Abstract
Polymerase chain reaction (PCR)-based analysis for detecting immunoglobulin heavy chain gene (IgH) rearrangements in lymphoproliferative disorders is well established. The presence of one or two discrete bands is interpreted as a monoclonal proliferation, whereas a smear pattern represents a polyclonal population. Prompted by our observation of discrete bands in histologically reactive processes with a relative paucity of B cells, we sought to determine whether low numbers of B cells in biopsy specimens could artifactually produce pseudomonoclonal bands. We performed IgH PCR analysis on serially diluted DNA samples from 5 B cell non-Hodgkin’s lymphomas (B-NHLs), 5 reactive lymph nodes, 5 reactive tonsils and 10 microdissected germinal centers from a lymph node with follicular hyperplasia. We also assessed multiple aliquots of DNA samples from small biopsy specimens of reactive lymphocytic processes from the stomach (5 cases). PCR products were evaluated using high resolution agarose or polyacrylamide gels, and DNA sequencing was performed on IgH PCR products from two reactive germinal centers, which yielded monoclonal bands of identical size. All 5 B-NHLs harboring monoclonal B cell populations yielded single discrete bands, which were maintained in all dilutions. By contrast, all of the reactive lesions with polyclonal patterns at 50 ng/μl starting template concentration showed strong pseudomonoclonal bands at dilutions of 1:1000 to 1:1500 in placental DNA. Two of the microdissected reactive germinal centers that showed bands of identical size on duplicate reactions were proven to have different IgH sequences by sequencing. We conclude that specimens containing low numbers of polyclonal B cells may produce pseudomonoclonal bands on IgH PCR analysis. IgH PCR analysis should be performed on multiple aliquots of each DNA sample, and only samples that yield reproducible bands of identical size can be reliably interpreted as monoclonal.
The utility of molecular techniques such as Southern blot hybridization (SBH) analysis and polymerase chain reaction (PCR) in the assessment of clonality in lymphoproliferative disorders is well established. Although SBH is a fairly sensitive and specific method, its utility as a routine diagnostic tool is hampered by its requirement for high quality DNA, its labor-intensive nature, and its consequent long turnaround time. On the other hand, the rapid and less labor-intensive PCR continues to gain popularity as a technique for the assessment of clonality in lymphoproliferative disorders. 1, 2 The results obtained with antigen receptor gene PCR correlate well with the results of SBH. 3, 4, 5
PCR-based assays have additional advantages over SBH analysis. PCR requires smaller quantities of DNA, and high molecular weight DNA is not necessary. Therefore, PCR has been applied to small biopsies and fixed paraffin-embedded tissues, which are generally unsuitable for SBH analysis. 6
PCR is extremely sensitive and can detect 1 positive cell in a background of 105 negative cells. Immunoglobulin heavy chain (IgH) PCR is capable of detecting 1 monoclonal B cell in a background of 102 to 103 polyclonal B cells. 7 The extreme sensitivity of PCR also constitutes a potential source for pitfalls in the interpretation of PCR-based antigen receptor gene rearrangement studies. Clearly, contamination is a frequent concern. Additionally, we have observed discrete bands in samples obtained from small biopsy specimens in which histological and immunophenotypic evaluation revealed a reactive process. We believe that the discrete bands generated in this situation are related to the paucity of B cells in the biopsy specimen and do not represent a true oligoclonal or monoclonal B cell population. For the purpose of this study, we refer to those bands as pseudomonoclonal bands.
We undertook this study to investigate the relationship between the relative numbers of B cells in polyclonal processes and the generation of pseudomonoclonal bands by IgH PCR. We have performed serial dilutions of DNA samples obtained from a variety of well characterized specimens, including reactive tonsils and lymph nodes and malignant B cell lymphomas, and assessed them for clonality using IgH PCR. In addition, we assessed cases of benign chronic active gastritis by IgH PCR. We believe that the results of this study have implications for the interpretation of the PCR-based studies for the assessment of clonal antigen receptor gene rearrangements (IgH or T cell receptor). We also provide some recommendations to aid in the identification of pseudoclonal bands to prevent their misinterpretation as bona fide monoclonal bands.
Materials and Methods
Clinical Samples
All of the clinical samples studied were obtained from formalin-fixed, paraffin-embedded tissue blocks selected from the files of the Department of Pathology of the University of Utah Health Sciences Center (Salt Lake City, UT). Five monoclonal B cell non-Hodgkin’s lymphomas were selected: one case each of lymph node biopsy specimens involved by chronic lymphocytic leukemia/small lymphocytic lymphoma, follicular lymphoma, mantle cell lymphoma, B cell lymphoblastic lymphoma, and diffuse large B cell lymphoma. Five cases each of reactive hyperplasia of the tonsils and lymph nodes were also included. We also assessed 5 cases of chronic active Helicobacter pylori-associated gastritis with florid follicle formation (ie, grade 2 lesion as defined by Wotherspoon and colleagues). 8 In addition, ten reactive germinal centers were microdissected from a case of follicular hyperplasia and subjected to IgH PCR analysis.
Laser Capture Microdissection
Laser capture microdissection of reactive germinal centers was performed on 5-μm-thick hematoxylin/eosin-stained sections from formalin-fixed, paraffin-embedded tissue as previously described, using a PixCell laser capture microscope (Arcturus Engineering, Santa Clara, CA). 9
DNA Extraction
DNA was extracted from whole sections of formalin-fixed, paraffin-embedded tissue according to standard procedures. 10 The approximate size of the tissue samples from which DNA was extracted was 2 cm × 1 cm × 5 μm (three sections each) for the tonsils and lymph node biopsies, 3 mm × 3 mm × 5 μm (three sections each) for the gastric biopsies, and 80 μm × 60 μm × 5 μm (150 cells) for each microdissected germinal center. DNA extraction from the microdissected reactive germinal centers was performed as previously described. 9
Serial Dilution of DNA Templates
DNA extracted from the whole sections of paraffin-embedded tissue were serially diluted in placental DNA from 1:2 to 1:1500, and then subjected to IgH PCR.
IgH PCR Analysis
IgH PCR analysis was performed on all samples using a previously described protocol and primers. 11 Briefly, amplification reactions were performed using a 1605 rapid air thermocycler (Idaho Technology, Idaho Falls, ID). A heminested approach was used using the VHA-FRIII (5′ ACA CGG C(C/T)G T(G/A)T ATT ACT GT 3′) and JHa (5′ ACC TGA GGA GAC GGT GAC C 3′) primers for the first round of PCR, which consisted of 45 cycles of denaturation (94°C for 0 seconds), annealing (55°C for 0 seconds) and extension (72°C for 1 second). Heminesting was performed by using 1 μl of the initial amplification product as the template in a second reaction consisting of 30 cycles of denaturation (94°C for 0 seconds), annealing (50°C for 0 seconds) and extension (72°C for 10 seconds) using an internal JH primer (5′ GTG ACC AGG GT(G/T/A/C) CCT TGG CCC CAG 3′) and the VHA-FRIII primer. This rapid cycle PCR assay is performed in glass capillary tubes that accommodate only 10 μl of reaction solution, providing a high surface-to-volume ratio and thus a high reaction efficiency. An initial template concentration of 50 ng per 10-μl reaction maintains a similar DNA template-to-reaction volume ratio of 5 to 10 ng/μl, as obtains in conventional PCR assays that use 500 ng to 1 μg in reaction volumes of up to 100 μl. In our system, utilization of higher starting DNA template-to-reaction volume ratios are inhibitory to rapid cycle amplification and result in PCR failure. Thus, 50 ng of DNA isolated from whole sections or 2 μl of the crude extract from the microdissected samples were used as the initial quantity of template DNA per reaction. PCR products were subjected to electrophoresis at 80V for 2 hours using ethidium bromide-stained 3.5% metaphor agarose gels (FMC BioProducts, Rockland, ME) and products detected by ultraviolet transillumination. A non-nested IgH-PCR approach was also used for assessing the serial dilutions from the reactive lymph nodes and tonsils. VHa-FRIII and JHa primers were used in a reaction with reagent concentrations as described for the first round of the heminested assay described above. The reaction consisted of 35 cycles of denaturation (94°C for 0 seconds), annealing (55°C for 0 seconds) and extension (72°C for 1 second). Each amplification was performed in duplicate and 10 μl of the PCR products were subjected to electrophoresis at 160V for 2 hours on 10% polyacrylamide gels (acrylamide:bisacrylamide = 29:1).
DNA Sequencing
The IgH PCR products from two microdissected germinal centers were subjected to bidirectional double-stranded DNA sequencing using IgH PCR primers and ABI PRISM 377 sequencer (PE Applied Biosystems, Foster City, CA). Comparison of sequences and identification of immunoglobulin heavy chain gene usage was performed using Lasergene software (DNASTAR, Inc., Madison, WI).
Results
IgH PCR in B Cell Lymphoma
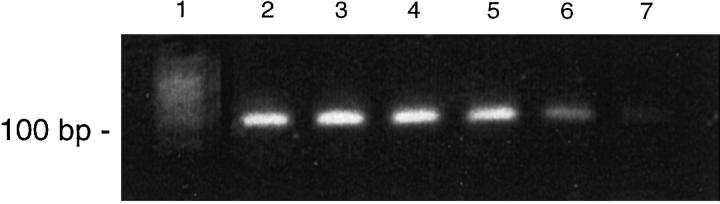
PCR assays to detect IgH gene rearrangements in all 5 B cell lymphomas yielded a single discrete product ranging from 80 to 250 bp. In each case, the monoclonal bands persisted in dilutions up to 1:1000 in placental DNA (ie, technical sensitivity = 0.1%; Figure 1 ). When diluted in polyclonal DNA from a reactive lymph node, monoclonal bands were reproducibly detectable only at a dilution of 1:50 (ie, diagnostic sensitivity = 2%).
Figure 1.
Immunoglobulin heavy chain (IgH) PCR in a monoclonal B cell process (malignant lymphoma). Ten microliters of PCR product were loaded in every lane. Serial dilutions were performed using 50 ng of template DNA as the starting quantity per reaction. Lane 1: polyclonal DNA control showing a smear pattern. Lane 2: undiluted sample at 50 ng/reaction showing a monoclonal band (∼100 bp). Lane 3: 1:10 dilution (5 ng/reaction). Lane 4: 1:100 dilution (0.5 ng). Lane 5: 1:500 dilution (0.25 ng). Lane 6: 1: 1000 dilution (0.05 ng). Lane 7: 1:1500 (0.033 ng). The monoclonal band (∼100 bp) is seen to persist at a dilution of 1:1000.
IgH PCR in Reactive Lymph Nodes and Tonsils
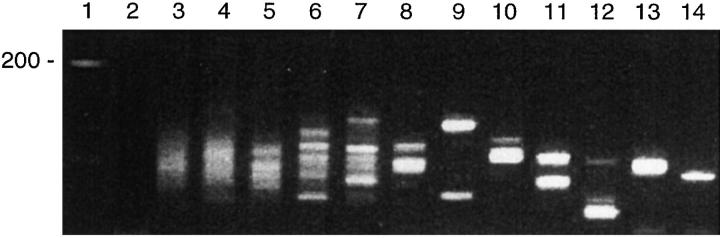
In the reactive lymph nodes and tonsils (5 each), the heminested PCR products showed a progressive tendency toward monoclonality at increasing dilutions. In all cases, an initial quantity of 50 ng of target DNA yielded a polyclonal smear pattern on 3.5% metaphor agarose gel electrophoresis. However, serial dilutions yielded oligoclonal bands at dilutions ranging from 1:20 (2.5 ng) to 1:500 (0.1 ng), and pseudomonoclonal bands at dilutions ranging from 1:200 (0.25 ng) to 1:1000 (0.05 ng). Figure 2 illustrates a case in which oligoclonal bands were detected at a dilution of 1:200 (0.25 ng) and pseudomonoclonal bands at 1:1000 dilution (0.05 ng). We also assessed a reactive lymph node and tonsil (1 each) using a non-nested IgH PCR assay and polyacrylamide gel electrophoresis. Using the non-nested assay and polyacrylamide gel electrophoresis, we detected oligoclonal bands at dilutions between 1:200 (0.25 ng) and 1:500 (0.1 ng) and pseudomonoclonal bands at 1:1000 (0.05 ng) and 1:1500 (0.033 ng; data not shown).
Figure 2.
IgH PCR in serial dilutions of DNA from a reactive lymph node. A progressive tendency toward the formation of pseudomonoclonal bands with incremental dilutions is evident. Lane 1: DNA molecular weight marker. Lane 2: negative (H20) control. Lane 3: a polyclonal smear pattern is seen in the undiluted DNA sample (50 ng). Lane 4: 1:10 dilution (5 ng), Lane 5: 1:100 dilution (0.5 ng). Lane 6: 1:200 dilution (0.25 ng). Lane 7: 1:300 dilution (0.17 ng). Lane 8: 1:400 (0.125 ng). Lane 9: 1:500 dilution (0.1 ng). Lane 10: 1:600 dilution (0.083 ng). Lane 11: 1:700 dilution (0.071 ng). Lane 12: 1:800 (0.0625 ng). Lane 13: 1:900 (0.056 ng). Lane 14: 1:1000 (0.05 ng).
IgH PCR in Microdissected Reactive Follicles
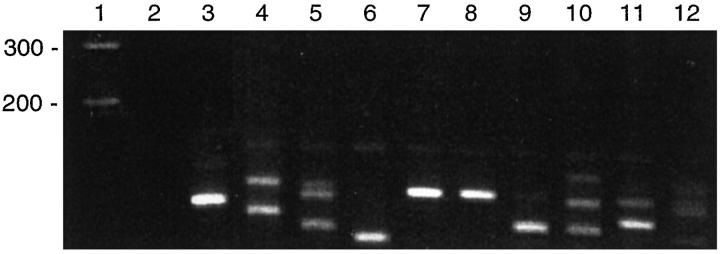
Ten reactive germinal centers from a lymph node biopsy specimen with follicular hyperplasia were microdissected and subjected to separate IgH PCR amplifications. Although the follicles varied in size, the follicles chosen for dissection contained approximately 150 cells in each 5-μm-thick section. Each microdissected follicle was suspended in 5 μl of digestion buffer, and 2 μl were utilized as the initial volume of template for PCR. Amplification was successful in all cases. Of the 10 microdissected follicles, pseudomonoclonal bands were generated in 5/10 (50%), oligoclonal bands in 2/10 (20%), and a polyclonal smear pattern in 1/10 (10%) follicles (Figure 3) . Two follicles yielded a pseudomonoclonal pattern with products of identical size (Figure 3) . However, DNA sequencing analysis revealed that the IgH sequences generated in both cases were completely unrelated (data not shown).
Figure 3.
IgH PCR on microdissected reactive germinal centers. Approximately 50 cells were used per reaction. Lane 1: molecular weight marker. Lane 2: negative (H20) control. Lane 3: positive (monoclonal) control. Lanes 5 and 10 show an oligoclonal pattern. Lanes 4, 6–9, and 11 show a pseudomonoclonal pattern (one or two bands). Lane 12 shows a polyclonal (smear) pattern. Note that lanes 7 and 8 show products of identical size even though the DNA used was extracted from separate germinal centers that were distant from one another.
IgH PCR in Chronic Gastritis
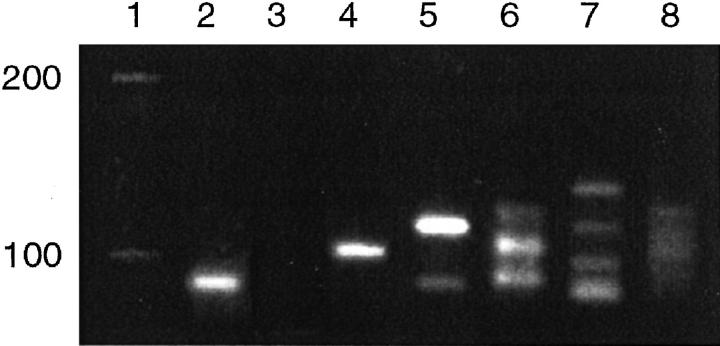
Pseudomonoclonal bands were generated in duplicate samples of 1/5 (20%) cases of chronic reactive gastritis. Similarly, 2/5 (40%) cases yielded oligoclonal bands that varied in size on duplicate reactions. A reproducible polyclonal smear pattern was generated in 2/5 (40%) cases. In the case of chronic gastritis with a pseudomonoclonal band, the monoclonal pattern was converted into an oligoclonal or a polyclonal pattern by increasing the number of whole sections (3 mm × 3 mm × 5 μm per section) from 3 to 7 sections per extraction (Figure 4) .
Figure 4.
IgH PCR performed on serially increasing numbers of whole sections of paraffin-embedded tissue from a case of reactive gastritis. There is a progressive tendency toward polyclonality as more serial sections putatively contributing more B cells are used for the DNA extraction. Lane 1: molecular weight marker. Lane 2: positive monoclonal (control). Lane 3: negative (H2O) control. Lane 4: pseudomonoclonal pattern (one discrete band) is obtained when using from DNA extracted from three 5-μm sections from the gastric biopsy specimen. Lane 5: a pseudomonoclonal pattern (two bands) is obtained with DNA from four 5-μm sections. Lanes 6 and 7: oligoclonal patterns obtained using five and six 5-μm sections respectively. Lane 8 shows a polyclonal smear pattern obtained using seven 5-μm tissue sections for DNA extraction.
Discussion
The establishment of monoclonality in lymphoproliferative processes has become an important adjunct in the diagnosis of malignant lymphoma. In particular, molecular methods have been useful in establishing the diagnosis of lymphoid neoplasia in specimens in which histological and immunophenotypic studies are inconclusive. 7 Whereas the criteria for determination of monoclonality by SBH analysis are relatively well established, 12 the parameters for distinguishing polyclonal from monoclonal lymphoid proliferations by PCR need further refinement. With the advent of less invasive techniques for the procurement of biopsy material and the consequent provision of smaller biopsy specimens to the diagnostic pathologist, the demand on molecular techniques for the resolution of diagnostic difficulties continues to increase.
PCR has provided enormous benefit because of its sensitivity and its amenability to use with smaller samples. Indeed, several studies have demonstrated the utility of IgH PCR analysis in the assessment of clonality in small tissue biopsy 13, 14 and fine needle aspiration specimens, 2, 15 and in small groups of cells microdissected from tissue sections 15, 16 or scraped from glass slides of cytologic material. 17 Although most of these studies have focused on the sensitivity of PCR-based methods for assessment of clonality, only a few reports have addressed issues relating to its specificity in a systematic fashion. Nevertheless, some studies have demonstrated instances of false positivity, particularly when small or microdissected specimens are used, 18 and in reactive conditions involving extranodal sites such as the stomach and salivary gland. 19, 20
The results of our study clearly show a relationship between a paucity of B cell targets and a tendency for generating pseudomonoclonal or pseudo-oligoclonal bands by IgH PCR. Using DNA obtained from reactive lymph nodes and tonsils, we found that polyclonal results were reliably obtained when the starting quantity of template DNA was 50 ng or more. Serial dilutions in placental DNA revealed pseudomonoclonal bands at dilutions beginning at 1:50 (1 ng) of template. Our results are consistent with those of a previous study which used dilutions of normal peripheral blood lymphocyte DNA. 2 These results collectively suggest that small amounts of polyclonal B cell DNA may generate pseudomonoclonal bands based on preferential amplification of the markedly diminished (restricted) IgH targets that are present in the highly dilute sample. Thus, a sufficient number of lymphocytes must be present in a sample to allow the production of heterogeneous IgH products in order that a polyclonal pattern may be obtained on gel electrophoresis. 2 Conversely, our data revealed that monoclonality could be scored reliably when DNA from a monoclonal B cell process is diluted at 1:1000 (0.05 ng) in placental DNA, but only at 1:50 (1 ng) in polyclonal B cell DNA.
An important distinguishing characteristic of the pseudomonoclonal bands is the absence of reproducibility in duplicate reactions. Our results confirm the observations of other investigators who have shown pseudomonoclonal bands of different molecular weight in parallel reactions of multiple aliquots of DNA from a polyclonal sample. 21 The inconstant nature of these bands is presumably reflective of the stochastic nature of the amplified IgH segments in the few B cells within the DNA sample. This phenomenon may explain the detection of a false positive monoclonal rearrangement in a case of colonic adenocarcinoma. 22
Using the heminested IgH PCR assay, we generated nonreproducible oligoclonal bands in 2/5 (40%) cases, a pseudomonoclonal band in 1/5 (20%) cases, and reproducible polyclonal bands in 2/5 (40%) cases of H. pylori-associated chronic active gastritis. In the pseudomonoclonal case, we were able to abolish the single discrete bands in subsequent reactions by increasing the number of whole tissue sections from which DNA was extracted, while maintaining the same overall concentration of DNA per reaction (50 ng). Although the non-nested assay also yielded nonreproducible pseudomonoclonal bands in the hyperplastic lymphoid tissues evaluated, the nonreproducible bands were initially detected at lower starting template concentrations than were obtained with the heminested assay. These results indicate that the heminested assay is more prone to the generation of pseudomonoclonal bands, especially at lower starting B cell template copy numbers.
Our studies show detection of pseudomonoclonal bands in polyclonal B cell populations diluted in placental DNA at a higher sensitivity (1:200–1:1000) than that of a monoclonal B cell population serially diluted with polyclonal B cell DNA (1:50). This is due in part to the fact that serial dilutions of the polyclonal B cells in placental DNA substantially reduce the number of B cell targets available for amplification, thus generating an artifactual oligoclonal B cell situation. By contrast, a monoclonal population serially diluted within a polyclonal background yields a scenario where the abundant polyclonal B cell targets effectively compete with the monoclonal population during amplification, and decrease the observer’s ability to discriminate the monoclonal population in an increasingly polyclonal background.
Nevertheless, the occurrence of pseudomonoclonal bands cannot be attributed entirely to the effect of limiting dilutions of polyclonal B cells. Based on the estimate of 6 pg of DNA per diploid cell, 23 we calculate that the lowest detection limit of our IgH PCR assay is 10 to 20 B cell genome equivalents. In other words, a limiting dilution down to one B cell will not be detectable by our assay, and hence will not yield a pseudomonoclonal band. An additional contributing factor to the detection of pseudomonoclonal or pseudo-oligoclonal bands is the existence of geographically restricted microclonal B cell populations within the specimen. In this regard, others have shown that non-reproducible pseudomonoclonal bands can also be generated in DNA samples obtained from specimens containing abundant lymphocytic aggregates. 19, 24
We recommend that for reliable assessment of IgH PCR-based clonality studies using DNA obtained from fixed, paraffin-embedded tissue extracted samples, no less than 50 ng of DNA should be used as the starting quantity of template DNA, of which B cells should account for at least 5 ng (10% of the total cells). We also recommend that multiple (at least two) aliquots of a DNA sample be subjected to IgH PCR analysis, and that specimens that produce pseudomonoclonal bands be categorized as indeterminate. Conversely, monoclonality may be scored if multiple aliquots and serial dilutions of a specimen yield one or two discrete and reproducible band(s) on IgH PCR analysis. In rare cases, reproducible pseudomonoclonal bands may be detected in reactive processes, particularly in microdissected material. It is worthwhile in these cases to use sequence-specific methods such as single strand conformational polymorphism analysis, denaturation gradient gel electrophoresis, or direct DNA sequencing to determine the relatedness of IgH PCR products from duplicate aliquots of the same sample. In light of the diagnostic significance of the assignment of monoclonality in a lymphoproliferative process, we believe that it is necessary to use clinical and histopathological parameters in conjunction with the results of molecular studies to ensure diagnostic accuracy.
Address reprint requests to Kojo S. J. Elenitoba-Johnson, M.D., Division of Anatomic Pathology, University of Utah Health Sciences Center, 50 North Medical Drive, Salt Lake City, UT 84132. E-mail: kojo.elenitobaj@path.med.utah.edu.
Footnotes
Performed at ARUP Institute for Clinical and Experimental Pathology, Salt Lake City, UT 84108.
References
- 1.McCarthy KP, Sloane JP, Wiedemann LM: Rapid method for distinguishing clonal from polyclonal B cell populations in surgical biopsy specimens. J Clin Pathol 1990, 43:429-432 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Wan JH, Sykes PJ, Orell SR, Morley AA: Rapid method for detecting monoclonality in B cell lymphoma in lymph node aspirates using the polymerase chain reaction. J Clin Pathol 1992, 45:420-423 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lehman CM, Sarago C, Nasim S, Comerford J, Karcher DS, Garrett CT: Comparison of PCR with southern hybridization for the routine detection of immunoglobulin heavy chain gene rearrangements. Am J Clin Pathol 1995, 103:171-176 [DOI] [PubMed] [Google Scholar]
- 4.Trainor KJ, Brisco MJ, Story CJ, Morley AA: Monoclonality in B-lymphoproliferative disorders detected at the DNA level. Blood 1990, 75:2220-2222 [PubMed] [Google Scholar]
- 5.Reed TJ, Reid A, Wallberg K, O’Leary TJ, Frizzera G: Determination of B-cell clonality in paraffin-embedded lymph nodes using the polymerase chain reaction. Diagn Mol Pathol 1993, 2:42-49 [PubMed] [Google Scholar]
- 6.Wan JH, Trainor KJ, Brisco MJ, Morley AA: Monoclonality in B cell lymphoma detected in paraffin wax embedded sections using the polymerase chain reaction. J Clin Pathol 1990, 43:888-890 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Medeiros L, Carr J: Overview of the role of molecular methods in the diagnosis of malignant lymphomas. Arch Pathol Lab Med 1999, 123:1189-1207 [DOI] [PubMed] [Google Scholar]
- 8.Wotherspoon AC, Doglioni C, Diss TC, Pan L, Moschini A, de Boni M, Isaacson PG: Regression of primary low-grade B-cell gastric lymphoma of mucosa- associated lymphoid tissue type after eradication of Helicobacter pylori. Lancet 1993, 342:575-577 [DOI] [PubMed] [Google Scholar]
- 9.Fend F, Quintanilla-Martinez L, Kumar S, Beaty MW, Blum L, Sorbara L, Jaffe ES, Raffeld M: Composite low grade B-cell lymphomas with two immunophenotypically distinct cell populations are true biclonal lymphomas: a molecular analysis using laser capture microdissection. Am J Pathol 1999, 154:1857-1866 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Forsthoefel KF, Papp AC, Snyder PJ, Prior TW: Optimization of DNA extraction from formalin-fixed tissue and its clinical application in Duchenne muscular dystrophy. Am J Clin Pathol 1992, 98:98-104 [DOI] [PubMed] [Google Scholar]
- 11.Lombardo JF, Hwang TS, Maiese RL, Millson A, Segal GH: Optimal primer selection for clonality assessment by polymerase chain reaction analysis. III. Intermediate and high-grade B-cell neoplasms. Hum Pathol 1996, 27:373-380 [DOI] [PubMed] [Google Scholar]
- 12.Cossman J, Uppenkamp M, Sundeen J, Coupland R, Raffeld M: Molecular genetics and the diagnosis of lymphoma. Arch Pathol Lab Med 1988, 112:117-127 [PubMed] [Google Scholar]
- 13.Calvert RJ, Evans PA, Randerson JA, Jack AS, Morgan GJ, Dixon MF: The significance of B-cell clonality in gastric lymphoid infiltrates. J Pathol 1996, 180:26-32 [DOI] [PubMed] [Google Scholar]
- 14.Sukpanichnant S, Vnencak-Jones CL, McCurley TL: Determination of B-cell clonality in paraffin-embedded endoscopic biopsy specimens of abnormal lymphocytic infiltrates and gastrointestinal lymphoma by polymerase chain reaction. Am J Clin Pathol 1994, 102:299-305 [DOI] [PubMed] [Google Scholar]
- 15.Pan LX, Diss TC, Peng HZ, Isaacson PG: Clonality analysis of defined B-cell populations in archival tissue sections using microdissection and the polymerase chain reaction. Histopathology 1994, 24:323-327 [DOI] [PubMed] [Google Scholar]
- 16.Kurosu K, Yumoto N, Mikata A, Taniguchi M, Kuriyama T: Monoclonality of B-cell lineage in primary pulmonary lymphoma demonstrated by immunoglobulin heavy chain gene sequence analysis of histologically non-definitive transbronchial biopsy specimens. J Pathol 1996, 178:316-322 [DOI] [PubMed] [Google Scholar]
- 17.Alkan S, Lehman C, Sarago C, Sidawy MK, Karcher DS, Garrett CT: Polymerase chain reaction detection of immunoglobulin gene rearrangement and bcl-2 translocation in archival glass slides of cytologic material. Diagn Mol Pathol 1995, 4:25-31 [DOI] [PubMed] [Google Scholar]
- 18.Zhou X-G, Sandvej K, Gregersen N, Hamilton-Dutoit SJ: Detection of clonal B cells in microdissected reactive lymphoproliferation: possible diagnostic pitfalls in PCR analysis of immunoglobulin heavy chain gene rearrangement. Mol Pathol 1999, 52:104-110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Torlakovic E, Cherwitz DL, Jessurun J, Scholes J, McGlennen R: B-cell gene rearrangement in benign and malignant lymphoid proliferations of mucosa-associated lymphoid tissue and lymph nodes. Hum Pathol 1997, 28:166-173 [DOI] [PubMed] [Google Scholar]
- 20.de Mascarel A, Dubus P, Belleannee G, Megraud F, Merlio JP: Low prevalence of monoclonal B cells in Helicobacter pylori gastritis patients with duodenal ulcer. Hum Pathol 1998, 29:784-790 [DOI] [PubMed] [Google Scholar]
- 21.Taylor JM, Spagnolo DV, Kay PH: B-cell target DNA quantity is a critical factor in the interpretation of B-cell clonality by PCR. Pathology 1997, 29:309-312 [DOI] [PubMed] [Google Scholar]
- 22.Ling FC, Clarke CE, Lillicrap D: Positive immunoglobulin gene rearrangement study by the polymerase chain reaction in a colonic adenocarcinoma. Am J Clin Pathol 1992, 98:116-119 [DOI] [PubMed] [Google Scholar]
- 23.Jeffreys AJ, Wilson V, Neumann R, Keyte J: Amplification of human minisatellites by the polymerase chain reaction: towards DNA fingerprinting of single cells. Nucleic Acids Res 1988, 16:10953-10971 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Saxena A, Moshynska O, Kanthan R, Bhutani M, Maksymiuk AW, Lukie BE: Distinct B-cell clonal bands in helicobacter pylori gastritis with lymphoid hyperplasia. J Pathol 2000, 190:47-54 [DOI] [PubMed] [Google Scholar]