradation in vivo and in vitro.
(A) Serum- and estrogen-deprived MCF-7 cells were treated with estradiol and 5% cFBS for 6 hours with or without immediate prior addition of MG132 or PP1, and ERα levels were assayed. Equal loading was confirmed by β-actin. ERα was precipitated, ERα complexes were resolved, and ubiquitylated ERα (Ub-ERα) was detected with anti-ubiquitin antibody. (B) For in vitro ERα ubiquitylation, recombinant ERα protein was reacted with E1 and E2, with or without E3 and with or without prior treatment of ERα with Src kinase (as described in Methods), for 60 minutes. ERα immunoprecipitates were resolved and blotted with anti-ubiquitin or anti-phosphotyrosine (pY) antibodies. The membrane was stripped and reprobed for ERα. (C) In vitro degradation of recombinant ERα used E1, E2, and E3 with or without prior incubation with Src and/or addition of 26S proteasome fraction as described in Methods.
Src activates ERα ubiquitylation and degradation in vitro.
To assay the effect of Src on ERα ubiquitylation and degradation in vitro, recombinant ERα was pretreated with Src kinase or mock treated, after which equal amounts of ERα were reacted with recombinant ubiquitin, ubiquitin-activating enzyme (E1), UbcH7 (E2), and E3 ubiquitin ligase supplied from asynchronous MCF-7 cell lysate. ERα was then precipitated and resolved, and ubiquitylated ERα was detected by immunoblotting with anti-ubiquitin antibody. In these assays, little ERα degradation occurred. Tyrosine phosphorylation of ERα was detected only in Src-treated samples. ERα ubiquitylation was enhanced by pre-treatment of the ERα with Src kinase (Figure 5B).
For ERα degradation, assay conditions were modified as described in Methods. Recombinant ERα was pretreated or not with Src kinase as above and then treated with the E1, E2, and E3 mixture with or without addition of 26S proteasome fraction. ERα degradation was minimal in assays with Src or 26S proteasome alone. When Src-pretreated ERα was incubated with E1, E2, and E3 together with 26S proteasome, ERα was completely degraded (Figure 5C).
cSrc is activated in ERα– primary breast cancers.
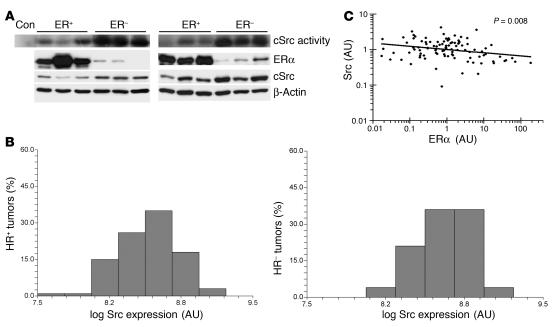
cSrc kinase activity was assayed in lysates from 18 ERα– and 22 ERα+ primary human breast cancers. The ERα status determined at diagnosis by LBA was verified by ERα immunoblotting. Blotting with β-actin verified equal loading and equal protein input in cSrc kinase assays. Elevated cSrc activity was observed in 78% (14 of 18) of ERα– breast cancers. In contrast, only 18% (4 of 22) of ERα+ tumors showed Src activity above that of nonspecific antibody controls (Figure 6A).
Figure 6. Elevation of cSrc activity and/or levels in ERα– primary breast cancers.
(A) Cryopreserved breast tumors were lysed, ERα status was verified by Western blot, and cSrc kinase activity was assayed. Equal loading was confirmed by β-actin. Blots are representative of 22 ERa+ and 18 ERa– tumors. (B and C) ERα and Src protein levels were quantitated by RPPA in 101 primary breast cancers as described in Methods. (B) Histogram distribution of log Src protein levels (arbitrary units) rounded to nearest log for HR+ (ERα+ and/or PR+) and HR– (ERα– and PR–) cancers. (C) Dot plot of Src and ERα protein values (expressed as log values, arbitrary units) in all cancers (P = 0.008).
Src and ERα levels are inversely correlated in primary human breast cancers.
To extend these findings, we quantitated ERα and Src protein expression by reverse phase tissue lysate array (RPPA) in 101 primary breast cancers using validated monospecific antibodies previously demonstrated to reflect Western blotting results with multiple tumor samples, providing a high-throughput quantitative analysis (50). Of 98 tumors in which the HR status was known, 68 were classified as positive for ERα and/or progesterone receptor (PR) by immunohistochemistry in pathology evaluation at diagnosis. ERα quantified by RPPA was significantly higher in pathologic ERα+ breast cancers (P < 0.001), as expected. Src protein was significantly higher in pathologic ERα– and PR– (P = 0.03) than in HR+ tumors (ERα+ and/or PR+). The distribution of Src values in HR+ and HR– cancers is shown in Figure 6B. When 68 HR+ tumors were compared with 23 “triple receptor-negative” tumors (ERα– and PR– by immunohistochemistry; HER2– by FISH), Src levels were highest in triple receptor-negative tumors (P = 0.02). In all tumors, quantified ERα and Src expression were inversely correlated (r = 0.26, P = 0.008; Figure 6C). In the subset of 68 HR+ tumors, there was also a statistically significant inverse correlation between quantified expression of ERα and Src (r = 0.30; P = 0.01).
Discussion
ERα– breast cancers have distinct gene expression profiles and are clinically more aggressive than are ERα+ cancers (12). The present study supports the hypothesis that, at least in a subset of ERα– breast cancers, Src activation may drive estrogen-dependent ERα proteolysis. ESR1 gene alterations are too infrequent to explain the lack of detectable ERα protein in up to one-third of breast cancers (6, 7). Early studies indicated that as many as 60%–70% of ERα– tumors express ESR1 mRNA (8–10). More sensitive QPCR demonstrated ESR1 mRNA in all of 52 ERα– primary breast cancers (51). In the present analysis, all of 200 ERα– breast cancers expressed ESR1 mRNA, with considerable variability and overlap in values in ERα+ and ERα– cancers. While mean ESR1 mRNA concentrations did not differ significantly between ERα+ and ERα– cancers, the modal distribution of ESR1 mRNA concentrations was lower in ERα– cancers. This is consistent with results from microarray studies that compared individual tumor ESR1 mRNA to reference cRNA pooled from ERα+ and ERα– tumors (11) or to the average signal from all tumors (12, 13) to reveal lower average ESR1 gene expression in ERα– versus ERα+ cancers. The variability in ESR1 mRNA levels and the discordance observed between ESR1 mRNA and protein in both tumor types point to important posttranscriptional controls of ERα levels.
Up to one-third of primary breast cancers show HER2/erbB-2 amplification, and a similar proportion has increased EGFR expression. Both are strongly associated with ERα– status (52, 53). Transfection of either EGFR or activated Her2 can reduce ERα levels in MCF-7 cells, and this has been attributed to MAPK activation (54). However, both of these receptors activate Src. In breast cancer cells, cSrc binds phosphorylated Her2 or EGFR, promoting synergistic activation to stimulate breast cancer cell proliferation and survival (49). Indeed, Src inhibitors impair Her2- and EGFR-driven mitogenesis (49, 55). Src is also transiently recruited to and activated by estrogen-bound ERα, leading to MAPK activation (22–25).
Src can phosphorylate ERα in vitro (56, 57). ERα phosphorylation by Src increases its affinity for estrogen (27), and may also affect ERα-coactivator binding and transcriptional activity (58, 59). The present study indicates that Src can drive expression of certain ERα target genes, which suggests the presence of an important feed-forward signaling loop involving estrogen, the ERα, and Src.
Crosstalk between liganded ERα and Src may not only regulate ERα transcriptional activity, but also activate ERα proteolysis. Inhibition of cellular Src impaired estrogen-mediated ERα ubiquitylation and ERα loss. Induced Src expression increased pS2 and GREB1 expression and ligand-activated ERα proteolysis. In breast cancer lines, increased Src activity correlated with a shortened ERα t1/2. In ERα+ as well as ERα– lines, proteasome inhibition increased ERα protein levels. Moreover, in both ERα+ and ERα– lines, estrogen withdrawal increased ERα levels and estrogen-stimulated ERα loss was impaired by Src inhibition. Because the Src inhibitor drug used affects not only Src, but other Src family kinases, we tested the effect of specific Src siRNA on ERα levels in the BT-20 line. Downregulation of cSrc expression using siRNA reduced estrogen-stimulated ERα loss in BT-20 cells. While we cannot exclude a possible contribution of other Src family kinases to estrogen-driven ERα proteolysis, this Src siRNA data supports a role for Src itself in this action.
Src kinase assays showed cSrc activation in a majority of primary ERα– tumors in a relatively small primary tumor set. In a larger group of over 100 primary breast cancers, Src protein levels correlated inversely with ERα in both ERα+ and ERα– tumors, as assayed by sensitive RPPA. ERα– cancers had higher Src levels than did ERα+ cancers, and this inverse statistical association was stronger in the subset of triple-negative compared with ERα+ tumors. These findings are consistent with our recent analysis of over 700 primary cancers in which the ERα– status correlated significantly (P < 0.001) with Src activation as detected by immunohistochemical staining for Y416-phosphorylated Src (A. Arnaout and J.M. Slingerland, unpublished observations). Although our data indicate that Src contributes to ERα regulation in breast cancers, there are clearly a number of tumors with high Src levels that retain ERα protein as well as tumors with low ERα levels that do not have high Src levels or activity. Thus, additional Src-independent mechanisms may regulate ERα protein levels. Tumors with very low ESR1 mRNA levels may reflect ERα promoter methylation (6).
cSrc appears to promote the ubiquitylation of ERα since cSrc inhibition impaired cellular ERα ubiquitylation and proteolysis in vivo. Moreover, ERα phosphorylation by Src increased both ERα ubiquitylation and 26S proteasome–mediated ERα degradation in vitro. These data support a model in which liganded ERα recruits cSrc or cSrc-dependent kinases, leading to phosphorylation events that facilitate ERα binding to coactivators and/or components of the proteolytic machinery. Ligand- and Src-activated ERα ubiquitylation may be linked to transcriptional activation of a subset of ERα-regulated genes. While our in vitro data support a direct effect, with Src phosphorylation of ERα promoting its ubiquitin-dependent degradation, Src may also have indirect effects to promote ERα degradation. Src or its downstream effectors may also affect ligand-activated ERα coactivator phosphorylation to regulate ERα degradation and transcriptional activity. SRC-3/AIB1 proteolysis accompanies estrogen-stimulated ERα activation (60). How specific Src-dependent ERα and/or coactivator phosphorylation events modulate the profile of coactivator binding, ERE selection, and ERα proteolysis will require further investigation. A recent report suggests that Src-mediated tyrosine phosphorylation may also regulate androgen receptor function (61).
Signaling pathways that activate many transcription factors, including c-Jun, c-Myc, and E2F-1, also trigger their ubiquitin-dependent degradation (48), thereby limiting transactivator function. Ubiquitylation is required for transcriptional activity of certain transcription factors (48, 62). Transcription factor ubiquitylation may influence coactivator/repressor binding (48), with coactivators subsequently enhancing ubiquitylation of certain transcription factors (63). Ligand-mediated proteolysis regulates turnover of most nuclear receptors (reviewed in ref. 64). Several ERα coactivators are also known to be ubiquitin ligases (43–45) or proteasome subunits (65).
In some models (40, 41), but not all (66, 67), proteasome inhibition decreases estrogen-ERα transcriptional activity despite an increase in ERα abundance. ESR1 mutations that impair coactivator binding abrogate ligand-stimulated ERα degradation (41). Thus, co-activator binding may regulate not only transcriptional activity but also ligand mediated ERα degradation. ERα cycles on and off the ERE (40, 68). Ligand increases the duration of ERα-ERE binding and modifies ubiquitin ligase binding (40). Proteasome inhibition has been shown to dissociate ubiquitin-bound ERα from ERE motifs and reduce ERα transcriptional activity. Thus, for a subset of ERα-driven genes, ERα ubiquitylation and transcription may be closely linked.
Cell type– and promoter-specific differences affect how ERα proteolysis influences target gene expression (66, 67). In one study, proteasome inhibition increased expression of cellular pS2 and CTSD but decreased PR expression (67). In certain promoter contexts, ligand-activated ERα may escape ubiquitylation and proteasomal degradation and yet remain functional. While proteolytic degradation of the ERα after ERE firing may allow reloading of the promoter, ubiquitylation and proteasomal degradation may potentially serve a more global role in regulating the abundance and overall activity of the ERα. Moreover, constitutive ERα activation could potentially lead to reduced ERα levels as a result of constitutive ERα proteolysis.
ERα phosphorylation by different signaling pathways could theoretically promote recruitment of different coactivators or ubiquitin pathway components, changing both the profile of ERα targets expressed and the rate of ligand-stimulated ERα proteolysis in different tissues. During breast cancer progression, Src activation may alter coactivator binding, shifting ERα transcriptional targets to profiles that promote oncogenic change. The present data do not allow us to estimate the contribution of Src-mediated ERα transcriptional activity and degradation to the overall oncogenic effect of Src in breast cancer. While we speculate that the effects of Src on ERα signaling crosstalk and transcriptional activity may make an important contribution to its oncogenicity, further work is required to tease out the specific contribution to breast carcinogenesis of Src acting on ERα.
ERα– breast cell lines are considered estrogen insensitive because they do not require estrogen for growth; coupled with the clinical observation that ERα– breast cancers do not respond to tamoxifen (2), this led to the belief that ERα– tumors are estrogen independent. Our data raise the concern that at least a subset of ERα– breast cancers, particularly those with oncogenic Src activation, may indeed be responsive to estrogen in vivo. Constitutive ERα proteolysis in at least a subset of ERα– cancers may not reflect extinguished ERα-dependent transcription, but rather indicate a shift to constitutive activation of different ERα transcriptional targets. The therapeutic implications of this work are potentially very significant and warrant further investigation.
Methods
Breast cancers used for ESR1 mRNA quantitation.
Cryopreserved primary invasive human breast cancers were obtained from the tumor repository of the Sunnybrook Health Sciences Center clinical ERα quantitation reference lab with approval from the Sunnybrook Health Sciences Center Review Ethics Board, Toronto, Ontario, Canada. One expert clinical biochemist performed all ERα cytosolic LBA (69). Concordance of ERα LBA with ERα immunohistochemistry was verified in 40 tumors.
RNA extraction and ESR1 mRNA quantitation.
mRNA was extracted from 300 macrodissected carcinomas (100 g) using TRI zol per the manufacturer’s instructions (Molecular Research Center). All RNAs were visualized on ethidium gels. A total of 250 tumor RNA samples with an OD260/OD280 greater than 1.3 and less than 2.1 from 50 ERα+ cancers (>30 fmol/μg protein by LBA) and 200 ERα– cancers (<10 fmol/μg protein by LBA) were analyzed. QPCR of human PBGD expression using the LightCycler hPBGD Housekeeping Gene Kit (Roche Applied Science) primer/hybridization mixture demonstrated similar expression and equal RNA quality in both groups (Student’s t test).
QPCR reactions used the LightCycler System (Roche Applied Science) and the QuantiTect SYBR Green RT-PCR kit (Qiagen). Primers are listed in Supplemental Methods. A standard curve for ESR1 mRNA quantitation was generated using serial dilutions of full-length human ERα cDNA plasmid PCMV5hER-α (provided by B. Katzenellenbogen, University of Illinois, Urbana, Illinois, USA). MCF-7 ESR1 mRNA was quantitated using the PCMV5hER-α plasmid standard curve. MCF-7 mRNA was run as a positive control in all tumor ESR1 mRNA QPCR reactions. Tumor ESR1 mRNA values ranged from 10 fg/μl to 1 μg/μl. Melting curve analysis ensured exclusion of primer dimmers from each analysis. ESR1 mRNA concentrations in ERα+ and ERα– cancers were compared by Student’s t test.
Sequencing of ERα cDNA PCR product.
All tumor ERα PCR products were visualized by gel electrophoresis. For a subset, PCR-amplified ERα cDNA was gel extracted with QIAquick Gel Extraction Kit (Qiagen), and 10 ng DNA was sequenced using 3.2 pmol of each ERα sequencing primer, Terminator Reaction Mix (ABI Prism dGTP BigDye Terminator v3.0 Cycle Sequencing Ready Reaction Kit; Applied Biosystems), and the ABI Prism 3100 Genetic Analyzer (Applied Biosystems).
Cell culture.
MCF-7 cells were grown in 5% FBS, and estrogen was depleted in 5% cFBS for 48 hours as described previously (70). For depletion of both growth factors and estrogen, cells were transferred to 0.1% cFBS for 48 hours. The ERα– BT-20 and BT549 cell lines and the weakly ERα+ MDA-MB-361 cell line (provided by S. Parsons) were grown in DMEM (49). The identity of ERα+ and ERα– lines was confirmed by karyotyping. To assay effects of growth factors on ERα levels, 10 nM β-estradiol with or without 5% FBS or 5% cFBS alone was added to MCF-7 cells that had been estrogen and growth factor depleted for 48 hours; ERα levels were assayed 1–6 hours later.
Plasmids and transfection.
Activated human cSrc vector, PCI-Src Y530F (from D. Fujita, University of Calgary, Calgary, Alberta, Canada) or empty PCI (10 μg) was transfected into MCF-7 cells using lipofectamine PLUSTM (Invitrogen). BT549 cells were transfected with PCMV5hER-α, and stable lines were cloned.
Construction of MCF-7 lines with inducible Src expression.
Src Y530F cDNA was cloned into pIND and transfected into the MCF-7 line with an integrated pVgRXR vector (Invitrogen). Src was induced with 2 μM PA. MCFpINDSrc2 cells had high Src induction 8–24 hours after induction with PA. This line was estrogen deprived as described above for 72 hours, and 2 μM PA was added or not for the last 24 hours of estrogen deprivation. Cells were then transferred to 0.1% cFBS, 10 nM estradiol was added for 6 hours, and ERα t1/2 was assayed by cycloheximide (CHX) chase as described below.
Flow cytometric analysis.
BrdU pulse labeling and flow cytometric analysis were performed as described previously (70).
Antibodies.
The ERα mAb H222 was supplied by G. Greene (University of Chicago, Chicago, Illinois, USA), ERα antibody HC-20 and anti-ubiquitin antibody P4D1 were from Santa Cruz Biotechnology Inc., and anti-Src mAb GD11 was from Upstate Biotechnology. Antibodies to MAPK, phosphosphorylated MAPK, total PKB, and activated PKB as well as anti-phosphotyrosine antibody P-tyr-102 were from Cell Signaling; antibody to β-actin was from Sigma-Aldrich.
Immunoblotting and CHX chase.
Cells were lysed in ice-cold D/RB buffer (50 mM HEPES, pH 7.5; 150 mM NaCl; 1 mM EDTA, pH 8.0; 2.5 mM EGTA, pH 8.0; 10% glycerol; 10 mM β-glycerophosphate; 1 mM NaF; 0.1% Tween-20; 1 mM PMSF; 0.1 mM Na2VO4; 0.5 mM DTT; and 0.02 mg/ml each of aprotinin, leupepsin, and pepstatin). Protein was quantitated by Bradford protein assay. Western blots used 20–100 μg protein per lane. The ERα t1/2 was determined by CHX chase, with addition of 100 μg CHX considered t = 0. Cells were lysed at the times indicated in Figures 2 and 4, and ERα was blotted. ERα protein was quantitated by densitometry from 3 experiments using ImageQuant imaging system (version 5.2; GE Healthcare).
Effects of MEK and PI3K inhibition on ERα stability.
To assay effects of MEK or PI3K inhibition on ERα levels, increasing concentrations of UO126 (0.1–10 μM; Promega) or LY294002 (0.5–8 μM; Promega) were added to asynchronous MCF-7 cells for 48 hours prior to immunoblotting or flow cytometry. Estrogen- and growth factor–depleted MCF-7 cells were treated with either 10 μM UO126 or 8 μM LY294002 for 30 minutes prior to stimulation with 17β-estradiol for 6 hours, followed by immunoblotting and flow cytometry.
Cellular Src kinase assays.
Cell lines or primary human breast cancers were lysed in ice-cold NP40 lysis buffer (70) with added 0.1 mM Na2VO4 and 1 mM EDTA (pH 8.0). Src was precipitated from 200 μg lysate, and Src kinase was assayed as described previously (71).
ERE luciferase assays.
MCF-7 cells were transfected with 500 ng of plasmid bearing 2 tandem ERE (2 × ERE luc), 50 ng phRL-TK luc, and 100 ng cSrc-Y530F using Lipofectamine Plus (Invitrogen) per the manufacturer’s instructions. Cells were treated with 10 μM PP1 and/or 10 nM estradiol for 4 hours prior to luciferase assays using dual-luciferase reporter assays (Promega) and Beckman Coulter LD400 Luminscence Detector.
QPCR of ERα target genes pS2 and GREB1.
MCFpINDSrc2 cells were maintained in 5% cFBS for 2 days before adding 2 μM PA for 24 hours to induce Src. The cells — with and without Src induction — were then treated with either 10 nM β-estradiol or 100 nM tamoxifen plus estradiol for 3 hours. Total RNA was isolated using TRI zol according to the manufacturer’s instructions (Invitrogen). cDNA synthesis was performed with 1 μg total RNA using iScript cDNA kit (Bio-Rad). QPCR was performed using icycleriQ PCR detection system (Bio-Rad) with 10 ng cDNA sample in iQ SyberGreen supermix (Bio-Rad). PCR conditions and primers are described in Supplemental Methods.
SiRNA-mediated inhibition of cSrc expression.
Dharmacon ON-TARGET plus SMARTpool siRNA reagent (Thermo Fisher Scientific) targeting cSrc and siCONTROL Non-Targeting siRNA Pools (Dharmacon; Thermo Fisher Scientific) were transfected into BT-20 cells cultured in media with 5% FBS using Lipofectamine 2000 (Invitrogen) for 4 hours. The cells were then estrogen deprived by transfer to media containing 5% cFBS for 48 hours. Cells were then treated with estradiol or not for 4 hours prior to lysis for analysis of ERα and Src levels by Western blotting.
Detection of ERα ubiquitylation in vivo.
MCF-7 cells were starved in 0.1% cFBS for 48 hours and then either maintained in 0.1% cFBS or transferred to 5% cFBS medium with 10 nM estradiol with or without 10 μM PP1 or 10 μM MG132. Six hours after estradiol addition, cells were lysed in 50 mM Tris (pH 7.5), 150 mM NaCl, 10% glycerol, 2 mM EDTA, 50 mM NaF, 1% NP40, and 1% SDS; boiled for 10 minutes; and centrifuged for 1 minute at 14,000 g at 22°C. Supernatant protein was quantitated, and ERα and β-actin were assayed by Western blot. To detect ubiquitylated ERα, ERα was immunoprecipitated from 500 μg lysate, resolved by SDS-PAGE, and transferred to nitrocellulose (Bio-Rad). The membrane was boiled in transfer buffer for 10 minutes and immunoblotted with antibody against ERα or ubiquitin.
In vitro ERα ubiquitylation assay.
Ubiquitylation assays used 40 ng recombinant ERα (Sigma-Aldrich), GST-ubiquitin-activating enzyme (E1), GST-ubiquitin-conjugating enzyme UbcH7 (E2), MCF-7 lysate (50 μg) as E3 source, and an energy regenerating solution (Boston Biochem) in 7.4 mM HEPES (pH 7.4), 5 mM KCl, and 1.5 mM MgCl2 for 60 minutes at 37°C. Prior to ubiquitylation assays, recombinant ERα was either incubated with 10 ng recombinant Src kinase (Upstate) or mock-treated for 5 minutes at 30°C in 7.4 mM HEPES (pH 7.4), 5 mM KCl, and 1.5 mM MgCl2. ERα was precipitated, and complexes were resolved and transferred to nitrocellulose (0.45 μm; Bio-Rad). The membrane was boiled for 10 minutes, and ERα and ubiquitylated ERα were detected as described above.
In vitro ERα degradation assay.
ERα degradation assays used E1, E2, and E3 as described above, with the following modifications. To catalyze in vitro degradation of ERα, 50 nM of 26S proteasome fraction (Boston Biochem) was added for 30 minutes at 37°C in 7.4 mM HEPES (pH 7.4), 5 mM KCl, 1.5 mM MgCl2, and 1 mM DTT. ERα was assayed by Western blot.
RPPA.
A total of 101 fresh-frozen primary breast tumors from the University of Texas MD Anderson Cancer Center Breast Tissue Tumor Bank were obtained with approval of the Institutional Review Board of the University of Texas MD Anderson Cancer Center. Tumors were macrodissected and lysed as described previously (50) and boiled in 1% SDS, and protein-rich supernatants were serially diluted manually. A robotic GeneTAC arrayer (Genomic Solutions) created arrays of 6 2-fold serial dilutions for each tumor lysate on nitrocellulose-coated glass slides (FAST Slides; Schleicher & Schuell). Arrayed slides were probed with ERα antibody (NeoMarkers) and Src (Upstate), and the signal was amplified using a DakoCytomation catalyzed system. A secondary antibody (anti-rabbit) was used as a starting point for signal amplification. The slides were scanned and each protein in each sample was assigned a relative quantification value in arbitrary units using MicroVigene software (version 2.0; Vigene Tech). All samples were normalized for protein loading as described previously (50, 72). NCSS software (2004 version; NCSS) was used for 2-tailed Student’s t tests and canonical correlation. ERα and/or PR were assayed by immunohistochemistry in pathology evaluation at diagnosis.
Statistics.
Differences in ESR1 and PBGD mRNA, quantitated by QPCR in the fresh frozen ERα+ and ERα– breast cancer samples, were analyzed statistically using the Student’s t test. The relationship between Src kinase activity and ERα protein status (positive or negative) in 40 primary breast cancers was determined using the Student’s t test. For the RPPA analysis, the arbitrary units expressing ERα and Src protein levels were converted to logarithms and the relationship between ERα and Src protein levels was analyzed using 2-tailed Student’s t tests and canonical correlation. P values of less than 0.05 were considered statistically significant.
Supplementary Material
Acknowledgments
We thank G. Greene for mAb H222 and D. Fujita for PCI-Src Y530F and S. Parsons for BT-20 and MDA-MB-361 cell lines. I. Chu was supported by a University of Toronto Medical Oncology Post-Graduate Award and by U.S. Department of Defense grant W81XWH-04-0392. A. Arnaout was supported by a Canadian Breast Cancer Foundation Physician Fellowship. J.M. Slingerland is supported by the Braman Family Breast Cancer Institute of University of Miami Sylvester Comprehensive Cancer Center.
Footnotes
Nonstandard abbreviations used: cFBS, charcoal-stripped FBS; CHX, cycloheximide; cSrc, cellular Src; ER, estrogen receptor; ERE, estrogen response element; HR, hormone receptor; LBA, ligand binding assay; PA, ponasterone A; PR, progesterone receptor; QPCR, quantitative real-time RT-PCR; RPPA, reverse phase tissue lysate array.
Conflict of interest: The authors have declared that no conflict of interest exists.
Citation for this article: J. Clin. Invest. 117:2205–2215 (2007). doi:10.1172/JCI21739
References
- 1.Jordan V.C. Studies on the estrogen receptor in breast cancer — 20 years as a target for the treatment and prevention of cancer. Breast Cancer Res. Treat. 1995;36:267–285. doi: 10.1007/BF00713399. [DOI] [PubMed] [Google Scholar]
- 2.Clarke M., et al. Tamoxifen for early breast cancer: an overview of the randomised trials. . Lancet. 1998;351:1451–1467. [PubMed] [Google Scholar]
- 3.Green S., et al. Cloning of the human oestrogen receptor cDNA. J. Steroid Biochem. 1986;24:77–83. doi: 10.1016/0022-4731(86)90035-x. [DOI] [PubMed] [Google Scholar]
- 4.Kuiper G.G., Enmark E., Pelto-Huikko M., Nilsson S., Gustafsson J.A. Cloning of a novel receptor expressed in rat prostate and ovary. Proc. Natl. Acad. Sci. U. S. A. 1996;93:5925–5930. doi: 10.1073/pnas.93.12.5925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Mosselman S., Polman J., Dijkema R. ER–beta: identification and characterization of a novel human estrogen receptor. FEBS Lett. 1996;392:49–53. doi: 10.1016/0014-5793(96)00782-x. [DOI] [PubMed] [Google Scholar]
- 6.Ferguson A.T., Davidson N.E. Regulation of estrogen receptor alpha function in breast cancer. Crit. Rev. Oncog. 1997;8:29–46. doi: 10.1615/critrevoncog.v8.i1.20. [DOI] [PubMed] [Google Scholar]
- 7.Roodi N., et al. Estrogen receptor gene analysis in estrogen receptor-positive and receptor-negative primary breast cancer. J. Natl. Cancer Inst. 1995;87:446–451. doi: 10.1093/jnci/87.6.446. [DOI] [PubMed] [Google Scholar]
- 8.Carmeci C., deConinck E.C., Lawton T., Bloch D.A., Weigel R.J. Analysis of estrogen receptor messenger RNA in breast carcinomas from archival specimens is predictive of tumor biology. Am. J. Pathol. 1997;150:1563–1570. [PMC free article] [PubMed] [Google Scholar]
- 9.Henry J.A., Nicholson S., Farndon J.R., Westley B.R., May F.E.B. Measurement of oestrogen receptor mRNA levels in human breast tumours. Br. J. Cancer. 1988;58:600–605. doi: 10.1038/bjc.1988.267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Garcia T., Lehrer S., Bloomer W.D., Schachter B. A variant estrogen receptor messenger ribonucleic acid is associated wity reduced levels of estrogen binding in human mammary tumors. Mol. Endocrinol. 1988;2:785–791. doi: 10.1210/mend-2-9-785. [DOI] [PubMed] [Google Scholar]
- 11.van ‘t Veer L.J., et al. Gene expression profiling predicts clinical outcome of breast cancer. Nature. 2002;415:530–536. doi: 10.1038/415530a. [DOI] [PubMed] [Google Scholar]
- 12.Gruvberger S., et al. Estrogen receptor status in breast cancer is associated with remarkably distinct gene expression patterns. Cancer Res. 2001;61:5979–5984. [PubMed] [Google Scholar]
- 13.West M., et al. Predicting the clinical status of human breast cancer by using gene expression profiles. Proc. Natl. Acad. Sci. U. S. A. 2001;98:11462–11467. doi: 10.1073/pnas.201162998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Perou C.M., et al. Molecular portraits of human breast tumours. Nature. 2000;406:747–752. doi: 10.1038/35021093. [DOI] [PubMed] [Google Scholar]
- 15.Sorlie T., et al. Gene expression patterns of breast carcinomas distinguish tumor subclasses with clinical implications. Proc. Natl. Acad. Sci. U. S. A. 2001;98:10869–10874. doi: 10.1073/pnas.191367098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Nilsson S., Gustafsson J.A. Estrogen receptor transcription and transactivation: basic aspects of estrogen action. Breast Cancer Res. 2000;2:360–366. doi: 10.1186/bcr81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Klinge C.M. Estrogen receptor interaction with co-activators and co-repressors. Steroids. 2000;65:227–251. doi: 10.1016/s0039-128x(99)00107-5. [DOI] [PubMed] [Google Scholar]
- 18.Tora L., et al. The human estrogen receptor has two independent nonacidic transcriptional activation functions. Cell. 1989;59:477–487. doi: 10.1016/0092-8674(89)90031-7. [DOI] [PubMed] [Google Scholar]
- 19.Danielian P.S., White R., Lees J.A., Parker M.G. Identification of a conserved region required for hormone dependent transcriptional activation by steroid hormone receptors. EMBO J. 1992;11:1025–1033. doi: 10.1002/j.1460-2075.1992.tb05141.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hall J.M., Couse J.F., Korach K.S. The multifaceted mechanisms of estradiol and estrogen receptor signaling. J. Biol. Chem. 2001;276:36869–36872. doi: 10.1074/jbc.R100029200. [DOI] [PubMed] [Google Scholar]
- 21.Coleman K.M., Smith C.L. Intracellular signaling pathways: nongenomic actions of estrogens and ligand-independent activation of estrogen receptors. Front. Biosci. 2001;6:D1379–D1391. doi: 10.2741/coleman. [DOI] [PubMed] [Google Scholar]
- 22.Migliaccio A., et al. Tyrosine kinase/p21ras/MAP-kinase pathway activation by estradiol receptor complex in MCF-7 cells. EMBO J. 1996;15:1292–300. [PMC free article] [PubMed] [Google Scholar]
- 23.Song R.X., et al. The role of Shc and insulin-like growth factor 1 receptor in mediating the translocation of estrogen receptor a to the plasma membrane. Proc. Natl. Acad. Sci. U. S. A. 2004;101:2076–2081. doi: 10.1073/pnas.0308334100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Migliaccio A., et al. Steroid-induced androgen receptor-oestradiol receptor beta-Src complex triggers prostate cancer cell proliferation. EMBO J. 2000;19:5406–5417. doi: 10.1093/emboj/19.20.5406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wong C.W., McNally C., Nickbarg E., Komm B.S., Cheskis B.J. Estrogen receptor-interacting protein that modulates its nongenomic activity-crosstalk with Src/Erk phosphorylation cascade. Proc. Natl. Acad. Sci. U. S. A. 2002;99:14783–14788. doi: 10.1073/pnas.192569699. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 26.Castoria G., et al. PI3-kinase in concert with Src promotes the S-phase entry of oestradiol-stimulated MCF-7 cells. EMBO J. 2001;20:6050–6059. doi: 10.1093/emboj/20.21.6050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Likhite V.S., Stossi F., Kim K., Katzenellenbogen B.S., Katzenellenbogen J.A. Kinase-specific phosphorylation of the estrogen receptor changes receptor interactions with ligand, DNA, and coregulators associated with alterations in estrogen and tamoxifen activity. Mol. Endocrinol. 2006;20:3120–3132. doi: 10.1210/me.2006-0068. [DOI] [PubMed] [Google Scholar]
- 28.Aronica S.M., Katzenellenbogen B.S. Stimulation of estrogen receptor-mediated transcription and alteration in the phosphorylation state of the rat uterine estrogen receptor by estrogen, cyclic adenosine monophosphate, and insulin-like growth factor-I. Mol. Endocrinol. 1993;7:743–752. doi: 10.1210/mend.7.6.7689695. [DOI] [PubMed] [Google Scholar]
- 29.Bunone G., Briand P.A., Miksicek R.J., Picard D. Activation of the unliganded estrogen receptor by EGF involves the MAP kinase pathway and direct phosphorylation. EMBO J. 1996;15:2174–2183. [PMC free article] [PubMed] [Google Scholar]
- 30.Kato S., et al. Activation of the estrogen receptor through phosphorylation by mitogen-activated protein kinase. Science. 1995;270:1491–1494. doi: 10.1126/science.270.5241.1491. [DOI] [PubMed] [Google Scholar]
- 31.Ignar-Trowbridge D.M., et al. Coupling of dual signaling pathways: epidermal growth factor action involves the estrogen receptor. Proc. Natl. Acad. Sci. U. S. A. 1992;89:4658–4662. doi: 10.1073/pnas.89.10.4658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Simoncini T., et al. Interaction of oestrogen receptor with the regulatory subunit of phosphatidylinositol-3-OH kinase. Nature. 2000;407:538–541. doi: 10.1038/35035131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Joel P.B., et al. pp90rsk1 regulates estrogen receptor-mediated transcription through phosphorylation of SER–167. Mol. Cell. Biol. 1998;18:1978–1984. doi: 10.1128/mcb.18.4.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Yudt M.R., et al. Function of estrogen receptor tyrosine 537 in hormone binding, DNA binding, and transactivation. Biochemistry. 1999;38:14146–14156. doi: 10.1021/bi9911132. [DOI] [PubMed] [Google Scholar]
- 35.Arnold S.F., Melamed M., Vorojeikina D.P., Notides A.C., Sasson S. Estradiol-binding mechanism and binding capacity of the human estrogen receptor is regulated by tyrosine phosphorylation. Mol. Endocrinol. 1997;11:48–53. doi: 10.1210/mend.11.1.9876. [DOI] [PubMed] [Google Scholar]
- 36.Weis K.E., Ekena K., Thomas J.A., Lazennec G., Katzenellenbogen B.S. Constitutively active human estrogen receptors containing amino acid substitutions for tyrosine 537 in the receptor protein. Mol. Endocrinol. 1996;10:1388–1398. doi: 10.1210/mend.10.11.8923465. [DOI] [PubMed] [Google Scholar]
- 37.Nirmala P.B., Thampan R.V. Ubiquitination of the rat uterine estrogen receptor: dependence on estradiol. Biochem. Biophys. Res. Commun. 1995;213:24–31. doi: 10.1006/bbrc.1995.2093. [DOI] [PubMed] [Google Scholar]
- 38.Nawaz Z., Lonard D.M., Dennis A.P., Smith C.L., O’Malley B.W. Proteasome-dependent degradation of the human estrogen receptor. Proc. Natl. Acad. Sci. U. S. A. 1999;96:1858–1862. doi: 10.1073/pnas.96.5.1858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Alarid E.T., Bakopoulos N., Solodin N. Proteasome-mediated proteolysis of estrogen receptor: a novel component in autologous Down-regulation. Mol. Endocrinol. 1999;13:1522–1534. doi: 10.1210/mend.13.9.0337. [DOI] [PubMed] [Google Scholar]
- 40.Reid G., et al. Cyclic, proteasome-mediated turnover of unliganded and liganded ERalpha on responsive promoters is an integral feature of estrogen signaling. Mol. Cell. 2003;11:695–707. doi: 10.1016/s1097-2765(03)00090-x. [DOI] [PubMed] [Google Scholar]
- 41.Lonard D.M., Nawaz Z., Smith C.L., O’Malley B.W. The 26S proteasome is required for estrogen receptor-alpha and coactivator turnover and for efficient estrogen reeptor-alpha transactivation. Mol. Cell. 2000;5:939–948. doi: 10.1016/s1097-2765(00)80259-2. [DOI] [PubMed] [Google Scholar]
- 42.Wijayaratne A.L., McDonnell D.P. The human estrogen receptor-alpha is a ubiquitinated protein whose stability is affected differentially by agonists, antagonists, and selective estrogen receptor modulators. J. Biol. Chem. 2001;276:35684–35692. doi: 10.1074/jbc.M101097200. [DOI] [PubMed] [Google Scholar]
- 43.Fan S., et al. BRCA1 inhibition of estrogen receptor signaling in transfected cells. Science. 1999;284:1354–1356. doi: 10.1126/science.284.5418.1354. [DOI] [PubMed] [Google Scholar]
- 44.Saji S., et al. MDM2 enhances the function of estrogen receptor alpha in human breast cancer cells. Biochem. Biophys. Res. Commun. 2001;281:259–265. doi: 10.1006/bbrc.2001.4339. [DOI] [PubMed] [Google Scholar]
- 45.Nawaz Z., et al. The Angelman syncrome-associated protein, E6-AP, is a coactivator for the nuclear hormone receptor superfamily. Mol. Cell. Biol. 1999;19:1182–1189. doi: 10.1128/mcb.19.2.1182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Thomas S.M., Brugge J.S. Cellular functions regulated by Src family kinases. Annu. Rev. Cell Dev. Biol. 1997;13:513–609. doi: 10.1146/annurev.cellbio.13.1.513. [DOI] [PubMed] [Google Scholar]
- 47.Rosen N., et al. Analysis of pp60c-src protein kinase activity in human tumor cell lines and tissues. J. Biol. Chem. 1986;261:13754–13759. [PubMed] [Google Scholar]
- 48.Tansey W.P. Transcriptional activation: risky business. Genes Dev. 2001;15:1045–1050. doi: 10.1101/gad.896501. [DOI] [PubMed] [Google Scholar]
- 49.Belsches-Jablonski A.P., et al. Src family kinases and HER2 interactions in human breast cancer cell growth and survival. Oncogene. 2001;20:1465–1475. doi: 10.1038/sj.onc.1204205. [DOI] [PubMed] [Google Scholar]
- 50.Sheehan K.M., et al. Use of reverse phase protein microarrays and reference standard development for molecular network analysis of metastatic ovarian carcinoma. Mol. Cell Proteomics. 2005;4:346–355. doi: 10.1074/mcp.T500003-MCP200. [DOI] [PubMed] [Google Scholar]
- 51.Iwao K., et al. Quantitative analysis of estrogen receptor-alpha and -beta messenger RNA expression in breast carcinoma by real-time polymerase chain reaction. Cancer. 2000;89:1732–1738. doi: 10.1002/1097-0142(20001015)89:8<1732::AID-CNCR13>3.0.CO;2-2. [DOI] [PubMed] [Google Scholar]
- 52.Pegram M.D., Pauletti G., Slamon D.J. HER–2/neu as a predictive marker of response to breast cancer therapy. Breast Cancer Res. Treat. 1998;52:65–77. doi: 10.1023/a:1006111117877. [DOI] [PubMed] [Google Scholar]
- 53.Nicholson S., et al. Epidermal growth factor receptor (EGFR) as a marker for poor prognosis in node-negative breast cancer patients: neu and tamoxifen failure. J. Steroid Biochem. Mol. Biol. 1990;37:811–814. doi: 10.1016/0960-0760(90)90424-j. [DOI] [PubMed] [Google Scholar]
- 54.Oh A.S., et al. Hyperactivation of MAPK induces loss of ERalpha expression in breast cancer cells. Mol. Endocrinol. 2001;15:1344–1359. doi: 10.1210/mend.15.8.0678. [DOI] [PubMed] [Google Scholar]
- 55.Biscardi J.S., et al. c-Src-mediated phosphorylation of the epidermal growth factor receptor on Tyr845 and Tyr1101 is associated with modulation of receptor function. J. Biol. Chem. 1999;274:8335–8343. doi: 10.1074/jbc.274.12.8335. [DOI] [PubMed] [Google Scholar]
- 56.Arnold S.F., Obourn J.D., Jaffe H., Notides A.C. Phosphorylation of the human estrogen receptor on tyrosine 537 in vivo and by Src family tyrosine kinases in vitro. Mol. Endocrinol. 1995;9:24–33. doi: 10.1210/mend.9.1.7539106. [DOI] [PubMed] [Google Scholar]
- 57.Arnold S.F., Obourn J.D., Yudt M.R., Carter T.H., Notides A.C. In vivo and in vitro phosphorylation of the human estrogen receptor. J. Steroid Biochem. Mol. Biol. 1995;52:159–171. doi: 10.1016/0960-0760(94)00166-j. [DOI] [PubMed] [Google Scholar]
- 58.Shah Y.M., Rowan B.G. The Src kinase pathway promotes tamoxifen agonist action in Ishikawa endometrial cells through phosphorylation-dependent stabilization of estrogen receptor (alpha) promoter interaction and elevated steroid receptor coactivator 1 activity. Mol. Endocrinol. 2005;19:732–748. doi: 10.1210/me.2004-0298. [DOI] [PubMed] [Google Scholar]
- 59.Sisci D., et al. Fibronectin and type IV collagen activate ERalpha AF-1 by c-Src pathway: effect on breast cancer cell motility. Oncogene. 2004;23:8920–8930. doi: 10.1038/sj.onc.1208098. [DOI] [PubMed] [Google Scholar]
- 60.Shao W., Keeton E.K., McDonnell D.P., Brown M. Coactivator AIB1 links estrogen receptor transcriptional activity and stability. Proc. Natl. Acad. Sci. U. S. A. 2004;101:11599–11604. doi: 10.1073/pnas.0402997101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Guo Z., et al. Regulation of androgen receptor activity by tyrosine phosphorylation. Cancer Cell. 2006;10:309–319. doi: 10.1016/j.ccr.2006.08.021. [DOI] [PubMed] [Google Scholar]
- 62.Salghetti S.E., Caudy A.A., Chenoweth J.G., Tansey W.P. Regulation of transcriptional activation domain function by ubiquitin. Science. 2001;293:1651–1653. doi: 10.1126/science.1062079. [DOI] [PubMed] [Google Scholar]
- 63.Fukuchi M., et al. Ligand-dependent degradation of Smad3 by a ubiquitin ligase complex of ROC1 and associated proteins. Mol. Biol. Cell. 2001;12:1431–1443. doi: 10.1091/mbc.12.5.1431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Nawaz Z., O’Malley B.W. Urban renewal in the nucleus: is protein turnover by proteasomes absolutely required for nuclear receptor-regulated transcription? Mol. Endocrinol. 2004;18:493–499. doi: 10.1210/me.2003-0388. [DOI] [PubMed] [Google Scholar]
- 65.vom Baur E., et al. Differential ligand-dependent interactions between the AF-2 activating domain of nuclear receptors and the putative transcriptional intermediary factors mSUG1 and TIF1. EMBO J. 1996;15:110–124. [PMC free article] [PubMed] [Google Scholar]
- 66.Alarid E.T., Preisler–Mashek M.T., Solodin N.M. Thyroid hormone is an inhibitor of estrogen-induced degradation of estrogen receptor-alpha protein: estrogen-dependent proteolysis is not essential for receptor transactivation function in the pituitary. Endocrinology. 2003;144:3469–3476. doi: 10.1210/en.2002-0092. [DOI] [PubMed] [Google Scholar]
- 67.Fan M., Nakshatri H., Nephew K.P. Inhibiting proteasomal proteolysis sustains estrogen receptor-alpha activation. Mol. Endocrinol. 2004;18:2603–2615. doi: 10.1210/me.2004-0164. [DOI] [PubMed] [Google Scholar]
- 68.Shang Y., Hu X., DiRenzo J., Lazar M.A., Brown M. Cofactor dynamics and sufficiency in estrogen receptor-regulated transcription. Cell. 2000;103:843–852. doi: 10.1016/s0092-8674(00)00188-4. [DOI] [PubMed] [Google Scholar]
- 69.Hassapoglidou S., Diamandis E.P., Sutherland D.J.A. Quantification of p53 protein in tumor cell lines, breast tissue extracts and serum with time-resolved immunofluorometry. Oncogene. 1993;8:1501–1509. [PubMed] [Google Scholar]
- 70.Cariou S., et al. Down-regulation of p21WAF1/CIP1 or p27Kip1 abrogates antiestrogen-mediated cell cycle arrest in human breast cancer cells. Proc. Natl. Acad. Sci. U. S. A. 2000;97:9042–9046. doi: 10.1073/pnas.160016897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Egan C., et al. Activation of Src in human breast tumor cell lines: elevated levels of phosphotyrosine phosphatase activity that preferentially recognizes the Src carboxy terminal negative regulatory tyrosine 530. Oncogene. 1999;18:1227–1237. doi: 10.1038/sj.onc.1202233. [DOI] [PubMed] [Google Scholar]
- 72.Tibes R., et al. Reverse phase protein array: validation of a novel proteomic technology and utility for analysis of primary leukemia specimens and hematopoietic stem cells. Mol. Cancer Ther. 2006;5:2512–2521.. doi: 10.1158/1535-7163.MCT-06-0334. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.