Abstract
Human herpesviruses can cause significant morbidity and mortality in pediatric solid organ transplant recipients. It was hypothesized that viral burden quantification by polymerase chain reaction using an internal calibration standard could aid in distinguishing between viral disease and latency. Here we report the results of a 2-year prospective study of 27 pediatric solid organ (liver, kidney, or heart) transplant recipients in which multiple samples were analyzed for levels of all eight human herpesviruses by internal calibration standard-polymerase chain reaction. Herpes simplex viruses 1 and 2, varicella-zoster virus, and Kaposi’s sarcoma-associated herpesvirus were not detected in any of these samples. Human herpesvirus types 6 and 7 were detected in half of the patients, but were present at low levels, similar to those found in reference populations. Epstein-Barr virus (EBV) and cytomegalovirus (CMV) were detected in 89% and 56% of the patients, respectively. Viral burden analysis suggested distinct patient populations for CMV, with a natural cutoff of 10,000 viral targets/ml blood strongly associated with disease. In some cases, a dramatic increase in CMV levels preceded clinical evidence of disease by several weeks. EBV viral burden was relatively high in the only patient presenting with an EBV syndrome. However, two other patients without evidence of EBV disease had single samples with high EBV burden. Rapid reduction in both EBV and CMV burden occurred with antiviral treatment. These data suggest that viral burden analysis using internal calibration standard-polymerase chain reaction for CMV, and possibly other herpesviruses, is an effective method for monitoring pediatric transplant patients for significant herpesvirus infection and response to therapy.
Transplantation is being used as an effective treatment strategy for the correction of organ defects due to congenital malformation or the cytotoxic effects of chemicals and infectious agents. This therapeutic approach relies on the ability to shape the recipient’s immune system to accept the foreign organ. This has been greatly facilitated by the use of a variety of immunosuppressive drugs, including cyclosporin, FK506, prednisone, and mycophenolate, which suppress the cellular arm of the immune system. However, this approach to immunosuppression is associated with a serious side effect: an increased incidence of life-threatening diseases caused by infectious agents that are normally controlled by the immune systems of immunocompetent individuals. Among the agents that seriously affect immunocompromised individuals are the herpesviruses.
The eight human herpesviruses identified to date—herpes simplex viruses 1 and 2 (HSV1 and HSV2), varicella-zoster virus (VZV), Epstein-Barr virus (EBV), cytomegalovirus (CMV), human herpesvirus types 6 and 7 (HHV6 and HHV7), and Kaposi’s sarcoma-associated herpesvirus (KSHV or HHV8)—have been associated with significant morbidity and mortality in a variety of immunosuppressed patient populations. 1, 2, 3, 4, 5, 6, 7 For solid organ transplant recipients, localized infection can lead to inflammatory responses and tissue destruction in many different target organs, especially lung, liver, and gastrointestinal tract. For example, 13 to 30% of liver transplant recipients will develop pneumonia associated with CMV infection. 8 In many cases, herpesvirus infection targets the transplanted organ and contributes to organ rejection. 9, 10, 11, 12 For example, 17% of liver allograft recipients have been found to develop CMV-mediated hepatitis; in the high-risk subgroup (seronegative recipients with seropositive donors), the incidence of CMV disease approaches 50%. 9, 10, 11 In this case, initial evidence of infection often comes from the detection of elevated levels of liver enzymes in the circulation. Because elevated liver enzymes are also associated with immune-mediated organ rejection, histological evaluation of organ biopsy is often necessary to distinguish between these etiologies. 13 Finally, EBV appears to be unique among the herpesviruses in that it can also stimulate the proliferation of infected lymphocytes, in some cases leading to post-transplant lymphoproliferative disorder (PTLD), with many characteristics similar to malignant non-Hodgkin’s lymphoma. 5, 14, 15, 16, 17
Fortunately, a variety of virus-specific antiviral drugs and treatment approaches has been developed for patients with significant herpesvirus infection. Herpes simplex esophagitis is effectively treated with acyclovir. 18 Ganciclovir in combination with hyperimmune globulin is an effective therapeutic approach for CMV-mediated disease. 8, 19, 20 EBV-associated PTLD appears to be most effectively treated by tapering of the doses of the immunosuppressive drugs used to prevent transplant organ rejection. 17, 21 Because different viruses can give rise to similar organ pathologies, 22, 23, 24, 25, 26, 27, 28, 29 selection of the appropriate therapeutic approach involves accurate diagnosis of disease etiology.
Monitoring transplant recipients for significant herpesvirus infections has proved to be a diagnostic challenge for two reasons. First, the results of serology tests commonly used to diagnose viral infection can be dramatically influenced by the immunosuppressed state of the patient in ways that are not easily predicted. Second, there is a high prevalence of past infection by some of these viruses, which enter a latent state after primary infection, such that most humans are asymptomatic but continue to harbor latent virus. This is especially true for four of these viruses that cause significant problems for the transplant population: EBV, CMV, HHV6, and HHV7. Thus, sensitive techniques like polymerase chain reaction (PCR) to identify viral nucleic acids can often detect viral genomes in plasma and circulating lymphocytes of asymptomatic individuals. For these reasons, serology and standard PCR approaches have been problematic for the diagnosis of acute infections that are clinically relevant in this population. 30, 31, 32, 33
A number of groups have developed PCR-based approaches to quantify the number of specific nucleic acid targets. 34, 35, 36, 37, 38, 39 The approaches that appear to be the most accurate and precise use an internal calibration standard (ICS) that is included at known quantities in each reaction and amplified with the same primers as the viral target; these approaches have been termed ICS-PCR or competitive PCR. In theory, the development of these approaches will allow investigators to test the hypothesis that viral burden analysis in a clinical sample might be able to distinguish positive PCR results that are clinically relevant from those associated with viral latency in asymptomatic patients. Here we present the results of a 2-year prospective study of a cohort of pediatric solid organ transplant recipients in which all eight human herpesviruses were quantified in whole blood using ICS-PCR to determine whether viral burden analysis correlates with disease onset and/or response to antiviral therapy in this patient population.
Materials and Methods
Patient Population
A total of 264 blood samples were collected from 27 pediatric patients receiving a solid organ transplant at Children’s Medical Center (Dallas, TX) between September 1997 and September 1999. These included 16 liver, 6 kidney, and 5 heart transplant recipients. Blood samples were collected monthly while the patients were enrolled in the study. The median number of samples analyzed per patient was 8, with a range of 1 to 26 samples/patient. The median patient age at the time of transplant was 3 years, with a range of 5 months to 19 years. Patients received standard immunosuppressive therapy including cyclosporin or FK506 with or without mycophenolate. Some patients received acycolvir as an antiviral prophylactic agent. Upon diagnosis of a probable acute CMV infection using standard laboratory tests (antigenemia, serology, or histology), patients were usually treated with a combination of ganciclovir and CMV-hyperimmune globulin. None of the results from this study was used for patient management. The study was performed with approval by the local Institutional Review Board.
Reference Population
The reference population analyzed was composed of patients presenting to the Children’s Medical Center emergency room in September, 1999 who were having blood drawn as part of the routine diagnostic workup. Only patients with a normal CBC were included within this reference group. These patients ranged in age from 4 weeks to 17 years, with a median age of 4 years. Of the 51 patients analyzed, 25 had some evidence of acute infection, usually a fever.
Sample Preparation
Three milliliters of blood were collected in a purple-top collection tubes containing EDTA as the anticoagulant and stored at 4°C until processed (usually <24 hours, but never >72 hours). DNA was isolated from 0.2 ml of whole blood using the QIAamp DNA Mini Blood kit (QIAGEN Inc., Valencia, CA) according to the manufacturer’s current protocol.
ICS-PCR Procedure
The design and validation of the PCR procedure used to quantify the number of viral targets using an internal calibration standard, ICS-PCR, has been described in detail elsewhere. 35 Briefly, each PCR reaction mix (50 μl) contained 10 mmol/L Tris-HCl, pH 8.3, 50 mmol/L KCl, 1.5 mmol/L MgCl2, 0.001% gelatin, 200 μmol/L each of dATP, dCTP, and dGTP, 400 μmol/L dUTP, 20 pmoles of each oligonucleotide primer, 20 molecules of the HHVQ-1 ICS standard, 2.5 units of Taq polymerase (PE Applied Biosystems, Foster City, CA), and DNA from 10 μl of whole blood. Separate reaction vessels were prepared with primers specific for each of the eight human herpesviruses. The oligonucleotide primer sequences were published elsewhere 35 and are listed in Table 1 . A master mix was prepared that contained enough reagents for nine reactions without the oligonucleotide primers and then aliquoted into each of the eight PCR reaction tubes containing the virus-specific primers. Complete reactions were then placed in a GeneAmp PCR System 9600 thermocycler (PE Applied Biosystems) and amplified under the following conditions: 1 cycle of 2 minutes at 95°C, 36 cycles of 30 seconds at 94°C, 30 seconds at 65°C, and 1 minute at 72°C, followed by a final extension of 9 minutes at 72°C. After amplification, PCR products were separated by agarose gel electrophoresis, identified by staining with SYBR Gold (Molecular Probes, Eugene, OR) and quantified using a Fluorimager SI (Molecular Dynamics, Sunnyvale, CA).
Table 1.
HHVQ-1 Amplification Primers and PCR Product Sizes
| Virus | Primer set* | Target gene | 5′ Primer | 3′ Primer | Viral product† | Standard product† |
|---|---|---|---|---|---|---|
| HSV1 | A | TK3 | agcgtcttgtcattggcgaa | ttttctgctccaggcggact | 342 | 456 |
| HSV2 | B | pol | cgtcctggagtttgacagcg | cagcagcgagtcctgcacacaa | 445 | 564 |
| VZV | A | gene 71 | cgagtcagcctgacgatcta | tttggagacctagcaagcttcgttc | 304 | 415 |
| EBV | A | EBER | cccgcctacacaccaactat | agtctgggaagacaaccaca | 210 | 285 |
| CMV | A | gpB | tacccctatcgcgtgtgttc | ataggaggcgccacgtattc | 254 | 357 |
| HHV6 | B | BamHI frag. | gatccgacgcctacaaacac | taccgacatccttgacatattac | 249 | 330 |
| HHV7 | B | cgcatacaccaaccctactg | gactcattatggggatcgac | 264 | 352 | |
| KSHV | A | KS330 | agccgaaaggattccaccat | acatggacagatcgtcaagc | 274 | 375 |
Designation based on primer sets described in reference 35.
PCR product size in base pairs.
In this analysis, the limits of detection are largely determined by the amount of DNA that can be included in the PCR reaction without having an inhibitory effect on the amplification process, and by the ability to detect a specific fluorescent band above the background fluorescence in the gel. Under the conditions described, DNA isolated from 10 μl of whole blood can routinely be included in a 50 μl PCR reaction without evidence of amplification inhibition.
The impact of fluorescence detection on the overall limits of detection can be evaluated as follows. A weak band present anywhere in the gel is chosen as representing the minimum detectable fluorescence signal above background gel fluorescence in that experiment. The intensity of this weak band (WBI) can then be compared with the intensity of the band derived from the ICS standard (SBI) in each sample to calculate the limits of detection, in viral targets (VT) per milliliter, for each reaction according to the equation:
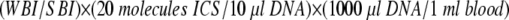 |
In the analysis presented in Figure 1A , the nonspecific weak band present in the HSV2 lane was chosen as representing the weakest signal detectable in this gel and compared with the intensity of the different standard bands. Quantitative fluorescence image analysis indicated that this blood sample from this patient contained <560 EBV VT/ml and <680 CMV VT/ml based on the intensities of these PCR product bands, indicating that these samples either lacked virus or had levels below the limits of detection for this assay.
Figure 1.
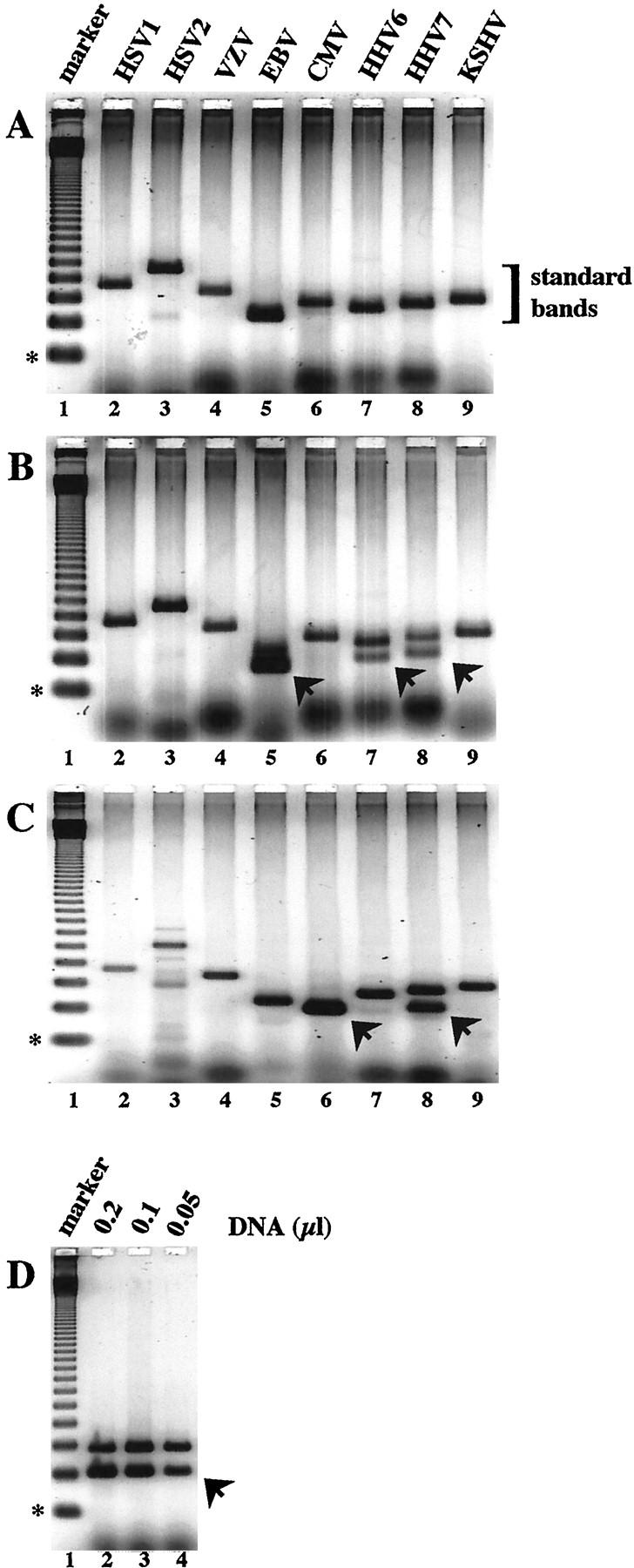
Quantitative ICS-PCR analysis of eight human herpesviruses in patient samples. PCR reactions containing purified DNA derived from 10 μl whole blood and 20 molecules of the HHVQ-1 ICS standard were amplified as described in Materials and Methods. DNA isolated from three different patients (A, B, and C) was amplified with primers specific for HSV-1 (lanes 2), HSV-2 (lanes 3), VZV (lanes 4), EBV (lanes 5), CMV (lanes 6), HHV-6 (lanes 7), HHV-7 (lanes 8), and KSHV (lanes 9). The sizes of the predicted PCR products derived from the HHVQ-1 standard and the specific viruses are given in Table 1 . PCR products derived from the HHVQ-1 ICS using the virus-specific primers are bracketed as standard bands. Arrowheads indicate PCR products derived from the viral targets. The weak bands observed with the HSV-2 primers in A and C, lanes 3 are nonspecific. D: DNA from the patient analyzed in C was re-analyzed by amplification with CMV-specific PCR primers and the amount of whole blood DNA indicated. In each panel, lane 1 contains a 123-bp DNA ladder as a size marker; the 123-bp fragment is indicated with an asterisk.
Results
ICS-PCR was used to quantify human herpesvirus nucleic acid targets in blood to determine whether viral burden measurements were useful in the management of pediatric transplant patients. In this method, an internal standard is included in each PCR reaction and coamplified with the specific herpesvirus in question using the same oligonucleotide primers. This approach was applied in a prospective study of 264 samples from 27 pediatric solid organ transplant patients to determine whether detection or quantification of virus in whole blood by PCR correlated with viral disease, whether changes in the level of virus would precede disease in individual patients, and whether changes in the levels of virus were found in response to antiviral therapy.
An example of the results from three representative patients is presented in Figure 1 . PCR reactions containing blood DNA from the first patient reveal single strong bands of amplification using primers specific for each of the eight human herpesviruses (Figure 1A) . The sizes of each of these bands indicate that these PCR products are derived from the HHVQ-1 standard (Table 1) . No virus-specific bands were seen. A weak band at ∼260 bp is found in the HSV2 lane. This band is due to nonspecific amplification, since it does not match the predicted size for the HSV2 viral genome (445 bp) and does not hybridize to an HSV2-specific probe (data not shown). Indeed, the size characteristics of the virus-specific bands provide an added layer of specificity to the analysis for all of the viruses.
For the second patient, the same constellation of standard bands is observed (Figure 1B) . In addition, three new bands are found below the standard bands in samples amplified with primers specific for EBV, HHV6, and HHV7. Each of these new bands is the size predicted for amplification of the specific viral target genome. Quantification is determined by comparing the fluorescence intensity of each product band. For example, the intensities of the two product bands using HHV7-specific primers are about the same. This reaction contained 20 molecules of HHVQ-1 and 10 μl of blood DNA. Thus, ∼20 HHV7 genomes were present in 10 μl of blood, or, when quantified accurately by fluorescence imaging, 1940 VT/ml. For HHV6, the viral band is weaker than the standard band and the level was calculated to be 1360 VT/ml; for EBV the viral band is stronger and the level was calculated to be 10,800 VT/ml.
For the third patient, virus-specific bands are found for CMV and HHV7 (Figure 1C) . In this case, amplification of the CMV viral target has precluded amplification of HHVQ-1 due to target competition, an indication of a high viral burden. In cases where only a viral band is found, a second series of samples are run containing dilutions of the blood DNA sample until bands of equal intensity are found (Figure 1D , lane 3), to achieve an accurate quantification. By this procedure, CMV viral burden was determined to be 1.7 × 105 VT/ml blood in this sample. Based on our previous validation studies, this simple approach has allowed us to accurately quantify all eight human herpesviruses in the 264 samples with CVs of <20%. 35
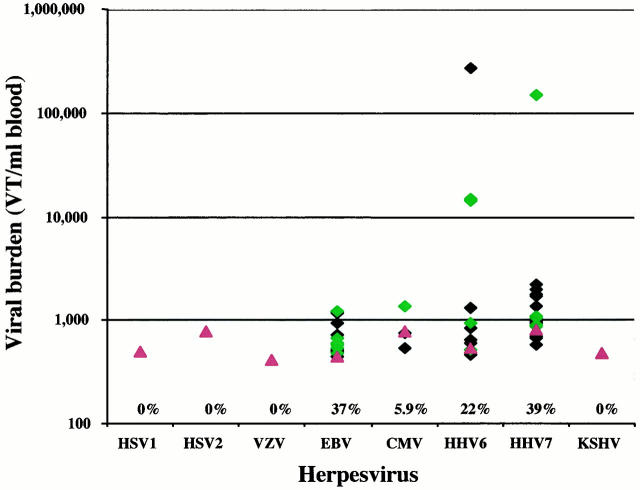
To understand the significance of viral burden determination with respect to disease, a suitable reference population was sought. Leftover blood samples from patients presenting to the Children’s Medical Center emergency room were used. Only blood samples from patients with normal CBCs were included. The HSV1, HSV2, VZV, and KSHV viruses were not detected in any of the 51 samples analyzed. For the other viruses, between 5.9 and 39% of the samples were positive for the different viral genomes (Figure 2) . In most cases, the levels of virus detected were close to the limits of detection of the assay. For EBV and CMV, viral burden was below 2000 VT/ml blood for all positive samples. For HHV6 and HHV7, the majority of positive samples had similar low levels. However, HHV6 viral burden in three samples and HHV7 viral burden in one sample appear to represent outliers. It is possible that each of these samples represents patients presenting with acute infection of these viruses, since each of these patients exhibited fever with unknown etiology or another clinical indication of infection. Unfortunately, none of these patients was evaluated at the time by serology testing to confirm the existence of an acute infection.
Figure 2.
Quantification of herpesvirus targets by ICS-PCR in a reference pediatric population. The levels of eight human herpesviruses in the blood of 51 pediatric emergency room patients was determined by ICS-PCR as described in Figure 1 . The diamonds indicate values (in VT/ml blood) for all samples that gave detectable virus-specific bands. The proportion of samples that gave detectable bands is indicated above each virus designation. Green diamonds are values from the subset of patients presenting with fever in this group. The red triangles indicate the average limits of detection for each virus in all negative samples analyzed in this study.
A total of 264 samples were prospectively collected from 27 transplant recipients (median of 8 samples/patient) and herpesvirus viral burden analyzed by ICS-PCR. As with the reference population, no HSV1, HSV2, VZV, or KSHV was detected in any sample. These likely represent true negative results, in that none of the patients developed any disease associated with these viruses during this study. In addition, each of these viruses has been detected in various disease settings in other studies using this method. For example, HSV1 has been detected in cerebrospinal fluid of herpes encephalitis patients; HSV2 has been detected in genital herpes swabs; VZV has been detected in vitreous fluid of a subset of patients with ocular disease; and KSHV has been detected in the blood of patients with Kaposi’s sarcoma and in effusion samples of patients with primary effusion lymphoma (X Bai, DB Dawson, and RH Scheuerman, unpublished results).
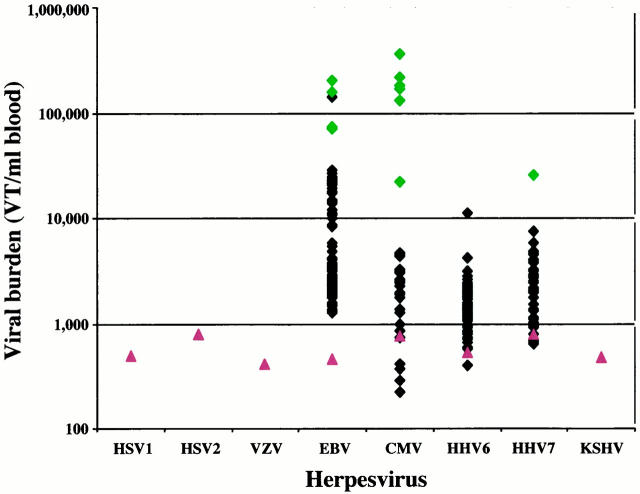
The majority of samples (63%) and patients (89%) contained detectable EBV targets (Table 2) . CMV, HHV6, and HHV7 were detected in 13%, 28%, and 17% of samples, respectively, and in more than half of the patients. For each of these four viruses, viral burden was relatively low for the vast majority of samples, with median values ranging from 1380 to 2540 VT/ml blood (Table 2) . Most of the values clustered around these median levels (Figure 3) . However, careful examination of the scatter plots suggests that for CMV and EBV, bimodal and trimodal distribution of values appear, with a significant minority of samples clustering at much higher levels.
Table 2.
Prevalence of PCR Positive Samples and Virus Levels
| Virus | Positive samples, number (%)* | Positive patients, number (%)† | Median viral burden‡ | Range of viral burden‡ | Proposed viral burden cutoff‡§ | Samples above cutoff, number (%)¶ | Patients above cutoff, number (%)** | Median viral burden of samples above cutoff‡ | Range of viral burden for samples above cutoff‡ |
|---|---|---|---|---|---|---|---|---|---|
| HSV1 | 0 (0%) | NA | NA | NA | ND | NA | NA | NA | NA |
| HSV2 | 0 (0%) | NA | NA | NA | ND | NA | NA | NA | NA |
| VZV | 0 (0%) | NA | NA | NA | ND | NA | NA | NA | NA |
| EBV | 166 (63%) | 24 (89%) | 2300 | 460–200,000 | 30,000 | 6 (3.6%) | 3 (13%) | 150,000 | 72,000–200,000 |
| CMV | 34 (13%) | 15 (56%) | 2540 | 220–360,000 | 10,000 | 8 (24%) | 4 (27%) | 175,000 | 22,000–360,000 |
| HHV6 | 75 (28%) | 15 (56%) | 1380 | 400–11,340 | 10,000 | 1 (1.3%) | 1 (6.7%) | 11,340 | NA |
| HHV7 | 46 (17%) | 14 (52%) | 2120 | 640–25,400 | 10,000 | 1 (2.2%) | 1 (7.1%) | 25,400 | NA |
| KSHV | 0 (0%) | NA | NA | NA | ND | NA | NA | NA | NA |
A sample was considered positive if a virus-specific PCR product was detected.
A patient was considered positive if a virus-specific PCR product was detected in at least one sample.
Viral burden is given as the number of viral genome targets detected by ICS-PCR per ml whole blood (VT/ml blood).
Viral burden cutoffs were determined by examination of data presented in Figures 2 and 3 , as described in the text.
Values indicate the number of sample above the proposed viral burden cutoff. Percent is the proportion of PCR positive samples with viral burden above the cutoff.
Values indicate the number of patients with at least one sample above the proposed viral burden cutoff. Percent is the proportion of positive patients with at least one sample above the viral burden cutoff.
Figure 3.
Quantification of herpesvirus targets by ICS-PCR in pediatric solid organ transplant patients. The levels of eight human herpesviruses in 264 blood samples from 27 pediatric solid organ transplant recipients was determined by ICS-PCR as described in Figure 1 . The diamonds indicate values (in VT/ml blood) for all samples that gave detectable virus-specific bands. The proportion of samples and patients with positive results for each virus can be found in Table 2 . The green diamonds indicate the levels in samples from patients with clinical or laboratory evidence of an acute viral infection within 5 weeks of the date of sample harvest (see text for details). The red triangles indicate the average limits of detection for each virus in all negative samples analyzed in this study.
To investigate the clinical significance of these values, each patient’s chart was reviewed for evidence of viral disease from standard clinical or laboratory evaluation. Four patients (15%) were diagnosed with CMV disease by serology, histology, or other laboratory tests consistent with non-autoimmune-mediated tissue destruction. Samples analyzed within 5 weeks of diagnosis of CMV disease are indicated with green diamonds in the CMV column of Figure 3 . Viral levels in these samples associated with CMV disease appear to form a separate cluster of values and suggest a cutoff of 10,000 VT/ml blood to identify clinically relevant levels for CMV.
A similar cutoff may be operative for HHV7, since the one value >10,000 was found in a patient presenting with a raised reddened area on the abdomen 10 days after ganciclovir treatment had been discontinued and developing stuffiness and congestion shortly thereafter. A single sample gave viral burden of slightly >10,000 for HHV6; however, no evidence of an acute viral infection was evident in this patient at the time.
For EBV, 22% of the positive samples gave levels >10,000 VT/ml, suggesting that this cutoff might not be a good predictor of EBV-associated disease. Only a single patient (3.7%) had evidence of lymphadenopathy suggestive of an EBV syndrome. Results from samples harvested from this patient close to dates in which radiographic studies demonstrated enlarged lymph nodes are indicated with green diamonds in the EBV column of Figure 3 . Again, these samples cluster at the highest levels, >30,000 VT/ml. Of the two other high positive EBV samples, one occurred in a patient presenting with nausea, vomiting, and diarrhea of unknown etiology, perhaps associated with an acute EBV infection. A second cluster of values between 8000 and 30,000 VT/ml blood was also observed. However, many of these samples came from patients with no evidence of EBV-associated disease.
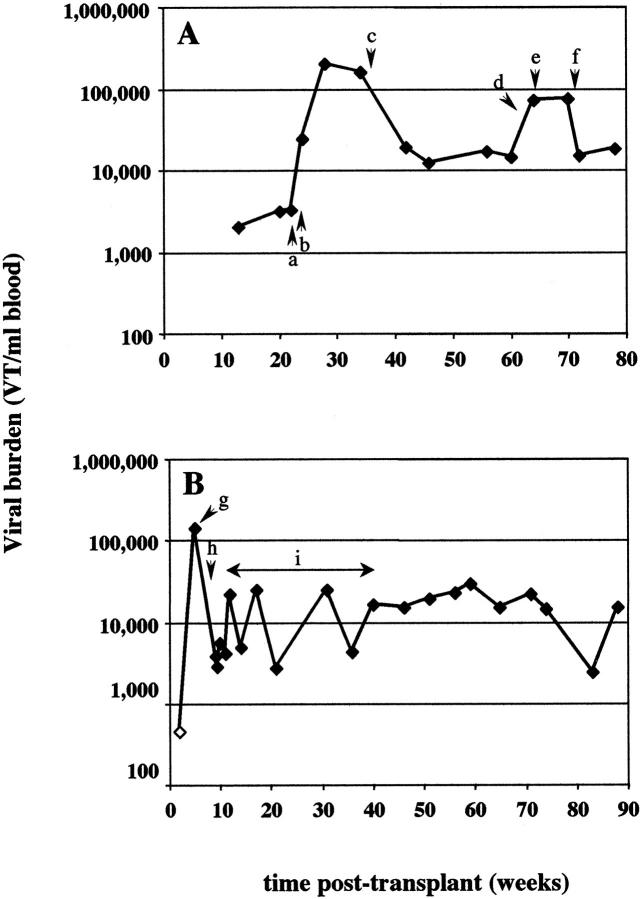
For CMV, a strong correlation was found between high viral burden determined by ICS-PCR and concurrent disease. With multiple samples from most patients, it was also possible to determine whether ICS-PCR could be used to predict impending disease or to monitor the efficacy of antiviral therapy. For three of the four patients, CMV viral burden >10,000 VT/ml was detected 2 to 4 weeks before clinical and standard laboratory evidence of disease (Figure 4 , A and B, and data not shown). In all three patients from whom samples were obtained after diagnosis of CMV disease, treatment with antiviral agents ganciclovir and cytogam (Figure 4A and 4C) or a change in the level of immunosuppression (Figure 4B) was associated with a rapid and sustained drop in viral burden.
Figure 4.
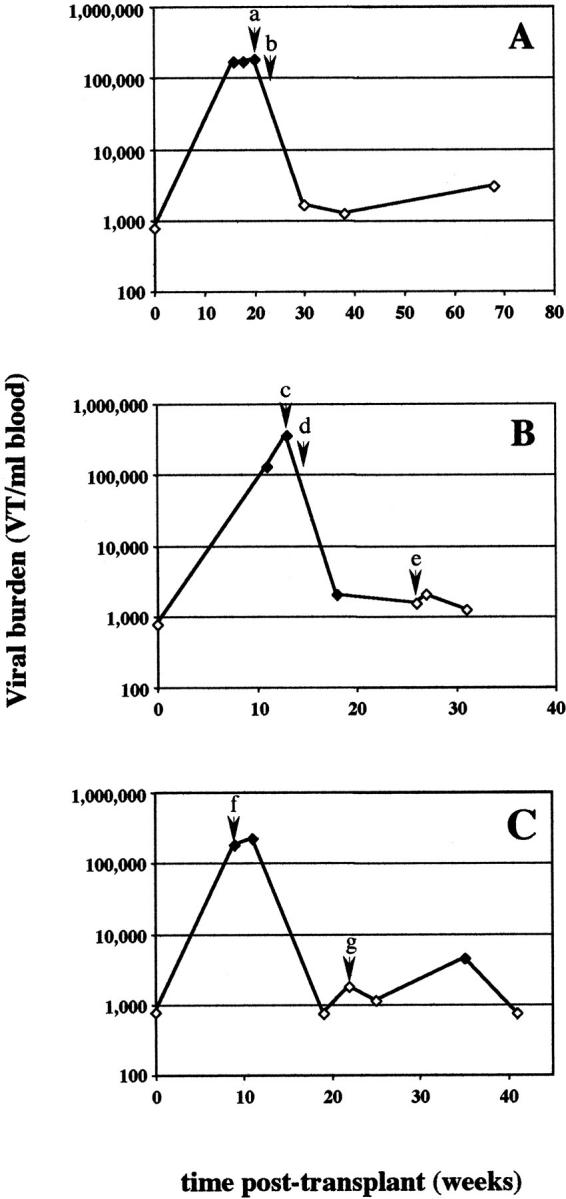
CMV levels in sequential samples from selected patients with evidence of acute infection. CMV levels were determined by ICS-PCR in multiple samples from three patients who developed clinical disease apparently associated with an acute CMV infection. Evidence for CMV infection included changes in CMV serology, histological detection of CMV inclusions, or other evidence of tissue damage. CMV levels are plotted against the time the sample was harvested after organ transplantation. Open symbols indicate samples in which no viral band was detected; the viral burdens indicated represent the limits of detection for that PCR reaction. The significant clinical findings are as follows. A: Patient 1. a: High creatinine levels were noted during routine laboratory studies. b: Cells containing viral inclusion bodies were identified in kidney biopsies leading to a diagnosis of CMV nephritis. Retinitis was also detected by clinical examination. The levels of cyclosporin were lowered; mycophenolate treatment was discontinued; treatment with ganciclovir and cytogam was begun. B: Patient 6. c: Elevated liver enzymes were noted during retrospective chart review. d: Cyclosporin and prednisone levels were reduced. e: Acyclovir treatment was begun. C: Patient 7. f: Patient was admitted to the hospital with complaint of recurrent fever. CMV serology was positive in the patient that was previously negative. Lymphopenia was noted. Patient was switched from acyclovir to ganciclovir and cytogam. Mycophenolate treatment was discontinued. g: Evidence of acute cellular rejection (tubulitis) from kidney biopsy was noted. Patient was treated with mycophenolate.
In this study, one patient (Patient 17) exhibited signs of lymphadenopathy by radiographic evaluation consistent with an EBV syndrome (Figure 5A , points c, e, and f). Each of these occurrences was accompanied by elevated EBV levels close to 100,000 VT/ml blood as determined by ICS-PCR. The initial increase in EBV levels was associated with an increase in immunosuppressive therapy to treat possible acute rejection. Interestingly, the levels of EBV appeared to drop in response to ganciclovir and cytogam, an anti-CMV combination. Two other patients gave a single sample positive for EBV above the proposed 30,000 VT/ml cutoff. In one of these patients (Patient 5), the high positive sample was taken ∼6 weeks posttransplant when the patient presented with nausea, vomiting, and diarrhea (Figure 5B) . The patient was admitted 1 month later with pneumonia and elevated liver enzymes. Although it is unclear if EBV was the etiological agent, the levels of CMV, HHV6, and HHV7 were low or negative during this period. At this point it is unclear whether this patient is at increased risk of developing EBV-mediated PTLD due to the sustained intermediate EBV levels at some point in the future. The second patient that had a single high positive sample for EBV (Patient 3) had an uneventful clinical course (data not shown). However, it is interesting to note that this sample also carried the highest HHV6 level seen in this study (11,340 VT/ml) and represents one of the few cases where parallel elevations in more than one virus were observed.
Figure 5.
Clinical correlations in selected patients with high EBV levels. EBV levels were determined by ICS-PCR in samples from two representative patients that showed relatively high EBV levels. EBV levels are plotted against the time the sample was harvested after organ transplantation. Open symbol indicates sample in which no viral band was detected; viral burden represents the limits of detection for that PCR reaction. The significant clinical findings are as follows. A: Patient 17. a: Cyclosporin levels were increased. b: Mild gastrointestinal symptoms developed; the patient was treated with intravenous prednisone. c: Mild lymphadenopathy was noted on chest X-ray. Treatment with ganciclovir and cytogam was begun. Immunosuppression was reduced slightly. d: Patient presented with gastroenteritis. e, f: CT scans show increasing lymph node size and number. B: Patient 5. g: Patient was admitted to hospital with complaints of nausea, vomiting, and diarrhea. h: Patient was admitted to hospital with pneumonia. Elevated liver enzymes were noted. High EBV and CMV IgG were detected by serological evaluation. i: Immunosuppression was tapered down threefold.
Discussion
In this prospective study, the clinical correlations of viral genome quantification by PCR for eight human herpesviruses were investigated in a cohort of pediatric solid organ transplant recipients. Although the HSV1, HSV2, VZV, and KSHV viruses were not detected in this cohort, the three γ herpesviruses (CMV, HHV6, and HHV7) and EBV were detected in a large proportion of the transplant patients. Quantification of these viruses was achieved by the ICS-PCR procedure in which an internal standard is included in each PCR reaction to control for PCR efficiency difference between samples. The distribution of viral burden levels (Figure 3) suggested that discrete patient populations were present. A comparison between the levels of viral targets and clinical evaluations indicates that viral burden correlates with disease.
Viral Burden Measurement Approaches
Several different approaches for PCR-based quantification have been described. A key requirement of an accurate approach is that it provide some evaluation of the quality of each sample analyzed. In some cases, amplification of a specific target is normalized against the amplification of a control target (eg, β-actin or GAPDH) using a different set of primers in either the same or different reaction mixes. On theoretical grounds, this approach is valid only if amplification of both specific and control targets occur with the same efficiency. 35, 40 Unfortunately, this is rarely the case. The data presented in Figure 1A provide a nice example of differences in amplification efficiencies occurring with different primers. A relatively strong band is observed in the sample amplified with EBV-specific primers, whereas a relatively weak band is observed in the sample amplified with VZV-specific primers. Yet both of these samples contain the exact same number of ICS targets included in the same master mix. If these samples were amplified for more cycles, the difference between the product amounts would continue to increase. In addition, different primer pairs are differentially affected by components of the reaction that could be variable between samples. For example, some primer pairs are very sensitive to differences in pH or Mg2+ concentrations between samples, whereas others are not. Thus, in our experience, for accurate quantification using PCR, the control target and the specific target must be amplified with the same primers in the same reaction.
Several other aspects of the ICS-PCR approach for target quantification are worth emphasizing. First, this approach yields quantification in absolute terms, expressed here as viral targets/ml blood. This allows for easy comparison of results between experiments and between laboratories. Second, the use of an internal standard obviates the need to run extra samples to generate a standard curve for quantification during each experiment. Third, the use of an internal standard helps to rule out false negative results due to sample inhibition; the presence of the standard product under conditions where a virus-specific product is not detected indicates that the negative result is real. However, the use of an internal standard cannot, in and of itself, rule out a failed DNA extraction. For samples containing nucleated blood cells, amplification of a genomic target can be used to evaluate DNA extraction. HHVQ-1 contains priming sites for the human RNA polymerase II large subunit gene that is used for this purpose.
The technique used here involves amplification in simple thermocyclers, agarose gel electrophoresis and fluorescence imaging. However, the approach could easily be adapted to a variety of different formats, including real-time PCR, TaqMan, or Invader approaches. From our experience, the key to any quantitative target amplification approach is the use of an internal standard, like the HHVQ-1 ICS, that can control for efficiency differences between reactions containing samples from different sources.
Predictive Value of Quantitative PCR
Many studies investigating the relationship between qualitative PCR detection of CMV and clinical disease have been reported. Several of these found that although the sensitivity and predictive value of a negative result were excellent, the specificity and predictive value of a positive result were disappointing. 30, 31, 32, 33, 41 We hypothesized that these results might relate to the presence of viral targets in latently infected cells that would not be associated with clinical disease. Further, we proposed that the level of virus in samples associated with disease would be higher than the levels associated with latency, and that the use of viral burden analysis would improve the predictive value of a PCR-based assay. Indeed, this appears to be the case for the patient population we studied. Four of the 27 patients (15%) followed here developed CMV disease, and yet 15 patients (56%) had at least one sample positive for CMV by PCR (Table 2) . For all of the samples analyzed, the sensitivity and predictive value of a negative result were both 100% by qualitative PCR (Table 3) . Unfortunately, the predictive value of a positive result was only 24%. However, examination of the data presented in Figure 3 suggested that CMV levels in this sample set were naturally segregated into two groups, with a cutoff of 10,000 VT/ml blood separating the two groups. If the predictive value is reanalyzed using this quantitative cutoff, both the specificity and predictive value of a positive result each reach 100%. Thus, quantitative PCR of CMV was found to correlate strongly with CMV disease in this patient population. Similar conclusions have been drawn from recent studies involving other immunosuppressed cohorts. 38, 42, 43, 44, 45, 46, 47, 48, 49, 50
Table 3.
Sensitivity, Specificity, and Predictive Value of Quantitative CMV Viral Burden Analysis
| Virus | True positive* | False positive | False negative | True negative | Sensitivity | Specificity | PPV | NPV |
|---|---|---|---|---|---|---|---|---|
| Qualitative CMV | 8 | 26 | 0 | 230 | 100% | 90% | 24% | 100% |
| Quantitative CMV at 10,000 VT/ml cutoff | 8 | 0 | 0 | 256 | 100% | 100% | 100% | 100% |
A sample was considered to be a true positive for a CMV-related disease if it gave a positive result or a value above the cutoff and was harvested within 5 weeks of evidence of CMV etiology by histology, serology, and/or non-immune-mediated tissue destruction.
Quantitative PCR for CMV appears to be an effective diagnostic tool in identifying patients with disease. In addition, PCR detection of DNA targets is less sensitive to sample handling than other diagnostic tests. In one study, PCR detection of CMV was unchanged even if the blood sample was stored at room temperature for 72 hours. 51 In contrast, positive shell vial results were found to decline progressively such that only 10% of the initial positive samples remained positive after 48 hours’ incubation. The CMV antigenemia test is widely used for diagnosis of disease in various immunosuppressed populations 52, 53, 54 and has largely replaced the standard viral culture and shell vial assays due to its increased sensitivity and predictive value. Several studies have found that CMV PCR compares favorable with CMV antigenemia in terms of both concordance and quantification 42, 47, 55, 56, 57, 58 (X Bai, RH Scheuermann, J Luby, K St. George, and C Rinaldo, unpublished findings). In some studies, PCR appeared to provide for earlier anticipation of disease 41, 57 and was superior for predicting recurrent disease. 50
The difference in predictive value between qualitative and quantitative PCR is also apparent in the analysis of EBV viral burden. 89% of the patients had at least one positive PCR result for EBV, and yet only one patient (3.7%) developed lymphadenopathy suggestive of EBV disease (Table 2) . For all of the samples analyzed, the specificity and positive predictive value of qualitative PCR were only 38% and 2.4%, respectively. Examination of the data in Figure 3 suggested that EBV levels could be segregated into three groups: those below 8000 VT/ml blood, those above 30,000 VT/ml, and those in between. Reanalysis of the data using these quantitative cutoffs would progressively improve the correlation with disease. However, since this analysis would be based on a single patient with evidence of an EBV syndrome, we feel that it would not be appropriate to calculate predictive values for this sample set.
Although these conclusions are based on positive samples from a single patient who developed lymphadenopathy consistent with an EBV syndrome, similar conclusions were suggested by data in previous studies. 35, 39, 59, 60, 61 In one of these studies, 35 the median EBV viral burden in ten patients diagnosed with PTLD was 440,000 VT/ml blood. The median EBV viral burden in samples from transplant patients without PTLD was 5400 VT/ml, and in a pediatric reference group it was 1200 VT/ml. In that study, a 30,000 VT/ml cutoff would have missed one PTLD patient out of ten (a false negative), and four of fourteen transplant samples without PTLD would be scored as false positives. Thus, in both of these studies, moderate to high EBV burden correlated with disease. However, in the past 2 years, three additional patients have presented with PTLD who were not part of this prospective study. Retrospective analysis of samples taken at the time of PTLD diagnosis revealed EBV levels in the range of 2000–4000 VT/ml blood, an order of magnitude lower than expected based on these other studies. At this point, it is unclear whether these patients represent a new trend in EBV levels associated with disease, whether the findings are related to changes in current immunosuppressive therapy approaches, or whether they are simply statistical outliers. Clearly, these recent findings warrant further investigation.
The prevalence of HHV6 and HHV7 in the reference and transplant populations was similar. Although seroprevalence of these two viruses approaches 100% in most adult populations, a wide range of values for detection of DNA in blood has been reported in the literature, with the most sensitive techniques reporting the highest values. 62, 63, 64, 65 From the data in this current study, the establishment of clinical cutoffs for HHV6 and HHV7 is problematic. It is safe to say that a positive PCR result is not indicative of viral etiology, in that low-level positive results are found in a substantial proportion of samples from the transplant population without any clinical evidence of viral disease and in samples from the reference group. However, applying a 10,000 VT/ml cutoff to the data from both groups (transplant and reference) reveals that five of the six samples above this cutoff are associated with some indication of an infectious disease process. Unfortunately, the absence of an independent measure of disease etiology for these two viruses makes it impossible to determine whether a significant correlation exists. Differences in HHV6 viral burden as measured by a semiquantitative PCR approach have been observed during seroconversion in pediatric patients hospitalized for febrile illness, 66 suggesting that viral levels do correlate with disease in some settings.
Because high viral burden is thought to be related to the level of immunosuppression, it appeared possible that elevation of one virus would be associated with elevated levels of others. Indeed, reactivation of HHV6 in transplant recipients experiencing primary CMV infection has been reported. 67, 68, 69 However, we found that this was not the case in our patient population. Although many of the eight samples with high CMV levels also demonstrated detectable EBV and/or HHV7 virus, they were present at low levels. HHV6 was not detected in any of these samples. Only one example of coordinated elevation was apparent in this pool of 264 samples; the one sample with HHV6 levels above 10,000 VT/ml also had elevated EBV (1.6 × 105). The levels of both viruses dropped in the next sample isolated 2 months later. The patient remained asymptomatic throughout this period.
In addition to the positive correlations between viral burden and disease for EBV and CMV, elevated levels were often detected well before clinical onset of disease. In four cases of CMV, elevated viral burden was found 0, 13, 28, and 28 days before diagnosis of CMV disease in the patients affected by standard laboratory evaluation. The ability to diagnose CMV disease earlier would allow antiviral therapy to be started earlier and would be expected to reduce the level of tissue destruction of the affected organ.
Finally, rapid reductions in both CMV and EBV levels were found with the onset of antiviral therapy. In each of the three patients analyzed after diagnosis of CMV disease, a two-log reduction in viral burden was observed in samples taken within 5 weeks after the onset of therapy to levels found in asymptomatic individuals. Similar results have been reported for other immunosuppressed populations. 34, 36, 45, 46, 50, 55, 70 Management of anti-CMV therapy using quantitative PCR could potentially reduce the incidence of therapy-related side effects and therapy costs if disease could be controlled with a more limited course of antiviral therapy. EBV levels were also found to respond to changes in medication in the patient with lympadenopathy. EBV-associated PTLD is usually treated by tapering immunosuppression. A side effect of this treatment approach is the increased risk of transplant organ rejection. The ability to monitor EBV levels closely by quantitative PCR as a surrogate marker of disease could be very useful in developing a therapeutic approach that would be effective at treating the viral disease while minimizing the risk of rejection.
In conclusion, as a result of this 2-year prospective study of pediatric solid organ transplant recipients, viral burden analysis by ICS-PCR appears to be an excellent diagnostic tool for patient monitoring. By applying quantitative viral burden cutoffs (10,000 VT/ml blood for CMV and 30,000 VT/ml blood for EBV), the prevalence of false positive results can be effectively reduced to give better specificity and positive predictive value for both viruses. Added benefits also include the abilities to predict the onset of clinical disease several weeks in advance and to monitor antiviral therapeutic response with a simple, cost-effective blood test. The ability to monitor all eight herpesviruses simultaneously is another significant advantage over the current approaches in which each virus is evaluated by a different diagnostic approach, eg, antigenemia, histology, serology, or viral culture. In addition, the technique used here is amenable to further modification to facilitate high throughput analysis and more rapid turnaround time, including the possible use of real-time PCR, which combines the PCR amplification and product analysis steps, or the multiplexing of multiple targets in single PCR reactions.
Acknowledgments
We thank Dr. R. Domiati-Saad for critical review of this manuscript and D. Haas, T. Romano, N. Simonds, and J. Wells for their help with obtaining patient samples. We also acknowledge the support of scientists at Biosource International, Inc., especially Drs. K. J. Reagan, C. Cabradilla, B. Shuman, and N. Stollar, for helpful advice in the development of the ICS-PCR technique.
Address reprint requests to Richard H. Scheuermann, Ph.D., Department of Pathology, U.T. Southwestern Medical Center, 5323 Harry Hines Blvd., Dallas, TX 75390-9072. E-mail: scheuerm@utsw.swmed.edu.
References
- 1.Kanj SS, Sharara AI, Clavien PA, Hamilton JD: Cytomegalovirus infection following liver transplantation: review of the literature. Clin Infect Dis 1996, 22:537-549 [DOI] [PubMed] [Google Scholar]
- 2.Allen RD, Chapman JR: A Manual of Renal Transplantation. 1994:pp 217-237 Edward Arnold, London
- 3.Maddrey WC, Sorrell MF: Infectious Disease Problems. 1995:pp 367-398 Appleton & Lange, Norwalk, CT,
- 4.Wang FZ, Dahl H, Linde A, Brytting M, Ehrnst A, Ljungman P: Lymphotropic herpesviruses in allogeneic bone marrow transplantation. Blood 1996, 88:3615-3620 [PubMed] [Google Scholar]
- 5.Craig FE, Gulley ML, Banks PM: Posttransplantation lymphoproliferative disorders. Am J Clin Pathol 1993, 99:265-276 [DOI] [PubMed] [Google Scholar]
- 6.Morgan G, Superina RA: Lymphoproliferative disease after pediatric liver transplantation. J Pediatr Surg 1994, 29:1192-1196 [DOI] [PubMed] [Google Scholar]
- 7.Birkeland SA, Hamilton DS, Sandvej K, Andersen HM, Bendtzen K, Moller B, Jorgensen KA: EBV-induced post-transplant lymphoproliferative disorder (PTLD). Transplant Proc 1995, 27:3467-3472 [PubMed] [Google Scholar]
- 8.George MJ, Snydman DR, Werner BG, Dougherty NN, Griffith J, Rohrer RH, Freeman R, Jenkins R, Lewis WD: Use of ganciclovir plus cytomegalovirus immune globulin to treat CMV pneumonia in orthotopic liver transplant recipients. The Boston Center for Liver Transplantation CMVIG-Study Group. Transplant Proc 1993, 25:22-24 [PubMed] [Google Scholar]
- 9.Stratta RJ, Shaefer MS, Markin RS, Wood RP, Kennedy EM, Langnas AN, Reed EC, Woods GL, Donovan JP, Pillen TJ, Duckworth RM, Shaw BW, Jr: Clinical patterns of cytomegalovirus disease after liver transplantation. Arch Surg 1989, 124:1443-1449 [DOI] [PubMed] [Google Scholar]
- 10.Paya CV, Hermans PE, Wiesner RH, Ludwig J, Smith TF, Rakela J, Krom RA: Cytomegalovirus hepatitis in liver transplantation: prospective analysis of 93 consecutive orthotopic liver transplantations. J Infect Dis 1989, 160:752-758 [DOI] [PubMed] [Google Scholar]
- 11.Sayage LH, Gonwa TA, Goldstein RM, Husberg BS, Klintmalm GB: Cytomegalovirus infection in orthotopic liver transplantation. Transplant Int 1989, 2:96-101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hornef MW, Bein G, Fricke L, Steinhoff J, Wagner HJ, Hinderer W, Sonneborn HH, Kirchner H: Coincidence of Epstein-Barr virus reactivation, cytomegalovirus infection, and rejection episodes in renal transplant recipients. Transplantation 1995, 60:474-480 [DOI] [PubMed] [Google Scholar]
- 13.Demetris AJ, Lasky S, Van TD, Starzl TE, Dekker A: Pathology of hepatic transplantation: a review of 62 adult allograft recipients immunosuppressed with a cyclosporine/steroid regimen. Am J Pathol 1985, 118:151-161 [PMC free article] [PubMed] [Google Scholar]
- 14.Hanto DW, Frizzera G, Purtilo DT, Sakamoto K, Sullivan JL, Saemundsen AK, Klein G, Simmons RL, Najarian JS: Clinical spectrum of lymphoproliferative disorders in renal transplant recipients and evidence for the role of Epstein-Barr virus. Cancer Res 1981, 41:4253-4261 [PubMed] [Google Scholar]
- 15.Hanto DW, Frizzera G, Gajl PK, Simmons RL: Epstein-Barr virus, immunodeficiency, and B cell lymphoproliferation. Transplantation 1985, 39:461-472 [DOI] [PubMed] [Google Scholar]
- 16.Ho M, Miller G, Atchison RW, Breinig MK, Dummer JS, Andiman W, Starzl TE, Eastman R, Griffith BP, Hardesty RL, Bahnson HT, Hakala TR, Rosenthal JT: Epstein-Barr virus infections and DNA hybridization studies in posttransplantation lymphoma and lymphoproliferative lesions: the role of primary infection. J Infect Dis 1985, 152:876-886 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Basgoz N, Preiksaitis JK: Post-transplant lymphoproliferative disorder. Infect Dis Clin North Am 1995, 9:901-923 [PubMed] [Google Scholar]
- 18.Mosimann F, Cuenoud PF, Steinhauslin F, Wauters JP: Herpes simplex esophagitis after renal transplantation. Transplant Int 1994, 7:79-82 [DOI] [PubMed] [Google Scholar]
- 19.Reed EC, Bowden RA, Dandliker PS, Lilleby KE, Meyers JD: Treatment of cytomegalovirus pneumonia with ganciclovir and intravenous cytomegalovirus immunoglobulin in patients with bone marrow transplants. Ann Intern Med 1988, 109:783-788 [DOI] [PubMed] [Google Scholar]
- 20.Emanuel D, Cunningham I, Jules EK, Brochstein JA, Kernan NA, Laver J, Stover D, White DA, Fels A, Polsky B, Castro-Malaspina H, Peppard JR, Bartus P, Hammerling U, O’Reilly RJ: Cytomegalovirus pneumonia after bone marrow transplantation successfully treated with the combination of ganciclovir and high-dose intravenous immune globulin. Ann Intern Med 1988, 109:777-782 [DOI] [PubMed] [Google Scholar]
- 21.Starzl TE, Nalesnik MA, Porter KA, Ho M, Iwatsuki S, Griffith BP, Rosenthal JT, Hakala TR, Shaw BJ, Hardesty RL, Jaffe R, Bahnson HT: Reversibility of lymphomas and lymphoproliferative lesions developing under cyclosporin-steroid therapy. Lancet 1984, 1:583-587 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Morris DJ, Littler E, Arrand JR, Jordan D, Mallick NP, Johnson RW: Human herpesvirus 6 infection in renal-transplant recipients (letter). N Engl J Med 1989, 320:1560-1561 [PubMed] [Google Scholar]
- 23.Dubedat S, Kappagoda N: Hepatitis due to human herpesvirus-6 (letter). Lancet 1989, 2:1463-1464 [DOI] [PubMed] [Google Scholar]
- 24.Asano Y, Yoshikawa T, Suga S, Yazaki T, Kondo K, Yamanishi K: Fatal fulminant hepatitis in an infant with human herpesvirus-6 infection (letter). Lancet 1990, 335:862-863 [DOI] [PubMed] [Google Scholar]
- 25.Carrigan DR, Drobyski WR, Russler SK, Tapper MA, Knox KK, Ash RC: Interstitial pneumonitis associated with human herpesvirus-6 infection after marrow transplantation. Lancet 1991, 338:147-149 [DOI] [PubMed] [Google Scholar]
- 26.Drobyski WR, Knox KK, Majewski D, Carrigan DR: Brief report: fatal encephalitis due to variant B human herpesvirus-6 infection in a bone marrow-transplant recipient. N Engl J Med 1994, 330:1356-1360 [DOI] [PubMed] [Google Scholar]
- 27.Lautenschlager I, Hockerstedt K, Linnavuori K, Taskinen E: Human herpesvirus-6 infection after liver transplantation. Clin Infect Dis 1998, 26:702-707 [DOI] [PubMed] [Google Scholar]
- 28.Ratnamohan VM, Chapman J, Howse H, Bovington K, Robertson P, Byth K, Allen R, Cunningham AL: Cytomegalovirus and human herpesvirus 6 both cause viral disease after renal transplantation. Transplantation 1998, 66:877-882 [DOI] [PubMed] [Google Scholar]
- 29.Griffiths PD, Ait KM, Bearcroft CP, Clark DA, Quaglia A, Davies SE, Burroughs AK, Rolles K, Kidd IM, Knight SN, Noibi SM, Cope AV, Phillips AN, Emery VC: Human herpesviruses 6 and 7 as potential pathogens after liver transplant: prospective comparison with the effect of cytomegalovirus. J Med Virol 1999, 59:496-501 [DOI] [PubMed] [Google Scholar]
- 30.Lo CY, Ho KN, Yuen KY, Lui SL, Li FK, Chan TM, Lo WK, Cheng IK: Diagnosing cytomegalovirus disease in CMV seropositive renal allograft recipients: a comparison between the detection of CMV DNAemia by polymerase chain reaction and antigenemia by CMV pp65 assay. Clin Transplant 1997, 11:286-293 [PubMed] [Google Scholar]
- 31.Hebart H, Schroder A, Loffler J, Klingebiel T, Martin H, Wassmann B, Gerneth F, Rabenau H, Jahn G, Kanz L, Müller CA, Einsele H: Cytomegalovirus monitoring by polymerase chain reaction of whole blood samples from patients undergoing autologous bone marrow or peripheral blood progenitor cell transplantation. J Infect Dis 1997, 175:1490-1493 [DOI] [PubMed] [Google Scholar]
- 32.Abecassis MM, Koffron AJ, Buckingham M, Kaufman DB, Fryer JP, Stuart J, Stuart FP: Role of PCR in the diagnosis and management of CMV in solid organ recipients: what is the predictive value for development of disease and should PCR be used to guide antiviral therapy? Transplant Proc 1997, 29:800-801 [DOI] [PubMed] [Google Scholar]
- 33.Benedetti E, Mihalov M, Asolati M, Kirby J, Dunn T, Raofi V, Fontaine M, Pollak R: A prospective study of the predictive value of polymerase chain reaction assay for cytomegalovirus in asymptomatic kidney transplant recipients. Clin Transplant 1998, 12:391-395 [PubMed] [Google Scholar]
- 34.Drouet E, Colimon R, Michelson S, Fourcade N, Niveleau A, Ducerf C, Boibieux A, Chevallier M, Denoyel G: Monitoring levels of human cytomegalovirus DNA in blood after liver transplantation. J Clin Microbiol 1995, 33:389-394 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Bai X, Hosler G, Rogers BB, Dawson DB, Scheuermann RH: Quantitative polymerase chain reaction for human herpesvirus diagnosis and measurement of Epstein-Barr virus burden in posttransplant lymphoproliferative disorder. Clin Chem 1997, 43:1843-1849 [PubMed] [Google Scholar]
- 36.Imbert-Marcille B, Cantarovich D, Ferre AV, Richet B, Soulillou JP, Billaudel S: Usefulness of DNA viral load quantification for cytomegalovirus disease monitoring in renal and pancreas/renal transplant recipients. Transplantation 1997, 63:1476-1481 [DOI] [PubMed] [Google Scholar]
- 37.Reagan KJ, Cabradilla C, Shuman B, Stollar N, Laudermann J, Bai X, Hosler G, Scheuermann RH: Analytical performance of a quantitative CMV DNA detection method. Scholz M Rabenau HF Doerr HW Cinatl J, Jr eds. CMV-Related Immunopathology. 1998, :pp 252-261 Karger, Basel [Google Scholar]
- 38.Ferreira-Gonzalez A, Fisher RA, Weymouth LA, Langley MR, Wolfe L, Wilkinson DS, Garrett CT: Clinical utility of a quantitative polymerase chain reaction for diagnosis of cytomegalovirus disease in solid organ transplant patients. Transplantation 1999, 68:991-996 [DOI] [PubMed] [Google Scholar]
- 39.Baldanti F, Grossi P, Furione M, Simoncini L, Sarasini A, Comoli P, Maccario R, Fiocchi R, Gerna G: High levels of Epstein-Barr virus DNA in blood of solid-organ transplant recipients and their value in predicting posttransplant lymphoproliferative disorders. J Clin Microbiol 2000, 38:613-619 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Scheuermann RH, Bauer SR: PCR-based mRNA quantification using an internal standard: A method for the analysis of oncogene expression. Methods Enzymol 1993, 218:446-473 [DOI] [PubMed] [Google Scholar]
- 41.Tong CY, Cuevas L, Williams H, Bakran A: Use of laboratory assays to predict cytomegalovirus disease in renal transplant recipients (published erratum appears at J Clin Microbiol 1999, 37: 881). J Clin Microbiol 1999, 36:2681-2685 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Boeckh M, Boivin G: Quantitation of cytomegalovirus: methodologic aspects and clinical applications. Clin Microbiol Rev 1998, 11:533-554 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Evans PC, Soin A, Wreghitt TG, Alexander GJ: Qualitative and semiquantitative polymerase chain reaction testing for cytomegalovirus DNA in serum allows prediction of CMV related disease in liver transplant recipients. J Clin Pathol 1998, 51:914-921 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Gor D, Sabin C, Prentice HG, Vyas N, Man S, Griffiths PD, Emery VC: Longitudinal fluctuations in cytomegalovirus load in bone marrow transplant patients: relationship between peak virus load, donor/recipient serostatus, acute GVHD and CMV disease. Bone Marrow Transplant 1998, 21:597-605 [DOI] [PubMed] [Google Scholar]
- 45.Roberts TC, Brennan DC, Buller RS, Gaudreault KM, Schnitzler MA, Sternhell KE, Garlock KA, Singer GG, Storch GA: Quantitative polymerase chain reaction to predict occurrence of symptomatic cytomegalovirus infection and assess response to ganciclovir therapy in renal transplant recipients. J Infect Dis 1998, 178:626-635 [DOI] [PubMed] [Google Scholar]
- 46.Aitken C, Barrett MW, Millar C, Templeton K, Thomas J, Sheridan F, Jeffries D, Yaqoob M, Breuer J: Use of molecular assays in diagnosis and monitoring of cytomegalovirus disease following renal transplantation. J Clin Microbiol 1999, 37:2804-2807 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Humar A, Gregson D, Caliendo AM, McGeer A, Malkan G, Krajden M, Corey P, Greig P, Walmsley S, Levy G, Mazzulli T: Clinical utility of quantitative cytomegalovirus viral load determination for predicting cytomegalovirus disease in liver transplant recipients. Transplantation 1999, 68:1305-1311 [DOI] [PubMed] [Google Scholar]
- 48.Sia IG, Wilson JA, Espy MJ, Paya CV, Smith TF: Evaluation of the COBAS AMPLICOR CMV MONITOR test for detection of viral DNA in specimens taken from patients after liver transplantation. J Clin Microbiol 2000, 38:600-606 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Tong CY, Cuevas LE, Williams H, Bakran A: Comparison of two commercial methods for measurement of cytomegalovirus load in blood samples after renal transplantation. J Clin Microbiol 2000, 38:1209-1213 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Weinberg A, Hodges TN, Li S, Cai G, Zamora MR: Comparison of PCR, antigenemia assay, and rapid blood culture for detection and prevention of cytomegalovirus disease after lung transplantation. J Clin Microbiol 2000, 38:768-772 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Roberts TC, Buller RS, Gaudreault KM, Sternhell KE, Garlock K, Singer GG, Brennan DC, Storch GA: Effects of storage temperature and time on qualitative and quantitative detection of cytomegalovirus in blood specimens by shell vial culture and PCR. J Clin Microbiol 1997, 35:2224-2228 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Landry ML, Ferguson D: Comparison of quantitative cytomegalovirus antigenemia assay with culture methods and correlation with clinical disease. J Clin Microbiol 1993, 31:2851-2856 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Lautenschlager I, Hockerstedt K, Salmela K: Quantitative CMV-antigenemia test in the diagnosis of CMV infection and in the monitoring of response to antiviral treatment in liver transplant recipients. Transplant Proc 1994, 26:1719-1720 [PubMed] [Google Scholar]
- 54.Murray BM, Brentjens J, Amsterdam D, Myers J, Gray V, Pawlowski I, Schewegler K, Singh JP, Venuto RC: The cytomegalovirus-antigenemia assay in the diagnosis of posttransplant cytomegalovirus infection. J Am Soc Nephrol 1994, 4:1615-1622 [DOI] [PubMed] [Google Scholar]
- 55.Hebart H, Einsele H, Klein R, Fischer I, Buhler S, Dietz K, Jahn G, Berg PA, Kanz L, Muller CA: CMV infection after allogeneic bone marrow transplantation is associated with the occurrence of various autoantibodies and monoclonal gammopathies. Br J Haematol 1996, 95:138-144 [DOI] [PubMed] [Google Scholar]
- 56.Wirgart BZ, Claesson K, Eriksson BM, Brundin M, Tufveson G, Totterman T, Grillner L: Cytomegalovirus (CMV) DNA amplification from plasma compared with CMV pp65 antigen (ppUL83) detection in leukocytes for early diagnosis of symptomatic CMV infection in kidney transplant patients. Clin Diag Virol 1996, 7:99-110 [DOI] [PubMed] [Google Scholar]
- 57.Evans MJ, Edwards SY, Myers J, Wendt A, Povinelli D, Amsterdam D, Rittenhouse DK, Armstrong D, Murray BM, Greenberg SJ, Riepenhoff TM: Polymerase chain reaction assays for the detection of cytomegalovirus in organ and bone marrow transplant recipients. Immunol Invest 1997, 26:209-229 [DOI] [PubMed] [Google Scholar]
- 58.Tanabe K, Tokumoto T, Ishikawa N, Koyama I, Takahashi K, Fuchinoue S, Kawai T, Koga S, Yagisawa T, Toma H, Ota K, Nakajima H: Comparative study of cytomegalovirus (CMV) antigenemia assay, polymerase chain reaction, serology, and shell vial assay in the early diagnosis and monitoring of CMV infection after renal transplantation. Transplantation 1997, 64:1721-1725 [DOI] [PubMed] [Google Scholar]
- 59.Rowe DT, Qu L, Reyes J, Jabbour N, Yunis E, Putnam P, Todo S, Green M: Use of quantitative competitive PCR to measure Epstein-Barr virus genome load in the peripheral blood of pediatric transplant patients with lymphoproliferative disorders. J Clin Microbiol 1997, 35:1612-1615 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Rogers BB, Sommerauer J, Quan A, Timmons CF, Dawson DB, Scheuermann RH, Krisher K, Atkins C: Epstein-Barr virus polymerase chain reaction and serology in pediatric post-transplant lymphoproliferative disorder: three-year experience. Pediatr Dev Pathol 1998, 1:480-486 [DOI] [PubMed] [Google Scholar]
- 61.Niesters HG, van EJ, Fries E, Wolthers KC, Cornelissen J, Osterhaus AD: Development of a real-time quantitative assay for detection of Epstein-Barr virus. J Clin Microbiol 2000, 38:712-715 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Wilborn F, Schmidt CA, Zimmermann R, Brinkmann V, Neipel F, Siegert W: Detection of herpesvirus type 6 by polymerase chain reaction in blood donors: random tests and prospective longitudinal studies. Br J Haematol 1994, 88:187-192 [DOI] [PubMed] [Google Scholar]
- 63.Di Luca D, Dolcetti R, Mirandola P, De RV, Secchiero P, Carbone A, Boiocchi M, Cassai E: Human herpesvirus 6: a survey of presence and variant distribution in normal peripheral lymphocytes and lymphoproliferative disorders. J Infect Dis 1994, 170:211-215 [DOI] [PubMed] [Google Scholar]
- 64.Cuende JI, Ruiz J, Civeira MP, Prieto J: High prevalence of HHV-6 DNA in peripheral blood mononuclear cells of healthy individuals detected by nested-PCR. J Med Virol 1994, 43:115-118 [DOI] [PubMed] [Google Scholar]
- 65.Hoang MP, Rogers BB, Dawson DB, Scheuermann RH: Quantitation of 8 human herpesviruses in peripheral blood of human immunodeficiency virus-infected patients and healthy blood donors by polymerase chain reaction. Am J Clin Pathol 1999, 111:655-659 [DOI] [PubMed] [Google Scholar]
- 66.Chiu SS, Cheung CY, Tse CY, Peiris M: Early diagnosis of primary human herpesvirus 6 infection in childhood: serology, polymerase chain reaction, and virus load. J Infect Dis 1998, 178:1250-1256 [DOI] [PubMed] [Google Scholar]
- 67.Herbein G, Strasswimmer J, Altieri M, Woehl JM, Wolf P, Obert G: Longitudinal study of human herpesvirus 6 infection in organ transplant recipients. Clin Infect Dis 1996, 22:171-173 [DOI] [PubMed] [Google Scholar]
- 68.Dockrell DH, Prada J, Jones MF, Patel R, Badley AD, Harmsen WS, Ilstrup DM, Wiesner RH, Krom RA, Smith TF, Paya CV: Seroconversion to human herpesvirus 6 following liver transplantation is a marker of cytomegalovirus disease. J Infect Dis 1997, 176:1135-1140 [DOI] [PubMed] [Google Scholar]
- 69.DesJardin JA, Gibbons L, Cho E, Supran SE, Falagas ME, Werner BG, Snydman DR: Human herpesvirus 6 reactivation is associated with cytomegalovirus infection and syndromes in kidney transplant recipients at risk for primary cytomegalovirus infection. J Infect Dis 1998, 178:1783-1786 [DOI] [PubMed] [Google Scholar]
- 70.Toyoda M, Carlos JB, Galera OA, Galfayan K, Zhang X, Sun Z, Czer LS, Jordan SC: Correlation of cytomegalovirus DNA levels with response to antiviral therapy in cardiac and renal allograft recipients. Transplantation 1997, 63:957-963 [DOI] [PubMed] [Google Scholar]