Abstract
Myxoid liposarcoma (LS), the most common subtype of LS, is known to be characterized by the specific t(12;16) resulting in a TLS-CHOP fusion in almost all cases. We wished to address the following questions: (i) Is this genetic hallmark also present in other types of LS with predominant myxoid change? (ii) What is the proportion of cases with the variant EWS-CHOP fusion? (iii) What is the optimal approach for Southern blot detection of TLS breakpoints? We identified 59 LS characterized histologically by >90% myxoid component, in which frozen tissue tumor was available for DNA extraction. These 59 LS with myxoid features were divided into 2 groups: 42 LS with classic myxoid/round cell appearance (myxoid LS) and 17 well-differentiated LS (WDLS) with a predominant (>90%) myxoid component. Within the myxoid LS group, 29 tumors were low grade and 13 high grade (>20% round cell component). Among the 17 predominantly myxoid WDLS, there were 15 low grade and 2 focally high grade tumors. In addition, we selected as control group, 20 LS of other histological types with minimal or no myxoid change (17 WDLS and 3 pleomorphic LS) and 13 myxofibrosarcomas. Southern blot analysis was performed in all cases using a CHOP cDNA probe, and in all CHOP rearranged cases using a TLS cDNA probe. Probe/enzyme combinations for Southern blot analysis were CHOP exon 3–4 cDNA probe with BamHI or SacI, TLS exon 3–6 cDNA probe with BclI. All 42 cases of myxoid LS showed a CHOP rearrangement and 38 of them also had a TLS rearrangement. Among the 4 myxoid LS without Southern blot evidence of TLS rearrangement, 1 showed an EWS-CHOP fusion by Southern blotting and reverse transcriptase-polymerase chain reaction and in another case, reverse transcriptase-polymerase chain reaction detected a TLS-CHOP fusion transcript. None of the predominantly myxoid WDLS and none of the tumors included in the control group showed rearranegements with CHOP probe. In addition, 12 predominantly myxoid WDLS, 10 other LS, and 5 myxofibrosarcoma from the control group were also tested for TLS rearrangement; all were negative. The TLS-CHOP fusion is highly sensitive and specific for the entity of classic myxoid/round cell LS. Other types of LS, even with a predominant myxoid component, lack the TLS-CHOP rearrangement, confirming that they represent a genetically distinct group of LS. The prevalence of the EWS-CHOP variant fusion was approximately 2% in this series. The optimal enzyme for TLS genomic breakpoint detection is BclI.
Myxoid liposarcoma (LS) is the most common subtype of LS and occurs predominantly in the extremities 1, 2 A subset of cases show histological progression to round cell histology, which is associated with a significantly poorer prognosis. 2 The karyotypic hallmark of myxoid LS is the t(12;16)(q13;p11), present cytogenetically in >90% of the cases. 3 The translocation leads to the fusion of the CHOP and TLS genes at 12q13 and 16p11, respectively, and the generation of a TLS-CHOP hybrid protein. 4, 5, 6 In 4 cases of myxoid LS, a variant chromosomal translocation has been described, t(12;22), in which CHOP fuses instead with EWS, a gene highly related to TLS. 7, 8
Different cytogenetic features and their underlying molecular alterations define distinct entities among LS. A strong and specific association of the t(12;16) with myxoid LS has been confirmed by most cytogenetic and molecular analyses. 4, 9, 10, 11, 12, 13 The same translocation is present in pure round cell LS and combined myxoid and round cell LS, confirming the biological continuum between these two forms of LS, proposed on histopathological grounds. 9, 11, 12, 13 In contrast, well-differentiated LS (WDLS) and pleomorphic LS contain no specific recurrent translocation. Instead, WDLS are characterized by cytogenetic evidence of gene amplification (giant marker chromosomes, ring chromosomes, double minutes), whereas pleomorphic LS typically show highly complex karyotypes. 10, 14, 15 Nonetheless, a single recent report has suggested that TLS-CHOP fusion transcripts may also be present in pleomorphic LS and WDLS. 16 The existence of a mixed tumor with combined features of myxoid LS and WDLS has been proposed based on cases of LS showing histological features of both. 11 For instance, up to one-third of WDLS show some degree of myxoid change. 15 Furthermore, rare cases of myxoid LS have shown apparent dedifferentiation, a phenomenon typically associated with WDLS. 17 These problematic tumors showing combined features of myxoid LS and WDLS, most often seen in the retroperitoneum, have not been systematically studied for the presence of the t(12;16). Thus the issue of the histological specificity of the TLS-CHOP fusion and the broader question of the biological relationship among different forms of LS have not been completely resolved. Aside from these biological and nosological questions, the differential diagnosis of myxoid LS can be sometimes quite difficult, due to overlapping morphological features with other myxoid neoplasms, myxofibrosarcoma in particular.
In the present study, we sought to address these issues by systematically studying myxoid LS, WDLS, pleomorphic LS, WDLS with extensive myxoid change, and myxofibrosarcomas for the TLS-CHOP rearrangement by Southern blotting. The TLS genomic breaks are clustered in introns 5 and 7, 18, 19 whereas the vast majority of CHOP breakpoints occur in intron 1 or immediately upstream of exon 1; 19 in both genes, the breakpoint regions are relatively small (<10 kb), allowing Southern blot analysis. We have therefore also evaluated the optimal approach for Southern blot detection of TLS breakpoints. As part of this study, we also provide the first estimate of the prevalence of the variant EWS-CHOP fusion in a large group of myxoid LS, a datum germane to the interpretation of TLS-CHOP molecular diagnostic assays in myxoid LS.
Materials and Methods
Study Group and Histopathological Data
We identified 59 LS characterized histologically by >90% myxoid areas (including areas of round cell change), operated at Memorial Sloan-Kettering Cancer Center (MSKCC) between 1987 and 1998, in which frozen tumor was available for DNA extraction. Patient materials were procured under MSKCC protocol 90–049 approved by our Institutional Review Board. The 59 LS with myxoid features were divided into two groups: 42 LS with classic myxoid/round cell appearance (myxoid LS) and 17 well-differentiated LS with a predominant (>90%) myxoid change (predominantly myxoid WDLS). Within the myxoid LS group, 28 tumors arose in the extremities, 4 in the pelvic girdle, and 3 in the retroperitoneum, and 7 were multifocal. Histologically, 29 of the myxoid LS were low grade (Figure 1A) and 13 were high grade (defined as >20% round cell component). The predominantly myxoid WDLS tumors were located in the following axial sites: retroperitoneum (13 cases), pelvis (2 cases), intraabdominal (1 case), and mediastinum (1 case). No cases of this histological type presented in the extremities. The majority of predominantly myxoid WDLS were histologically low grade (15 cases), showing focal areas (by the study criteria <10%) of lipoma-like or sclerosing LS, diagnostic of WDLS (Figure 1B) . In the remaining 2 cases of predominantly myxoid WDLS areas of dedifferentiation into a high grade spindle cell sarcoma were intermixed with the myxoid component.
Figure 1.
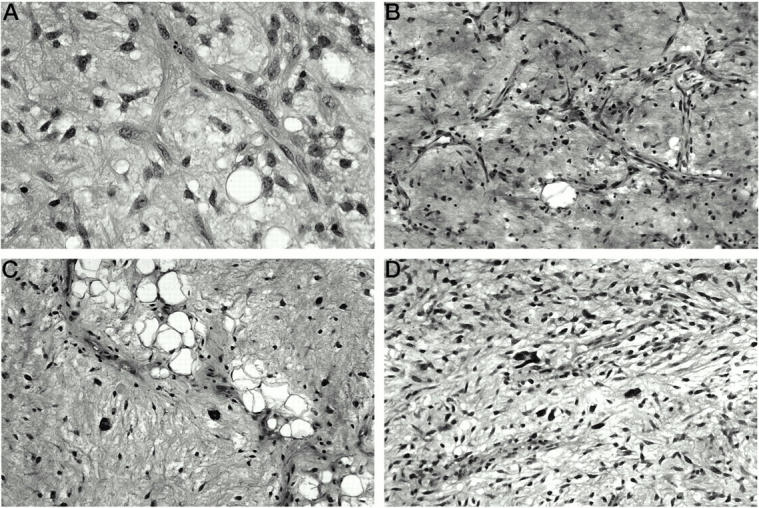
A: Classic myxoid LS, low grade, with small, uniform tumor cells in a background of myxoid stroma and delicate “chicken-wire” type vascular network. Of note is the absence of the pleomorphic giant tumor cells characteristic of WDLS with myxoid changes. B: WDLS, predominantly myxoid. Although the myxoid background and the branching vasculature might suggest the diagnosis of classic myxoid LS, the tumor cells are larger and less uniform. C: WDLS, predominantly myxoid, with focal areas of lipoma-like LS and scattered tumor giant cells. D: Myxofibrosarcoma, low grade. Most of the tumor cells are uniform, but predominantly spindly, in contrast with the round to oval appearance of myxoid LS. Rare pleomorphic tumor cells are scattered within the myxoid stroma, which helps in distinguishing this tumor from classic myxoid LS.
In addition, we selected a control group of 20 LS of other histological types (17 WDLS and 3 pleomorphic LS) and 13 myxofibrosarcomas for which adequate frozen tumor was available for molecular analysis. The 17 cases of WDLS included in the control group showed lesser or absent myxoid change, as follows: 6 cases with 25 to 50% myxoid change, 5 cases with <25%, and 6 cases with no myxoid change (Figure 1C) . Of these 17 WDLS, 13 were uniformly histologically low grade and 4 were focally high grade, and most were located in the retroperitoneum (15 cases). The other 2 WDLS were in the lower extremity. In addition to the WDLS, 3 cases of high grade pleomorphic LS of the extremities were also available for molecular analysis.
As the main differential diagnosis of myxoid LS in the extremities is with myxofibrosarcoma, we also included 13 cases of this tumor type in the control group (Figure 1D) to assess whether some cases in this morphological category are genetically related to myxoid LS. There were 5 low grade and 8 high grade myxofibrosarcomas, of which 10 arose in the lower extremity and 3 in the shoulder/axillary area.
Southern Blot Analysis
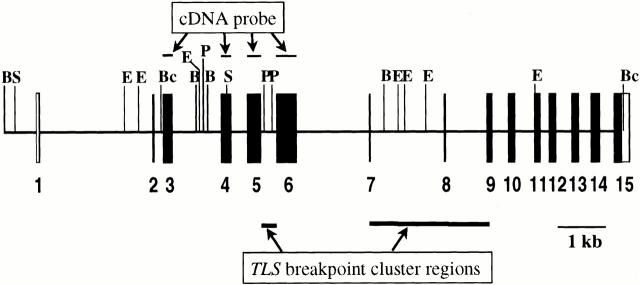
DNA was isolated from snap-frozen tumor tissue stored at −70°C using a standard organic extraction protocol. Southern blot analysis was performed on genomic DNA, digested in all cases with at least two restriction enzymes, separated by 0.7% agarose gel electrophoresis, transferred onto nylon membranes, and hybridized with radiolabeled probes. The CHOP and TLS probes were partial cDNA clones, derived from a full length TLS-CHOP cDNA (LPS41; gift of David Ron, New York University Medical Center, New York, NY). 5 The CHOP probe consisted of a 753-bp PstI-XhoI fragment corresponding to exons 3 and 4. This probe is expected to detect all CHOP rearrangements in genomic DNA digested with BamHI or SacI. 4, 20 Extra bands, probably cross-hybridizing bands, were occasionally observed in samples digested with SacI and probed with the CHOP probe, and migrated at 5.5 and 3.5 kb. The TLS cDNA probe was a 780 bp XbaI-BglII fragment including exons 1 to 6 of TLS. According to the restriction enzyme site analysis of the complete genomic sequence of TLS (GenBank no. AF071213), this probe covers the entire TLS breakpoint region in BclI-digested DNA (Figure 2) . Although the TLS cDNA fragment used as a probe included exons 1 and 2, these span only 90 and 25 bases, respectively, and are therefore too short to hybridize reliably with the corresponding restriction fragments (as confirmed by the lack of these germline fragments on Southern blots). The TLS probe used was thus effectively equivalent to an exon 3 to 6 cDNA fragment, as depicted in Figure 2 . In certain cases, mainly where the primary restriction enzymes were uninformative, other enzymes were used, such as EcoRI and HindIII for CHOP and PstI and BamHI for TLS cDNA probes. No polymorphisms were observed with the TLS probe, but it was associated with a background of multiple weak cross-hybridizing bands in certain samples. The EWS probe was a previously described 741-bp partial cDNA probe which hybridizes to exons 6 to 12 of EWS, covering in EcoRI and HindIII-digested DNA the entire genomic breakpoint cluster region. 21
Figure 2.
TLS genomic map with selected restriction enzyme sites (B, BamHI; E, EcoRI; Bc, BclI; P, PstI; S, SacI). The map is based on the complete genomic sequence of TLS (GenBank no. AF071213 17 ). Exons encoding translated sequences are shown as filled boxes, and the 3′ untranslated portion of exon 15 is shown as an empty box. The TLS breakpoint cluster regions are based on references 18 and 19. The TLS probe is described in the text (see Methods). Partial restriction maps of the CHOP gene have been previously published. 4 6 12 20
Results
Classic Myxoid LS Group
Among 59 LS with >90% myxoid histology, 42 cases were classified as classic myxoid LS. All of these 42 myxoid LS showed a CHOP rearrangement, and in 38 of them (90%) TLS rearrangement was also demonstrated (Table 1) . Regarding the CHOP analysis, in 35 cases (83%) the CHOP rearrangement was detected with both BamHI and SacI (Figure 3) , in 3 cases only with BamHI (germline pattern with SacI), and in 3 cases only with SacI (germline pattern in with BamHI in 1 case and inadequate DNA for BamHI digest in 2 cases). Finally, one case that was germline with BamHI and had inadequate DNA for SacI had shown rearrangements previously with two other restriction enzymes (EcoRI and HindIII). Regarding the detection of TLS rearrangements, 32 of 37 cases (86%) studied with BclI digests were positive (Figure 4) . In 6 additional cases, TLS rearrangement was detected with other restriction enzymes, including EcoRI, BamHI, and PstI (see Table 1 ). Among the 4/42 cases that were not rearranged with TLS probe, 1 case could not be further tested due to degraded DNA, while 3 had sufficient DNA for further Southern blot analysis which showed a germline pattern with 4 additional restriction enzymes (BamHI, SacI, EcoRI, HindIII). Unfortunately, karyotypic data were not available in any of these 4 TLS-germline cases. One of the latter was tested for TLS-CHOP by reverse transcriptase-polymerase chain reaction (RT-PCR) and found to have the relatively uncommon fusion of TLS exon 8 to CHOP exon 2 (results not shown). The remaining 2 cases of myxoid LS germline for TLS in 5 different restriction digests were tested for EWS rearrangement. One case showed a rearranged EWS band in HindIII-digested tumor DNA. RT-PCR analysis confirmed the presence of the EWS-CHOP fusion in this case, which is presented in more detail elsewhere. 22 This left one TLS-germline CHOP-rearranged case in which the putative CHOP translocation could not be further defined at the molecular level.
Table 1.
Summary of Results
| Histologic group | CHOP | TLS | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Overall result | Results by restriction digest | Overall result | Results by restriction digest | |||||||
| BamHI and SacI digests* | BamHI | SacI | HindIII | EcoRI | BclI | BamHI | PstI | |||
| Myxoid LS (n = 42) | 42 R | 35 R | 38 R | 38 R | 1 R | 1 R | 38/42 R | 32 R | 3 R | 1 R |
| 2 G | 3 G | 5 G | ||||||||
| 2 F | 1 F | 2 F | ||||||||
| Predominantly myxoid WDLS (n = 17) | 17 G | 17 G | 17 G | 17 G | 6 G | 3 G | 12 G | 10 G | 2 G | 2 G |
| WDLS with minor or no myxoid areas (n = 17) | 17 G | 14 G | 17 G | 14 G | 8 G | 4 G | 9 G | 2 G | 6 G | 2 G |
| 1 F | ||||||||||
| Pleomorphic LS (n = 3) | 3 G | 3 G | 3 G | 3 G | 2 G | ND | 1 G | 1 G | ND | ND |
| Myxofibrosarcoma (n = 13) | 13 G | 12 G | 13 G | 12 G | 7 G | 4 G | 5 G | 4 G | ND | 1 G |
LS, liposarcoma; WDLS, well-differentiated LS; R, rearranged; G, germline; F, failure; ND, not done.
Except for this column, which shows the number of cases with identical results for both enzymes, in cells where the number of cases is less than n, the remaining cases were not studied with the enzyme in question, unless otherwise specified.
Includes 2 cases that showed TLS rearrangement in EcoRI digests only; for details on 4 cases without detectable TLS rearrangement, see Results.
Figure 3.
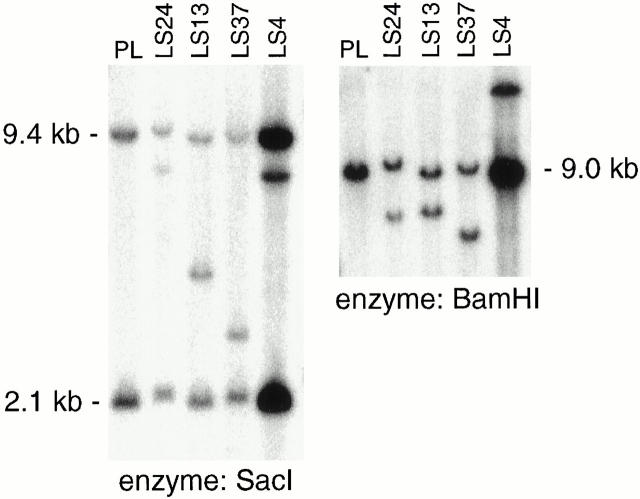
CHOP rearrangement analysis by Southern blotting. Rearranged bands are seen in both SacI- and BamHI-digested genomic DNA in four cases of classic myxoid LS (LS4, LS13, LS24, LS37) using a CHOP exon 3 and 4 cDNA probe (see Methods).
Figure 4.
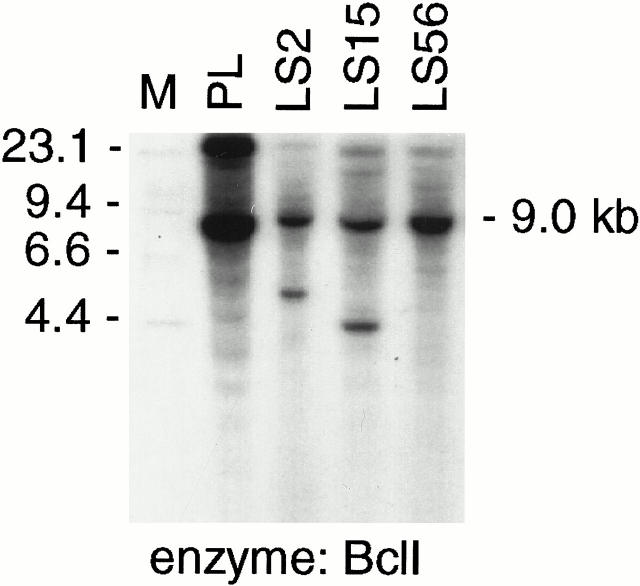
TLS rearrangement analysis by Southern blotting. Rearranged bands are seen in BclI-digested genomic DNA in two cases of classic myxoid LS (LS2 and LS15), but not in a case of predominantly myxoid WDLS (LS56) using a TLS exon 3–6 cDNA probe (see Methods and Figure 2 ).
Predominantly Myxoid WDLS Group
Among the WDLS with >90% myxoid histology, all of the cases tested had a germline pattern with CHOP probe by both BamHI and SacI digests. In addition, 12 of 17 cases were also tested for TLS rearrangements and showed a germline profile: 8 by BclI digest only, 2 by both BclI and PstI, and 2 by BamHI digest only.
Other WDLS and Pleomorphic LS
The other lipomatous tumors composing the control group, including LS with variable myxoid component (25 to 50%) or with absent or focal myxoid change had a germline pattern with CHOP probe: 14/17 cases with both BamHI and SacI and 3/17 cases only with BamHI (technical failure with SacI digest). In 9 cases, TLS was also studied and was not rearranged in any case. All 3 pleomorphic LS had a germline pattern for CHOP using both BamHI and SacI, and in 2/3 also with HindIII. Southern blot analysis with the TLS cDNA probe was performed in 1 of the 3 pleomorphic LS and showed a germline pattern in BclI-digested DNA.
Myxofibrosarcomas
All myxofibrosarcomas had a germline profile with CHOP (12/13 cases with both BamHI and SacI, and 1/13 with BamHI, EcoRI, and HindIII). Five cases were also tested for TLS and showed no genomic rearrangement.
Discussion
The CHOP gene encodes a member of leucine zipper transcription factor family implicated in adipocyte differentiation and growth arrest. 23, 24 In myxoid LS, most of the CHOP gene is fused to the 5′ portion of TLS (for translocated in liposarcoma), 6 also known as FUS (for fusion). 5 The fusion gene encodes a protein that consists of the amino terminus of TLS fused to the full-length CHOP-coding region. The oncogenic effect of TLS-CHOP may be mediated at least in part by inhibition of pre-adipocyte differentiation. 25, 26, 27
The TLS genomic breaks are clustered in introns 5 and 7 18 (Figure 2) , 19 whereas the vast majority of CHOP breakpoints occur in intron 1 or immediately upstream of exon 1. 19, 20 In the cases where the CHOP genomic break is upstream of exon 1, the latter is systematically spliced out of the mRNA, presumably because it would result in loss of the reading frame. By RT-PCR, approximately two-thirds of cases contain a hybrid transcript in which TLS exon 5 is fused to CHOP exon 2 (type II transcript), whereas in about one-third, the TLS-CHOP transcript also contains exons 6 and 7 of TLS (type I transcript). 12, 13, 20
The extensive homology between TLS and EWS suggest that the two genes are closely related and may have originated from a common ancestor gene. 28 Thus, it is not entirely surprising that in rare cases of myxoid LS, the EWS gene at 22q12 is an alternative translocation partner of CHOP. 7, 8 It has been previously difficult to estimate the prevalence of the EWS-CHOP fusion in myxoid LS. In our study approximately 2% (1 of 42) of myxoid LS contain EWS-CHOP instead of TLS-CHOP. In a smaller previous study (n = 20), EWS-CHOP appeared to make up 5% of myxoid LS. 29
The distinction between myxoid LS and other subtypes of myxoid sarcomas is important in predicting the biological behavior and, hence, planning the management of these patients. More than other types of LS or other myxoid sarcomas of the extremities, myxoid LS are prone to metastasize to soft tissue locations, such as retroperitoneum, opposite extremity, and axilla. 2, 30 Furthermore, myxoid LS is the predominant histological type among soft tissue sarcomas with multifocal presentation. 31 In a significant number of cases of myxoid LS, about 10 to 20%, patients present clinically with either synchronous or metachronous multifocal tumors. 22 In a recent study addressing the question of clonality by molecular analysis in six patients who presented with either synchronous or metachronous multifocal myxoid LS, we confirmed the monoclonal origin of these multifocal tumors, establishing the metastatic nature of distant soft tissue lesions in these cases. 22
The characterization of the specificity and sensitivity of genomic rearrangements of TLS and CHOP genes for myxoid LS is needed to apply this test in the routine diagnostic work-up of myxoid tumors. Our results support the complete specificity and high sensitivity of CHOP rearrangement, due either to the TLS-CHOP or EWS-CHOP fusion, for the entity of myxoid/round cell LS. The sensitivity appears limited only by technical factors such as the quantity and quality of genomic DNA available, and the occurrence of rare cases in which detection of the rearrangement may require enzymes other than BamHI and SacI. In terms of specificity, the absence of CHOP or TLS gene rearrangements in 15 cases of myxofibrosarcoma in the present series is in keeping with the findings of Nilbert et al, 32 in which 41 cases of malignant fibrous histiocytoma (MFH), including 13 of myxoid type, had a germline pattern by Southern blot analysis of CHOP.
Recently, Willeke et al 16 reported TLS-CHOP fusion transcripts detected by nested RT-PCR in several cases of WDLS and pleomorphic LS. The TLS-CHOP fusion transcripts detected in these cases were mostly unusual in structure, 16 unlike the common fusions of TLS exon 5 or 7 to CHOP exon 2. These findings are in disparity with several previous RT-PCR studies 9, 12, 13 and the large cytogenetic study reported by the CHAMP group. 11, 14 In the latter study, none of the 264 cases of various types of adipose tumors, other than myxoid LS, exhibited the t(12;16), including 19 cases of atypical lipomatous tumors with myxoid changes. The results of our study confirm the consistency of CHOP and TLS genomic rearrangements in a large group of myxoid LS by Southern blotting. The presence of these molecular alterations appears to be highly specific for the myxoid and round cell variants of LS, and none of the other histological types tested for either CHOP or TLS genes showed genomic rearrangements.
Overwhelming evidence points to the specificity of different cytogenetic changes for particular types of LS. Notably, the t(12;16)(q13.3;p11.2) is present in >95% of myxoid and round cell LS. 9, 14 About 80% of WDLS show ring or marker chromosomes. 14 These are derived from 12q13–15, including amplified most typically, rearranged HMGI-C, and, perhaps secondarily, MDM2, CDK4, and CHOP. Nonetheless, occasional reports of apparent mixed-type LS (WDLS + myxoid LS) and a recent description of 3 cases of myxoid LS with areas of dedifferentiation into a high grade nonlipogenic component resembling MFH (akin to dedifferentiation in WDLS) has led to the suggestion of a closer relationship between these two types of LS than previously accepted. 17 We should note, however, that although dedifferentiation has been associated with a limited number of sarcoma types, it is clearly not sufficiently specific to establish biological relationships between the sarcoma types in which it has been observed. In these studies, no molecular or cytogenetic data were available.
Some cytogenetic reports have, however, seemed to contradict the clear distinction between myxoid LS and WDLS. Complex karyotypes, including ring and/or marker chromosomes, have been described in three cases of apparent mixed myxoid and well-differentiated LS. 33, 14 In a different cytogenetic study including 31 LS samples, tumors from three of nine patients with a histological diagnosis of myxoid LS lacked the t(12;16). 10 In one of these patients, the tumor samples had a cytogenetic phenotype resembling that of WDLS with telomeric associations, large markers, and ring chromosomes, whereas in the other two cases, the karyotype was more akin to pleomorphic LS. 10 We believe that in many, if not all, of these instances of mixed myxoid + well-differentiated LS 14, 33 or translocation-negative myxoid LS, 10 the tumors analyzed may have represented predominantly myxoid WDLS or pleomorphic LS with myxoid change, as supported by the cytogenetic data in the respective reports. The presence of microscopic foci of lipoma-like or sclerosing areas, characteristic of WDLS, constitutes sufficient histological evidence, in our opinion, to exclude the diagnosis of myxoid LS, as supported by our molecular analysis showing the consistent absence of CHOP or TLS genomic rearrangements in such tumors. The present results thus reinforce the concept that myxoid LS and WDLS are distinct entities and suggest that most cases that seem to contradict this sharp distinction might be due to incomplete histopathological or molecular/cytogenetic analysis. Biologically, it is of some interest to speculate on the possible basis for the preferential occurrence of the TLS-CHOP rearrangement in extremities, compared to retroperitoneum.
Acknowledgments
We thank Debbie MacDougall for assistance with artwork and Kin Kong and Allyne Manzo for photographic work.
Address reprint requests to Marc Ladanyi, M.D., Department of Pathology, Memorial Sloan-Kettering Cancer Center, 1275 York Avenue, New York, NY 10021. E-mail: ladanyim@mskcc.org.
Footnotes
Supported in part by grant CA47179 (to M. F. B.) from the National Institutes of Health. M. H. was supported through the Student Research Program of the Society for Pediatric Research and the American Pediatric Society.
References
- 1.Enzinger FM, Weiss SW: Liposarcoma (chapter 17). Enzinger FM Weiss SW eds. Soft Tissue Tumors 3rd ed. 1995:431-466 Mosby St. Louis, MO
- 2.Kilpatrick SE, Doyon J, Choong PF, Sim FH, Nascimento AG: The clinicopathologic spectrum of myxoid and round cell liposarcoma: a study of 95 cases. Cancer 1996, 77:1450-1458 [DOI] [PubMed] [Google Scholar]
- 3.Heim S, Mitelman F: Cancer Cytogenetics 2nd ed. 1995:484-489 Wiley-Liss New York
- 4.Aman P, Ron D, Mandahl N, Fioretos T, Heim S, Arheden K, Willen H, Rydholm A, Mitelman F: Rearrangement of the transcription factor gene CHOP in myxoid liposarcomas with t(12;16)(q13;p11). Genes Chromosomes Cancer 1992, 5:278-285 [DOI] [PubMed] [Google Scholar]
- 5.Crozat A, Aman P, Mandahl N, Ron D: Fusion of CHOP to a novel RNA-binding protein in human myxoid liposarcoma. Nature 1993, 363:640-644 [DOI] [PubMed] [Google Scholar]
- 6.Rabbitts TH, Forster A, Larson R, Nathan P: Fusion of the dominant negative transcription regulator CHOP with a novel gene FUS by translocation t(12;16) in malignant liposarcoma. Nat Genet 1993, 4:175-180 [DOI] [PubMed] [Google Scholar]
- 7.Panagopoulos I, Hoglund M, Mertens F, Mandahl N, Mitelman F, Aman P: Fusion of the EWS and CHOP genes in myxoid liposarcoma. Oncogene 1996, 12:489-494 [PubMed] [Google Scholar]
- 8.Dal Cin P, Sciot R, Panagopoulos I, Aman P, Samson I, Mandahl N, Mitelman F, Van Den Berghe H, Fletcher CD: Additional evidence of a variant translocation t(12;22) with EWS/CHOP fusion in myxoid liposarcoma: clinicopathological features J Pathol 1997, 182:437-4419306965 [Google Scholar]
- 9.Knight JC, Renwick PJ, Dal Cin P, Van Den Berghe H, Fletcher CDM: Translocation t(12;16)(q13;p11) in myxoid liposarcoma and round cell liposarcoma: molecular and cytogenetic analysis Cancer Res 1995, 55:24-27 [PubMed] [Google Scholar]
- 10.Sreekantaiah C, Karakousis CP, Leong SP, Sandberg AA: Cytogenetic findings in liposarcoma correlate with histopathologic subtypes. Cancer 1992, 69:2484-2495 [DOI] [PubMed] [Google Scholar]
- 11.Tallini G, Akerman M, Dal Cin P, de Wever I, Fletcher CD, Mandahl N, Mertens F, Mitelman F, Rosai J, Rydholm A, Sciot R, Van Den Berghe H, Van den Ven W, Vanni R, Willen H: Combined morphologic and karyotypic study of 28 myxoid liposarcomas: implications for a revised morphologic typing (a report from the CHAMP Group). Am J Surg Pathol 1996, 20:1047-1055 [DOI] [PubMed] [Google Scholar]
- 12.Kuroda M, Ishida T, Horiuchi H, Kida N, Uozaki H, Takeuchi H, Tsuji K, Imamura T, Mori S, Machinami R, Watanabe T: Chimeric TLS/FUS-CHOP gene expression and the heterogeneity of its junction in human myxoid and round cell liposarcoma. Am J Pathol 1995, 147:1221-1227 [PMC free article] [PubMed] [Google Scholar]
- 13.Hisaoka M, Tseiji S, Morimitsu Y, Hashimoto H, Shimajiri S, Komiya S, Ushijima M: Detection of TLS/FUS-CHOP fusion transcripts in myxoid and round cell liposarcomas by nested reverse transcription-polymerase chain reaction using archival paraffin-embedded tissues. Diagn Mol Pathol 1998, 7:96-101 [DOI] [PubMed] [Google Scholar]
- 14.Fletcher CD, Akerman M, Dal Cin P, de Wever I, Mandahl N, Mertens F, Mitelman , Rosai J, Rydholm A, Sciot R, Tallini G, Van Den Berghe H, Van de Ven W, Vanni R, Willen H: Correlation between clinicopathological features and karyotype in lipomatous tumors: a report of 178 cases from the Chromosomes and Morphology (CHAMP) Collaborative Study Group Am J Pathol 1996, 148:623-630 [PMC free article] [PubMed] [Google Scholar]
- 15.Rosai J, Akerman M, Dal Cin P, de Wever I, Fletcher CD, Mandahl N, Mertens F, Mitelman F, Rydholm A, Sciot R, Tallini G, Van Den Berghe H, Van de Ven W, Vanni R, Willen H: Combined morphologic and karyotypic study of 59 atypical lipomatous tumors: evaluation of their relationship and differential diagnosis with other adipose tissue tumors (a report of the CHAMP Study Group) Am J Surg Pathol 1996, 20:1182-1189 [DOI] [PubMed] [Google Scholar]
- 16.Willeke F, Ridder R, Mechtersheimer G, Schwarzbach M, Duwe A, Weitz J, Lehnert T, Herfarth C, von Knebel D: Analysis of FUS-CHOP fusion transcripts in different types of soft tissue liposarcoma and their diagnostic implications. Clin Cancer Res 1998, 4:1779-1784 [PubMed] [Google Scholar]
- 17.Mentzel T, Fletcher CD: Dedifferentiated myxoid liposarcoma: a clinicopathological study suggesting a closer relationship between myxoid and well-differentiated liposarcoma. Histopathology 1997, 30:457-463 [DOI] [PubMed] [Google Scholar]
- 18.Panagopoulos I, Mandahl N, Mitelman F, Aman P: Two distinct FUS breakpoint clusters in myxoid liposarcoma and acute myeloid leukemia with the translocations t(12;16) and t(16;21). Oncogene 1995, 11:1133-1137 [PubMed] [Google Scholar]
- 19.Kanoe H, Nakayama T, Hosaka T, Murakami H, Yamamoto H, Nakashima Y, Tsuboyama TNT, Ron D, Sasaki MS, Toguchida J: Characteristics of genomic breakpoints in TLS-CHOP translocations in liposcarcomas suggest the involvement of Translin and topoisomerase II in the process of translocation. Oncogene 1999, 18:721-729 [DOI] [PubMed] [Google Scholar]
- 20.Panagopoulos I, Mandahl N, Ron D, Hoglund M, Nilbert M, Mertens F, Mitelman F, Aman P: Characterization of the CHOP breakpoints and fusion transcripts in myxoid liposarcomas with the 12;16 translocation. Cancer Res 1994, 54:6500-6503 [PubMed] [Google Scholar]
- 21.Ladanyi M, Lewis R, Garin-Chesa P, Rettig WJ, Huvos AG, Healey JH, Jhanwar SC: EWS rearrangement in Ewing’s sarcoma and peripheral neuroectodermal tumor: molecular detection and correlation with cytogenetic analysis and MIC2 expression. Diag Mol Pathol 1993, 2:141-146 [PubMed] [Google Scholar]
- 22.Antonescu CR, Elahi A, Healey JH, Brennan MF, Lui MY, Lewis JJ, Jhanwar SC, Woodruff JM, Ladanyi M: Monoclonality of multifocal myxoid liposarcoma: confirmation by analysis of TLS-CHOP or EWS-CHOP rearrangements Clin Cancer Res press)(in 2000, [PubMed]
- 23.Ron D, Habener JF: CHOP, a novel developmentally regulated nuclear protein that dimerizes with transcription factors C/EBP and LAP and functions as a dominant negative inhibitor of gene transcription. Genes Dev 1992, 6:439-453 [DOI] [PubMed] [Google Scholar]
- 24.Butterwith SC: Molecular events in adipocyte development. Pharmacol Ther 1994, 61:399-411 [DOI] [PubMed] [Google Scholar]
- 25.Kuroda M, Ishida T, Takanashi M, Satoh M, Machinami R, Watanabe T: Oncogenic transformation and inhibition of adipocytic conversion of preadipocytes by TLS/FUS-CHOP type II chimeric protein. Am J Pathol 1997, 151:735-744 [PMC free article] [PubMed] [Google Scholar]
- 26.Adelmant G, Gilbert JD, Freytag SO: Human translocation liposarcoma-CCAAT/enhancer binding protein (C/EBP) homologous protein (TLS-CHOP) oncoprotein prevents adipocyte differentiation by directly interfering with C/EBPbeta function. J Biol Chem 1998, 273:15574-15581 [DOI] [PubMed] [Google Scholar]
- 27.Ron D: TLS-CHOP and the role of RNA-binding proteins in oncogenic transformation. Curr Top Microbiol Immunol 1997, 220:131-142 [DOI] [PubMed] [Google Scholar]
- 28.Aman P, Panagopoulos I, Lassen C, Fioretos T, Mencinger M, Toresson H, Hoglund M, Forster A, Rabbitts TH, Ron D, Mandahl N, Mitelman F: Expression patterns of the human sarcoma-associated genes FUS and EWS and the genomic structure of FUS. Genomics 1996, 37:1-8 [DOI] [PubMed] [Google Scholar]
- 29.Panagopoulos I, Aman P, Mertens F, Mandahl N, Rydholm A, Bauer HF, Mitelman F: Genomic PCR detects tumor cells in peripheral blood from patients with myxoid liposarcoma. Genes Chromosomes Cancer 1996, 17:102-107 [DOI] [PubMed] [Google Scholar]
- 30.Gustafson P, Rydholm A, Willen H, Baldetorp B, Ferno M, Akerman M: Liposarcoma: a population-based epidemiologic and prognostic study of features of 43 patients, including tumor DNA content. Int J Cancer 1993, 55:541-546 [DOI] [PubMed] [Google Scholar]
- 31.Blair SL, Lewis JJ, Leung D, Woodruff J, Brennan MF: Multifocal extremity sarcoma: an uncommon and controversial entity. Ann Surg Oncol 1998, 5:37-40 [DOI] [PubMed] [Google Scholar]
- 32.Nilbert M, Mandahl N, Aman P, Rydholm A, Mitelman F: No rearrangements of the CHOP gene in malignant fibrous histiocytoma. Cancer Genet Cytogenet 1994, 72:155-156 [DOI] [PubMed] [Google Scholar]
- 33.Dal Cin P, Kools P, Sciot R, de Wever I, Van Damme B, Van de Ven W, Van Den Berghe H: Cytogenetic and fluorescence in situ hybridization investigation of ring chromosomes characterizing a specific pathologic subgroup of adipose tissue tumors Cancer Genet Cytogenet 1993, 68:85-90 [DOI] [PubMed] [Google Scholar]