Abstract
The human leukocyte antigens (HLA) encoded by genes within the major histocompatibility complex display an impressive degree of polymorphism. This variability is apparently maintained in human populations through the need to successfully display a wide range of processed foreign peptides to the T cell antigen receptor. The large number of alleles at the Class I and Class II loci pose a significant problem for molecular diagnosis. Knowledge of allele groups and specific alleles present in individuals has important implications in organ and stem cell transplantation and in disease association studies. Histocompatibility laboratories have transformed themselves during the past decade as they have adapted the techniques of molecular diagnostics to the challenge of identifying HLA alleles.
Histocompatibility laboratories support transplantation programs in several ways. They identify organ and stem cell recipient and donor human leukocyte antigens (HLA) for matching purposes, they screen waiting recipient sera for anti-HLA antibodies, and they perform cross-matches between recipient sera and donor cells just before a potential transplant procedure. Additional laboratory services may include microsatellite locus studies to assess the degree of donor engraftment following a bone marrow transplant, post-transplant serological and cellular studies to monitor the immune response and rejection, and allele identification for HLA-associated disease diagnosis and management. Recently, creative strategies for predicting rejection following renal transplantation via the quantification of mRNAs that are markers for cytotoxic T cell activity present within cells in urine samples have been described. 1
Perhaps no other clinical laboratory has undergone the marked changes in technology that the clinical histocompatibility laboratory has in the past decade. Serum screening has moved from HLA antigen targets on cell panels constructed from volunteers and commercially available frozen cells to flow cytometric detection of antibodies bound to microspheres coated with partially purified HLA antigens and to ELISA-based methods. 2 Cross-matching has transitioned from microlymphocytotoxicity-based assays assessing complement mediated cell lysis due to recipient anti-HLA antibodies coating donor cells to more sensitive flow cytometric-based tests. 3 Finally, HLA antigen detection by serological assays is being gradually replaced by nucleic acid-based methods for identification of HLA alleles. New technology for antibody screening and cross-matching has been described in the references cited above. The focus of this review will be the polymorphism, clinical significance, and methods for detection of HLA alleles within the major histocompatibility complex (MHC).
HLAallele identification could reasonably be proposed as the most complex contemporary problem in molecular diagnostics. First, more than 1300 alleles are now known to be present in worldwide populations at 12 expressed Class I and II loci. 4 The encoded polypeptides of these alleles differ from each other by one or more amino acid substitutions by what are effectively missense mutations. No other human genetic loci are as polymorphic as the HLA loci. For example, the HLA-B locus currently has more than 400 known alleles (Table 1) . Second, the steady description of new alleles plagues laboratories as allele calls made in the past for patients and donors become out of date. Third, laboratories are asked to provide allele identification at various levels of resolution for different clinical situations. High resolution allele level typing is required for unrelated bone marrow transplantation while serological or low resolution typing is adequate for renal transplantation.
Table 1.
HLA Loci and Known Alleles
| Genetic locus | Antigen or associated specificity | Number of known alleles |
|---|---|---|
| HLA-A | A1 to A80 | 214 |
| HLA-B | B7 to B81 | 425 |
| HLA-C | Cw1 to Cw10 | 108 |
| DRA | DR1 to DR18 | 2 |
| DRB1 | 289 | |
| DQA1 | DQ1 to DQ9 | 21 |
| DQB1 | 46 | |
| DPA1 | DPw1 to DPw6 | 19 |
| DPB1 | 94 |
The Major Histocompatibility Complex
The human major histocompatibility complex lies on the short arm of chromosome 6. Its complete genomic sequence is now available. 5 The MHC contains more than 200 genes and spans 4 megabases; 8 megabases if one includes the more telomeric Class I related gene HFE. 6 Among the genes within the MHC are more than 20 loci encoding proteins involved in binding and presentation of the peptide degradation products of proteins to the T cell antigen receptor. This review will focus on the most polymorphic of these loci, the Class I and II genes. However, other loci in the MHC may be polymorphic and participate at various levels in peptide antigen processing and presentation. 6
The Class I genes include HLA-A, -B, and -C and the non-classical MHC-Ib genes HLA-E, -F, and -G. HLA-E, -F, and -G are expressed at lower levels than the classical genes, do not display the extensive polymorphism of the HLA-A, -B, and -C genes, and appear to have more limited functions in the immune system. The Class II genes include 18 closely linked loci that code for the α and β chains of the class II molecules. The most clinically relevant of these are the DRA, DRB1, DQA1, DQB1, and DPA1, DPB1 genes encoding the DR, DQ, and DP heterodimers, respectively.
The Class I and the Class II genes are all members of the immunoglobulin gene family and have arisen via gene duplication and divergence from a common ancestor over time. They demonstrate significant sequence homology. This fact, and the reality that several of these genes have closely related pseudogenes with errors preventing successful transcription and translation necessitate care in designing polymerase chain reaction (PCR) primers for molecular diagnosis to ensure locus specific amplification.
Structure and Function of the Class I and II Molecules
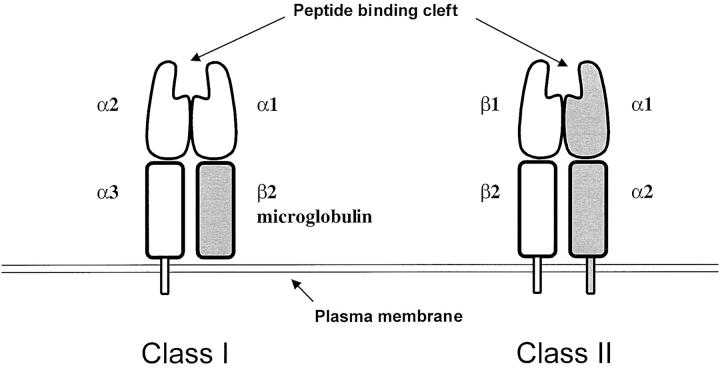
Class I and II molecules are heterodimeric, with variable extracellular and relatively constant transmembrane and intracytoplasmic domains. 7 Unlike the T cell antigen receptor and immunoglobulin genes, diversity is not achieved somatically, but through the maintenance of many alleles in populations. Any one individual will have one or two distinguishable alleles at a specific HLA locus. Class I molecules are composed of a transmembrane α chain encoded by the HLA-A, -B, or -C gene locus non-covalently bound to β-2 microglobulin, encoded by chromosome 12. The α-1 and α-2 domains contain variable amino acid sequences and form a cleft designed to bind peptides formed by protein degradation (Figure 1) .
Figure 1.
Structure of the Class I and II heterodimers.
Class II molecules are composed of an α chain non-covalently associated with a β chain encoded by the A and B gene loci, respectively, in the MHC. 7 For example the DP heterodimer is encoded by the DPA1 and DPB1 genes. For Class II, both the A and the B genes contribute to variable α-1 and β-1 domains that form a peptide binding cleft (Figure 1) .
Class I molecules are expressed in all nucleated cells and platelets, while Class II molecules are found primarily on antigen processing cells such as dendritic cells, B lymphocytes and macrophages, although they can be induced in several cell types.
The fundamental role of Class I and II molecules is to bind to self and non-self peptides and transport them to the plasma membrane of cells for recognition by the T cell antigen receptor. 8 Autoimmunity is avoided by several mechanisms to delete or suppress T cells which bind with high affinity to self peptides in the context of an individual’s Class I and II molecules. However, viral and bacterial peptides displayed by the Class I and II molecules to T cell antigen receptors generally result in an immune response. Class I molecules bind 8–10 amino acid peptides primarily resulting from proteosome degradation of cytoplasmic proteins and present these peptides to CD8+ cytotoxic T cells. Thus Class I molecules are a primary means of alerting T cells to virally infected cells. Class II molecules bind 13–25 amino acid peptides largely resulting from endosomal degradation of exogenous and endogenous proteins and present these peptides to CD4+ T helper cells. Class II molecules play an important role in eliciting immune responses to organisms such as pyogenic bacteria. CD8+ and CD4+ T cell activation by these two routes results in cell division and differentiation resulting in cellular and antibody mediated immune responses. 8
Nomenclature
Developing and maintaining a workable nomenclature for the numerous and rapidly accumulating Class I and II alleles is a challenging problem. Responsibility for developing standards for the data required to report a new allele and assignment of names for alleles rests with an international committee sponsored by the World Health Organization. 4, 9 The nomenclature for alleles is largely based on earlier serological names since the broad HLA antigen groups were originally defined based on their reaction with antisera in complement mediated microlymphocyotoxicity assays. Antisera were often isolated from women sensitized during pregnancy to the HLA specificities encoded by paternal haplotypes. Serological specificities are referred to as HLA-A1 through A80, HLA-B7 through B81; HLA-Cw1 through Cw10; HLA-DR1 through DR18, and HLA-DQ1 through DQ9 (Table 1) .
Once nucleic acid-based information regarding the sequence of alleles became available, a nomenclature complementary to the serological terms was devised (Table 2) . The first two digits of an allele name refer to the underlying serological specificity and the third and fourth digits indicate a specific allele sequence. For example, HLA-A*0205 and A*0210 are alleles encoding distinct polypeptides within the A2 serotype. These two alleles both encode the epitope recognized by the anti-A2 antisera but have 5 nucleotide differences elsewhere in exons 2–3 resulting in amino acid variations. Alleles within a serological group may vary from each other by a single or by several nucleotides. For Class II molecules, both the A and the B genes may contribute to antigen variability. Thus a DR15 serotype may be found in an individual with one of the DRB1*15 alleles such as DRB1*1501 and DRA*0101. When necessary, a fifth digit is used to identify silent polymorphisms and the sixth and seventh digits are used to denote variation occurring outside of coding regions, such as the promoter and introns. Null allele sequences which result in either no or reduced levels of functional HLA molecules because of transcription changes, aberrant RNA splicing, and frame shift and nonsense mutations, or in frame termination codons are designated by an allele number appropriate to the group and the letter N. Individuals with null alleles may have discrepancies between serological and DNA-based typing. This problem has clinical implications since an inappropriate donor may be sought if more than one laboratory using different techniques are involved in typing a recipient and potential donors.
Table 2.
HLA Allele Nomenclature
| Examples of alleles | Comment |
|---|---|
| A*24 and A*2404 | A*24 refers to any of 33 known alleles with closely related sequences encoding Class I antigens which usually react with A24 anti-sera. A*2404 is a specific allele within this group. |
| DRB1*0801 and*0805 | Differ in exon 2 at codon 74: At this position*0801 has CTG and*0805 has GCG encoding Leu and Ala, respectively. |
| A*01011 and*01012 | Differ by a silent polymorphism at codon 142: At this position*01011 has ATC and*01012 has ATT. Both encode Ile. |
| B*1501101 and*1501102N | The B*1501102N (null) allele has a 10-bp deletion near the 3′ end of intron 1. The mRNA is improperly spliced with a predicted truncated translated polypeptide. |
Nature of Class I and II Gene Polymorphism
Multiple alleles are found within most of the known serotypes, although a few serotypes (an example is DR9, DRB1*09012) are accounted for solely by a single allele. For example the B35 serotype has more than 39 alleles. HLA allele frequencies exhibit ethnic variation, with some alleles found widely distributed among populations and others almost exclusively within a particular ethnic group. The number of different phenotypes that are possible from all combinations of the known HLA alleles is greater than the earth’s population. However, the Class I and II loci reside on a relatively small region of chromosome 6 and specific haplotypes were apparently present at high frequencies in founding populations or were selected for or against by infectious organisms. In this setting, linkage disequilibrium results in a significant overrepresentation of certain haplotypes. 10 This fact makes it possible for organizations such as the National Marrow Donor Program to have a fairly high likelihood of finding a well matched donor for a Caucasian recipient with 3–4 million potential volunteer donors in its database. 11 Although HLA gene loci are closely linked, meiotic recombination may occur between loci. For example, the crossover rate is approximately 0.8% between the A and B loci and virtually zero between the DRB1 and DQB1 loci.
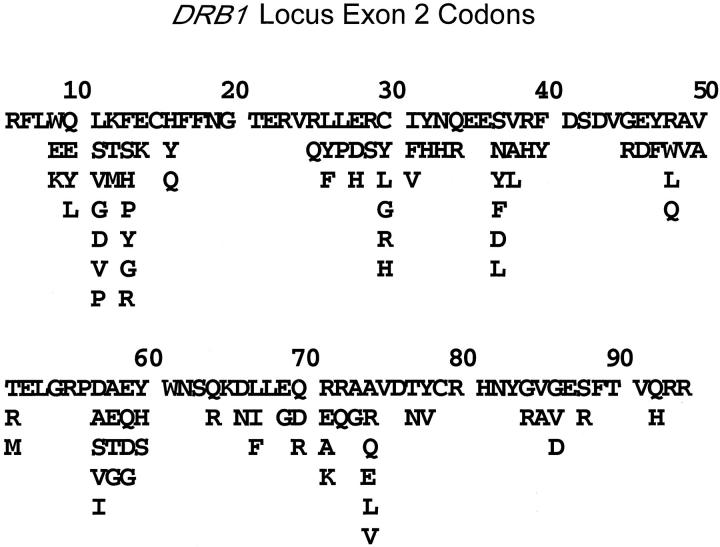
The great majority of the polymorphism found in the Class I and II genes occurs in the exons that encode the α-1 and α-2 (Class I, exons 2–3) and the α-1 and β-1 (Class II, exon 2) domains which bind processed peptides. 12 Some nucleotide positions in these exons are invariant, others may have two or three or even all four of the possible bases as possibilities (Figure 2) . Thus, some codons are constant while others display varying degrees of variability. Since the polymorphic exons are relatively short in length (about 250 nucleotides), they can easily be amplified in the PCR for molecular diagnostic studies.
Figure 2.
DRB1 Gene polymorphism. Single letter amino acid codes are shown for DRB1 exon 2 codons 6–94. About half of the positions are invariant while the remainder display polymorphism with a few codons encoding as many as seven different amino acids. For example, all DRB1 alleles have glycine encoded by position 20 while alleles may encode glycine, valine, or aspartic acid at codon 86. The 289 known DRB1 alleles arise through the many possible combinations of these polymorphisms. The polypeptide encoded by DRB1*0101 is shown on the top line.
Emergence of New Alleles
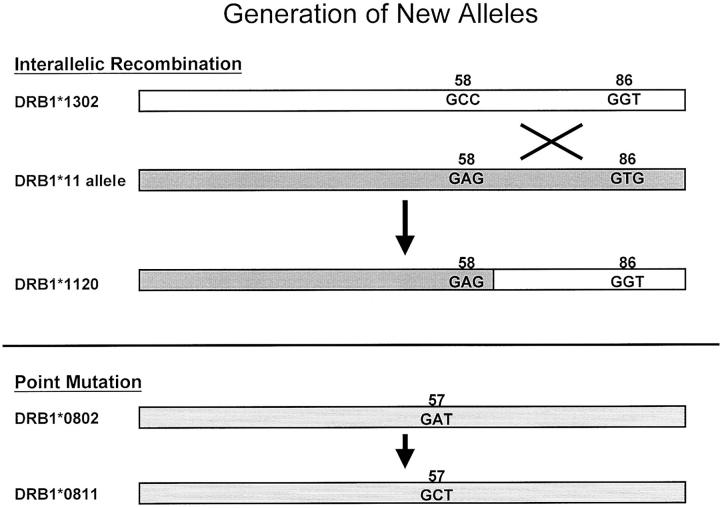
Ancestral HLA alleles that define specific serotypes likely predate the emergence of humans as a species and have correlates in primates. However, microvariation within serotypes and the creation of new alleles which are chimeras of two distinct serotypes may be more recent phenomena. For example, Bering Strait immigrants to the Americas more than 10,000 years ago probably bore the allele DRB1*0802 with high frequency. In present day native Americans, subtle variants of DRB1*0802 such as *0807 and *0811 are found as well as *0802. 13, 14 These variants are thought to have arisen in individuals with DRB1*0802 through mutations in germline cells. New alleles appear to emerge at a fairly high rate and become fixed in populations, in theory, if they provide a selective advantage in presenting peptides from infectious organisms. New alleles are generated via point mutation, recombination, and gene conversion-like events 12, 15 (Figure 3) .
Figure 3.
Class I and Class II alleles arise from existing alleles through several postulated mechanisms. 15 DRB1*1120 likely arose via interallelic recombination between DRB1*1302 and a DRB1*11 allele, 35 while DRB1*0811 probably is derived from DRB1*0802 by a point mutation. 14
Clinical Significance
Transplantation
Given the extensive polymorphism of the HLA loci and the fact that up to 10% of T cells will recognize any foreign HLA antigens with a subsequent vigorous immune response, it is remarkable that organ and stem cell transplantation can be successfully achieved. 8 Both antibody mediated and cellular responses are important in transplantation. 7 Mismatched HLA antigens may result in a strong cell mediated immune response as indicated above. These responses contribute to acute rejection in organ transplantation and graft versus host disease in stem cell transplantation. Transplant recipients may produce anti-HLA antibodies because of exposure to allogeneic HLA molecules during pregnancy, from previous transplants, and from donors of transfused blood components. Preformed anti-HLA antibodies may lead to hyperacute rejection of a donor kidney bearing the relevant HLA specificities. 16 Laboratories maintain serum screening programs and perform cross-matching tests to detect these antibodies.
Several medical and organizational strategies have made successful transplantation possible:
1. A relatively simple solution is to identify an HLA identical sibling, taking advantage of the fact that there is a one fourth chance of HLA identity with each sibling. Unfortunately, many potential transplant recipients do not have access to these donors.
2. Powerful inhibition of the immune response via direct suppression of T cell activation and other strategies have made organ transplantation possible even in the face of significant serological HLA mismatches with living related and unrelated and cadaver donors. 16
3. However, better HLA-A, -B, and -DR matching at a serological level generally increases the half life of the transplanted organ and decreases overall morbidity. 17 Thus cadaver donor kidneys are shared on a national basis with waiting recipients who have no mismatches.
4. Bone marrow transplantation poses special problems since the transfer of hematopoietic stem cells can result in graft-versus-host disease as well as rejection. Evidence is accumulating that survival decreases and the risk of severe graft-versus-host disease increases following stem cell transplantation when even subtle, allele level, donor-recipient HLA mismatches are present. 18, 19 Which mismatches are permissible and which HLA loci are relevant is still controversial and major retrospective studies are underway to address these questions. 20 In the absence of HLA identical siblings, finding donors of hematopoietic stem cells largely matched at an allele level is facilitated by the several million enrolled potential donors with known HLA types in the National Marrow Donor Program in the United States and similar programs in other countries. 11
Disease Association
A number of diseases are associated with specific HLA alleles and their encoded antigens. The pathophysiology of these autoimmune disorders is incompletely understood, although one hypothesis suggests that the Class I and II heterodimers encoded by specific HLA alleles have distinct abilities to present autoantigens to the T cell receptor in a manner leading to an aberrant immune response. 21
Some HLA allele-disease associations are quite strong and diagnostically useful. More than 95% of patients with narcolepsy have the DQB1*0602 allele, 22 while 25 to 30% of unaffected Caucasian populations have this allele. Similarly, 90% of patients with ankylosing spondylitis have B*27 alleles, while 5 to 10% of the normal population do. 21 The risk of developing ankylosing spondylitis for individuals with a B*27 group allele is about 3%, a relative risk 100-fold greater than in individuals without B*27. DQB1*0602 confers a similar magnitude risk of narcolepsy. Since these alleles are common in the population at large, HLA testing is most useful in helping to rule out narcolepsy or ankylosing spondylitis when these diagnoses are considered but the DQB1*0602 or B*27 alleles are not present.
HLA antigens associated with most other autoimmune disorders carry relative risks of disease that are lower than those seen in narcolepsy and ankylosing spondylitis. Thus, the association of DRB1*03/*04 with insulin-dependent diabetes mellitus and a shared DRB1*04 encoded epitope with severe rheumatoid arthritis have limited diagnostic utility, but provide insights into the pathological basis of these diseases. For complex disorders such as AIDS, HLA alleles may confer resistance or susceptibility to disease progression and there may be dosage effects. For example, recent studies have suggested that homozygosity at HLA loci and/or the presence of A*29, B*35-Cw*04, B*54, *55, and *56, and DRB1*11 are associated with more rapid progression, while HLA locus heterozygosity and/or the presence of B*14 and Cw*08 are associated with longer latency periods. 23, 24 This is a challenging area of inquiry given the complexity of these disorders and variations in the ethnicity of the populations studied, the subtypes of HIV-1 involved, and the definition of cases and controls.
Finally, the MHC includes genes associated with familial disorders as a consequence of the presence of pathological mutations. The CYP21 gene encodes the 21-hydroxylase enzyme involved in steroid hormone metabolism. CYP21 mutations are found in individuals with congenital adrenal hyperplasia. The HFE gene encodes a Class I-like polypeptide which associates with β-2 microglobulin. Rather than binding processed peptides, HFE interacts with the transferrin receptor to regulate iron stores. Mutations in HFE are found in genetic hemochromatosis. Because of linkage disequilibrium, individuals with congenital adrenal hyperplasia and genetic hemochromatosis have increased frequencies of specific HLA alleles. Direct CYP21 and HFE genotyping is preferred for the molecular diagnosis of these disorders. 25, 26
Methods for Detection of Class I and II Alleles
For many years, HLA antigens were detected by microlymphocytotoxicity assays originally developed by Terasaki. 27 Nucleic acid-based techniques have largely replaced serological assays because they allow identification of alleles not distinguished by serology and they are not compromised by reduced expression of HLA antigens on cells surfaces in disease states. DNA-based methods range from restriction fragment length polymorphism assays to direct DNA sequencing. Three major methods have emerged for HLA typing.
Sequence Specific Oligonucleotide Probe Hybridization (SSOPH)
Polymorphic regions of the Class I and Class II genes are amplified from genomic DNA with the PCR, using primers that anneal to 5′ and 3′ flanking regions that are conserved among individuals. Care must be taken in choosing primers that will result in roughly equal amplification of the two alleles in a heterozygous individual. Following PCR, the amplified DNA is used in standard dot-blot hybridization assays with sequence-specific oligonucleotide probes chosen to interrogate regions of polymorphism. 28 High stringency conditions allow detection of single nucleotide differences between alleles. Alleles are assigned on the basis of patterns of positive and negative hybridization reactions with oligonucleotides specific for a particular allele or sequence. SSOPH typing requires a substantial number of oligonucleotide probes to detect and distinguish among the large number of known Class I and II alleles; the exact number depends on whether allele level or serological level resolution is required. Alternatively, reverse line-blots employ multiple oligonucleotide probes specific for alleles of interest immobilized on a single membrane. 29 The development costs for reverse hybridization assays are substantial; thus these kits are generally available commercially rather than as locally developed reagants.
Sequence-Specific Primer PCR (SSP-PCR)
PCR primers are designed so that their 3′-most 1–2 nucleotides are complementary to base positions within Class I and II genes that differ for different alleles. 30 For a particular SSP-PCR, productive DNA amplification occurs if an allele perfectly complementary to the two primers chosen is present in genomic DNA. By choosing a series of primers assessing polymorphism at the relevant regions within exons 2–3 and exon 2 of the Class I and II genes, an HLA typing can be accomplished. SSP-PCR requires about 100 simultaneously performed PCR assays per patient to identify HLA-A, -B, and -DRB allele groups at serological equivalent resolution. More reactions are required if higher resolution typing is desired. In each reaction, a second set of primers directed at a non-HLA locus serves as a positive control. The presence or absence of PCR products of the correct size is then assessed by gel electrophoresis. Because of the large number of reactions and possible alleles, most laboratories use local or commercial software for analysis and calling alleles. SSP-PCR is most useful for serologic level resolution HLA typing and is widely used in laboratories supporting organ transplantation.
Direct DNA Sequencing of PCR Products
The SSOPH and SSP-PCR approaches above both suffer from the limitations of the need for large numbers of probes or primers and for frequent updating in response to newly published alleles. Direct sequencing is increasingly attractive as a general method for HLA typing. If amplification primers are carefully chosen to amplify all alleles present in a population, any known or currently unknown allele should be detectable by direct sequencing of the PCR products. Most alleles are uniquely defined by polymorphisms in exons 2–3 (Class I) or exon 2 (Class II); therefore, laboratories typically include at least these exons in their amplification products. Since the PCR approach used usually results in amplification of both alleles present at a locus, the electropherograms generated must be high quality to clearly display each position of heterozygosity. Advances in the ease of use of automated DNA-sequencing instruments, high quality DNA polymerases and fluorescent dyes for sequencing reactions, and powerful analysis software have made this method more attractive for clinical and research HLA allele identification. 31 DNA sequencing remains technically challenging and most laboratories continue to use the SSP-PCR and SSOPH methods described above. The development of non-electrophoretic methods is likely to make DNA sequencing practical for a larger fraction of clinical laboratories; however, the application of techniques such as sequencing via oligonucleotide arrays to HLA allele identification is in its infancy.
In choosing a method for HLA typing, laboratories must consider several factors. SSP-PCR and reverse SSOPH can be performed in a time (4–5 hours) appropriate for clinical situations such as cadaver renal transplantation. SSP-PCR and reverse SSOPH are useful for low and moderate typing volumes while forward SSOPH is attractive for very large clinical volumes or batched research typing. With considerable effort and numerous primers and probes, SSP-PCR and SSOPH can achieve allele level resolution; however, direct DNA sequencing is the most general and precise means of high resolution typing. Finally, serological typing remains useful for correlation of serological results with DNA-based results and in investigating the possibility of the presence of null alleles. 32
Assessing the Accuracy of HLA Typing Methods
The complexity of HLA allele identification necessitates care to ensure high quality results. Detailed and frequently updated standards for DNA-based HLA typing are maintained by the American Society for Histocompatibility and Immunogenetics (ASHI). 33 The joint College of American Pathologists (CAP)/ASHI and the South Eastern Organ Procurement Foundation programs offer proficiency testing to laboratories. More than 130 laboratories currently participate in the CAP/ASHI nucleic acid-based Class I and II surveys. Laboratories may type challenge samples at either serological equivalent or allele level resolution. In general, more than 95% of laboratories are able to accurately identify the intended types in each sample. The most difficult aspect of the surveys appears to be the allele level Class I challenges. The rapidly increasing number of known Class I alleles poses difficulties for laboratories attempting to unambiguously identify alleles at these loci.
To mimic the clinical testing environment, whole blood samples are provided to laboratories for proficiency testing. This requirement makes it logistically difficult to provide survey samples that test the ability of laboratories to detect unusual alleles or difficult heterozygous combinations. A number of laboratories participate in the University of California at Los Angeles DNA Exchange. This educational exchange provides DNA samples for testing representing a spectrum of common, rare, and newly described alleles from an extensive collection of cells. 34
Summary
Histocompatibility laboratories identify HLA alleles with technology that is similar to that used for other purposes in the field of molecular pathology. Few other molecular diagnostic problems approach the complexity of HLA allele identification in the setting of more than 1300 known alleles. Yet unambiguous definition of alleles is required for applications such as unrelated bone marrow transplantation and disease association studies. The continued development of automated methods for non-gel electrophoretic means of rapidly and reliably performing DNA sequencing will be of obvious interest to histocompatibility laboratories.
Address reprint requests to Thomas M. Williams, Department of Pathology, University of New Mexico Health Sciences Center, Room 337-BMSB, 915 Camino de Salud, NE, Albuquerque, NM 87131. E-mail: twilliams@salud.unm.edu.
References
- 1.Li B, Hartono C, Ding R, Sharma VK, Ramaswamy R, Qian B, Serur D, Mouradian J, Schwartz JE, Suthanthiran M: Noninvasive diagnosis of renal-allograft rejection by measurement of messenger RNA for perforin and granzyme B in urine. N Engl J Med 2001, 344:947-954 [DOI] [PubMed] [Google Scholar]
- 2.Gebel HM, Bray RA: Sensitization and sensitivity: defining the unsensitized patient. Transplantation 2000, 69:1370-1374 [DOI] [PubMed] [Google Scholar]
- 3.Bray RA: Flow cytometry crossmatching for solid organ transplantation. Methods Cell Biol 1994, 41:103-119 [DOI] [PubMed] [Google Scholar]
- 4.European Bioinformatics Institute, http://www.ebi.ac.uk/imgt/hla
- 5.Sanger Centre, http://www.sanger.ac.uk/HGP/Chr6/MHC/shtml
- 6.Rhodes DA, Trowsdale J: Genetics and molecular genetics of the MHC. Rev Immunogenet 1999, 1:21-31 [PubMed] [Google Scholar]
- 7.McCluskey J, Peh CA: The human leukocyte antigens and clinical medicine: an overview. Rev Immunogenet 1999, 1:3-20 [PubMed] [Google Scholar]
- 8.Janeway CA, Travers P: Antigen recognition by T lymphocytes. ed 3 Immunobiology, 1997, :pp 41-46 Garland Publishing, New York and London [Google Scholar]
- 9.Bodmer JG, Marsh SG, Albert ED, Bodmer WF, Bontrop RE, Dupont B, Erlich HA, Hansen JA, Mach B, Mayr WR, Parham P, Petersdorf EW, Sasazuki T, Schreuder GM, Strominger JL, Svejgaard A, Terasaki PI: Nomenclature for factors of the HLA system. Tissue Antigens 1998, 53:407-446 [DOI] [PubMed] [Google Scholar]
- 10.Alper CA, Awdeh Z, Yunis EJ: Conserved, extended MHC haplotypes. Exp Clin Immunogenet 1992, 9:58-72 [PubMed] [Google Scholar]
- 11.Kernan NA, Bartsch G, Ash RC, Beatty PG, Champlin R, Filipovich A, Gajewski J, Hansen JA, Henslee-Downey J, McCullough J: Analysis of 462 transplantations from unrelated donors facilitated by the National Marrow Donor Program. N Engl J Med 1993, 328:593-602 [DOI] [PubMed] [Google Scholar]
- 12.Little A-M, Parham P: Polymorphism and evolution of Class I and II genes and molecules. Rev Immunogenet 1999, 1:105-123 [PubMed] [Google Scholar]
- 13.Mack SJ, Erlich HA: HLA class II polymorphism in the Ticuna of Brazil: evolutionary implications of the DRB1*0807 allele. Tissue Antigens 1998, 51:41-50 [DOI] [PubMed] [Google Scholar]
- 14.Williams TM, Wu J, Foutz T, McAuley J, Troup GM: A new DRB1 allele (DRB1*0811) identified in Native Americans. Immunogenetics 1994, 40:314. [DOI] [PubMed] [Google Scholar]
- 15.Marsh SGE, Parham P, Barber LD: Evolution and anthropology of HLA. The HLA Facts Book. 2000, :pp 73-83 Academic Press, London [Google Scholar]
- 16.Suthanthiran M, Strom TB: Medical progress: renal transplantation. N Engl J Med 1994, 331:365-376 [DOI] [PubMed] [Google Scholar]
- 17.Takemoto SK, Terasaki PI, Gjertson DW, Cecka JM: Twelve years’ experience with national sharing of HLA-matched cadaveric kidneys for tansplantation. N Engl J Med 2000, 343:1078-1084 [DOI] [PubMed] [Google Scholar]
- 18.Petersdorf EW, Longton GM, Anasetti C, Martin PJ, Mickelson EM, Smith AG, Hansen JA: The significance of HLA-DRB1 matching on clinical outcome after HLA-A, B, DR identical unrelated donor marrow transplantation. Blood 1995, 86:1606-1613 [PubMed] [Google Scholar]
- 19.Sasazuki T, Juji T, Morishima Y, Kinukawa N, Kashiwabara H, Inoko H, Yoshida T, Kimura A, Akaza T, Kamikawaji N, Kodera Y, Takaku F: For the Japan Marrow Donor Program: Effect of matching of Class I alleles on clinical outcome after transplantation of hematopoietic stem cells from an unrelated donor. N Engl J Med 1998, 339:1177-1185 [DOI] [PubMed] [Google Scholar]
- 20.Hurley CK, Baxter-Lowe LA, Begovich AB, Fernandez-Vina M, Noreen H, Schmeckpeper B, Awdeh Z, Chopek M, Salazar M, Williams TM, Yunis E, Kitajima D, Kollman C, Shipp K, Splett J, Winden T, Johnson D, Ng J, Hartzman RJ, Hegland J: The extent of HLA Class II allele level disparity in unrelated bone marrow transplantation: analysis of 1,259 national marrow donor program donor-recipient pairs. Bone Marrow Transplant 2000, 25:385-393 [DOI] [PubMed] [Google Scholar]
- 21.HLA and Disease. Edited by R Lechler. London, Academic Press, 1994, pp 1–186
- 22.Pelin Z, Guilleminault C, Risch N, Grumet FC, Mignot E: HLA-DQB1*0602 homozygosity increases relative risk for narcolepsy but not disease severity in two ethnic groups: US Modafinil in Narcolepsy Multicenter Study Group. Tissue Antigens 1998, 51:96-100 [DOI] [PubMed] [Google Scholar]
- 23.Carrington M, Nelson GW, Martin MP, Kissner T, Vlahov D, Goedert JJ, Kaslow R, Buchbinder S, Hoots K, O’Brien SJ: HLA and HIV-1: Heterozygote advantage and B*35-Cw*04 disadvantage. Science 1999, 283:1748-1752 [DOI] [PubMed] [Google Scholar]
- 24.Hendel H, Caillat-Zucman S, Lebuanec H, Carrington M, O’Brien S, Andrieu J-M, Schachter F, Zagury D, Rappaport J, Winkler C, Nelson GW, Zagury J-F: New Class I and II HLA alleles strongly associated with opposite patterns of progression to AIDS. J Immunol 1999, 162:6942-6496 [PubMed] [Google Scholar]
- 25.White PC, Speiser PW: Congenital adrenal hyperplasia due to 21-hydroxylase deficiency. Endocrine Rev 2000, 21:245-291 [DOI] [PubMed] [Google Scholar]
- 26.Press RD, Flora K, Gross C, Rabkin JM, Corles CL: Hepatic iron overload. Direct HFE (HLA-H) mutation analysis vs quantitative iron assays for the diagnosis of hereditary hemochromatosis. Am J Clin Pathol 1998, 109:577-584 [DOI] [PubMed] [Google Scholar]
- 27.Terasaki PI, McClelland JD: Microdroplet assay of human serum cytotoxins. Nature 1964, 204:998-1000 [DOI] [PubMed] [Google Scholar]
- 28.Ng J, Hurley CK, Carter C, Baxter-Lowe LA, Bing D, Chopek M, Hegland J, Lee TD, Li TC, Hsu S, KuKuruga D, Mason JM, Monos D, Noreen H, Rosner G, Schmeckpeper B, Dupont B, Hartzman RJ: Large-scale DRB and DQB1 oligonucleotide typing for the NMDP registry: progress report from year 2. Tissue Antigens 1996, 47:21-26 [DOI] [PubMed] [Google Scholar]
- 29.Saiki RK, Walsh PS, Levenson CH, Erlich HA: Genetic analysis of amplified DNA with immobilized sequence-specific oligonucleotide probes. Proc Natl Acad Sci USA 1989, 86:6230-6234 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Olerup O, Zetterquist H: HLA-DR typing by PCR amplification with sequence specific primers (PCR-SSP) in 2 hours: an alternative to serological DR typing in clinical practice including donor-recipient matching in cadaveric transplantation. Tissue Antigens 1992, 39:225-235 [DOI] [PubMed] [Google Scholar]
- 31.Wu J, Griffith BB, Bassinger S, Moehlenkamp C, Brodie SG, Wu Y, Gribble GG, Troup GM, Williams TM: Strategies for unambiguous detection of allelic heterozygosity via direct DNA sequencing of PCR products: application to the HLA DRB1 locus. Mol Diagn 1996, 1:89-98 [DOI] [PubMed] [Google Scholar]
- 32.Schreuder GMT, Hurley CK, Marsh SGE, Lau M, Maiers M, Kollman C, Noreen H (on behalf of multiple contributors): The HLA dictionary 1999, a summary of HLA-A:-B, -C, -DRB1/3/4/5, -DQB1 alleles and their association with serologically defined HLA-A, -B, -C, -DR, and -DQ antigens. Tissue Antigens 1999, 54:409–437 [DOI] [PubMed]
- 33.American Society for Histocompatibility and Immunogenetics, http://www.ashi-hla.org
- 34.Lau M, Terasaki PI, Park MS: International Cell Exchange. Clin Transplants 1994, 1994:467-488 [PubMed] [Google Scholar]
- 35.Cizman BB, McKean ME, Kearns DJ, Heron SD, Wu J, Griffith BB, Argyros EG, Kamoun M, Zmijewski CM, Williams TM, Monos DS: DRB1*1120 allele; another example of the transition between the DR11 and DR13 families of alleles. Tissue Antigens 1996, 48:52-54 [DOI] [PubMed] [Google Scholar]