Abstract
We performed microsatellite analysis at chromosomal regions frequently altered in head and neck squamous carcinoma on matched saliva and tumor samples from 37 patients who had oral squamous carcinoma. The results were correlated with the cytologic findings and traditional clinicopathologic factors to assess the diagnostic and biological potential of these markers. Our data showed that 18 (49%) of the saliva samples and 32 (86%) of the tumors had loss of heterozygosity (LOH) in at least one of the 25 markers studied. In saliva, the combination of markers D3S1234, D9S156, and D17S799 identified 13 (72.2%) of the 18 patients with LOH in saliva (P < 0.001). For tumors, markers D3S1234, D8S254, and D9S171 together identified 27 (84.3%) of the 32 tumors with LOH at any of the loci tested (P < 0.001). Eleven (55%) of the 20 saliva samples with cytologic atypia and seven (35%) of the 17 specimens without atypia had LOH. Significant correlation between LOH in tumor at certain markers and smoking and alcohol use was found. Our results indicate that: 1) epithelial cells in saliva from patients with head and neck squamous tumorigenesis provide suitable material for genetic analysis; 2) combined application of certain markers improves the detection of genetic alteration in these patients; 3) clonal heterogeneity between saliva and matching tumor supports genetic instability of the mucosal field in some of these patients; and 4) LOH at certain chromosomal loci appears to be associated with smoking and alcohol consumption.
The oral cavity is an ideal site for screening individuals at high risk of developing head and neck squamous carcinoma (HNSC) because of the availability of cells shed in saliva and the convenience of visualizing and sampling lesions at these locations. 1, 2, 3 However, little progress has been made in the management of patients with oral squamous carcinoma due mainly to the nonspecific symptoms, minimal physical finding inpatients with early-stage cancer, and the lack of biological predictors of progression. 4 Identifying novel and reliable biogenetic markers for the biological assessment of squamous lesions may assist in early diagnosis and treatment of head and neck squamous tumorigenesis. Microsatellite DNA motifs consisting of highly polymorphic short tandem repeat sequences distributed throughout the genome have been widely and successfully used as markers for molecular analysis of tumorigenesis in head and neck and other neoplasms. 1, 5, 6, 7
Studies using microsatellite markers from different chromosomal arms in HNSC have shown that alterations at certain regions on chromosomes 3p, 9p, 17p and 18q to be associated with the development of these tumors. 1, 2, 6, 8, 9, 10, 11, 12 Although the timing and the order of these alterations are currently unknown, different studies have shown high incidence of loss of heterozygosity (LOH) in noninvasive lesions indicating an early association with tumorigenesis. 1, 6, 7 Analysis of selected microsatellite markers at these regions on epithelial cells from patients’ saliva is a convenient and non-invasive approach for molecular screening and early detection of this disease. 13, 14, 15, 16, 17, 18, 19, 20, 21, 22, 23, 24 In this study, we evaluated the diagnostic and biological implications of the alterations at several of the above mentioned microsatellite markers in prospectively collected saliva and tumor specimens from patients who had oral squamous cell carcinoma.
Materials and Methods
Forty matched histologically normal squamous mucosa and tumor tissues and saliva collected freshly from patients with primary untreated HNSC were prospectively acquired between 1996 and 1998 by the Department of Pathology at The University of Texas M. D. Anderson Cancer Center after obtaining patient consent. Ten saliva samples from normal individuals with negative history of cancer were also collected and used as a biological control. Histologically normal squamous mucosa from each cancer specimen was stripped from the farthest margin after frozen section evaluation and used as putative controls. Ficoll-Hypaque isolated lymphocytes from heparinized peripheral blood from all patients and 10 normal volunteer individuals were also obtained and immediately frozen; lymphocytes were used as control for saliva analysis from normal volunteers. Lymphocytes were used only if histologically matched mucosa was suspected to harbor alterations.
For all patients, tobacco and alcohol consumption histories were obtained from the epidemiology database. Patients’ demographic, pathological, and clinical information was collected retrospectively from pathology reports and patient records. The 10 normal saliva samples from healthy individuals were obtained from five nonsmokers and five current smokers.
Saliva Collection
After the mouth of each patient and normal subject was washed with sterile water, saliva specimens were collected in a sterile cap and transported immediately to the laboratory to be centrifuged at 1200 g for 5 minutes. The supernatants were decanted and the cell pellets were frozen at −80° in the same tubes. Three cases were eliminated for lack of sufficient DNA from saliva samples and the remaining 37 cases formed the cohort of materials for the analysis.
DNA Extraction
Extraction of DNA was performed using DNAzol (Molecular Research Center, Cincinnati, OH). Cells were lysed in DNAzol using a Tissue Tearor (Biospec Products, Bartlesville, OK) for fresh tissue, and a vortex homogenizer for saliva. The liberated DNA was precipitated with ethanol and resuspended in 10 mmol/L Tris, 1 mmol/L EDTA.
Microsatellite Analysis
Aliquots of DNA were subjected to standard polymerase chain reaction (PCR) analysis using primers for the following loci: D3S656, D3S1293, D3S1234, D3S1217, D3S1261, D8S254, D8S261, LPL-tet, D8S298, D8S283, D9S104, D9S156, D9S168, D9S171, D9S199, D17S513, TP53, D17S799, CHRNB1, D17S122, D18S46, D18S363, D18S35, D18S39, and D18S41 (Research Genetics, Huntsville, AL). The loci were chosen based on the frequency of their alteration in head and neck tumors. Microsatellite location on the chromosomal arms was determined based on the latest map in the Genetic Location Database of the University of Southampton, U. K.
One primer was end-labeled using γ32P-ATP and T4 polynucleotide kinase. PCR was performed in a 20 μl volume with 10 ng of genomic DNA, 10 mmol/L Tris-HCL (pH 8.3), 50 mmol/L KCl, 2.5 mmol/L MgCl2, 0.001% gelatin, 0.5 μmol/L of each unlabeled primer, 0.01 μmol/L labeled primer, 0.2 mmol/L dNTPs, 5% dimethyl sulfoxide (Sigma Chemical, St. Louis, MO), and 0.5 units of Amplitaq Gold DNA polymerase (Perkin-Elmer, Norwalk, CT).
The amplification process consisted of: (a) an initial 10-minute denaturation step at 94°; (b) 35 cycles of denaturation at 94° for 30 seconds, annealing at 55–60° for 1 minute, and elongation at 72° for 1 minute; and (c) a final elongation at 72° for 5 minutes. Sequencing stop buffer was added to the reactions. PCR products were then denatured at 94° for 5 minutes and quickly chilled, and 4–6 μl was loaded on 7% acrylamide-urea sequencing gels containing 32% formamide. Electrophoresis was performed at 80 W for 2–4 hours, depending on the fragment size. The gels were dried and exposed to Hyperfilm-MP (Amersham, Arlington Heights, IL).
Evaluations were conducted by two independent observers who visually scored the pattern and the band intensity between normal tissue, tumor, and saliva specimens from each case before the identification of the patients. LOH was defined by the presence of an allelic band difference between nonmalignant epithelium and tumor or saliva of more than 50%. Visually suspicious cases were subjected to densitometry, and a difference of >30% was scored as LOH. Instability was defined as the appearance of a novel band that was not seen in the normal control.
Acridine Orange Flow Cytometry
Disaggregated cells were adjusted to 1.0 × 106 cells/ml. A cytospin preparation was evaluated for quality and cellular integrity. Cells were subsequently stained with acridine orange according to the two-step procedure of Traganos et al. 25 Ploidy status was defined by the DNA index, which represents the ratio of the relative G0/G1 stemline portion of the tumor samples to that of the normal peripheral blood lymphocytes. Diploid DNA is defined by a single G0/GI peak with a DNA index of 1.0, and DNA aneuploidy is defined by the presence of one or more additional stemlines to the right (hyperdiploid, DI > 1.0) or the left (hypodiploid, <1.0) of the G0/G1 diploid peak. A near diploid (hypodiploid or hyperdiploid) DI was determined after mixing the test sample with lymphocyte controls. The coefficient of variation (CV) of DNA diploid and aneuploid stemline ranged from 2.1 to 5.4 with a mean of 3.6 ± 1.2 and 2.8 to 6.1 with a mean of 4.9 ± 1.6, respectively.
Statistical Methods
All correlations were performed using a two-tailed Fisher’s exact test.
Results
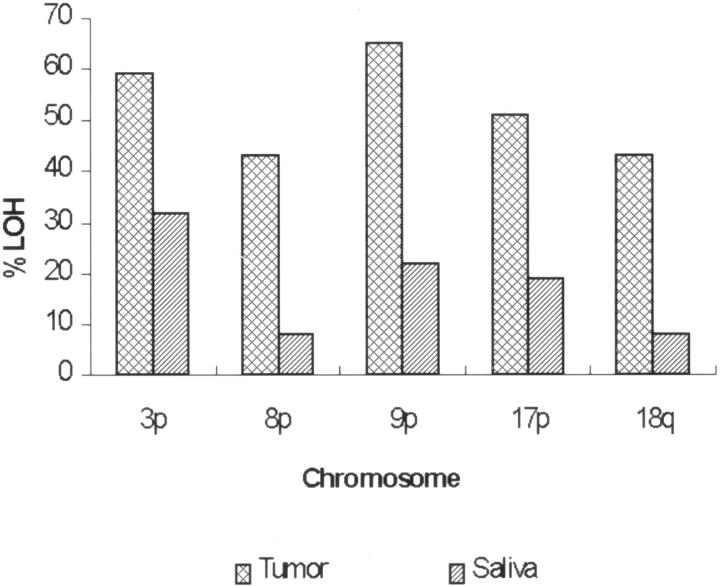
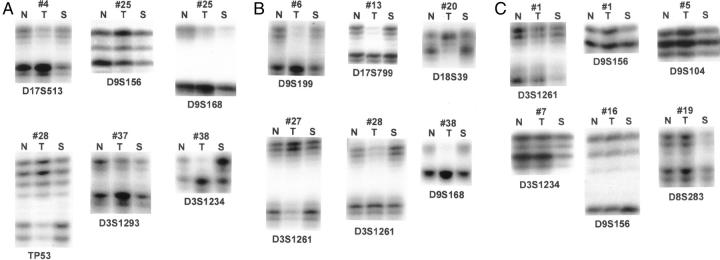
Table 1 presents the clinicopathologic and epidemiological characteristics of the 37 study subjects. None of the histologically normal squamous mucosas used as a control showed any alterations at the microsatellite sites tested. Accordingly, no further analysis of the lymphocytes from these patients were performed. The incidence of LOH at individual markers and its occurrence per chromosomal arm for both tumor and corresponding saliva samples are presented in Table 2 and Figure 1 . Chromosomes 9p, 3p, and 17p showed the highest incidence of LOH in both tumor and saliva. We also investigated the use of small combinations of markers to improve the detection of genetic alterations in both saliva and tumor samples as a likely approach for future clinical applications. Figure 2 illustrates LOH found in tumor and saliva (A), tumor alone (B), and saliva alone (C).
Table 1.
Clinicopathological and Epidemiological Characteristics of Patients with Oral Squamous Carcinoma
| Case no.* | Sex | Grade | Site | Size (cm) | Stage | Sm† | Alcohol HX | DI | PI |
|---|---|---|---|---|---|---|---|---|---|
| 1 | M | MD | Tongue | 4.9 | II | Yes | Yes | 1 | 7 |
| 2 | F | PD | FOM | n/a | U | No | No | 1 | 10 |
| 3 | M | MD | Palate | 4 | I | Yes | Yes | 1 | 13 |
| 4 | M | WD | Tongue | 2.5 | II | Quit | Yes | 1.67 | 6 |
| 5 | M | WD | Tongue | 2.5 | II | Quit | Quit | 1.82 | 9 |
| 6 | F | PD | Palate | 5.5 | U | Yes | None | 1 | 3 |
| 7 | M | MD | Tongue | 2 | II | Quit | None | 1.82 | 17 |
| 8 | F | MD | Tongue | 0.9 | III | Yes | Social | 1.9 | 8 |
| 9 | M | MD | Tongue | 4 | III | Yes | Social | 1 | 5 |
| 11 | M | MD | Tongue | n/a | III | Yes | Social | 1 | 13 |
| 12 | M | MD | Tongue | 4.6 | III | Yes | Quit | 1 | 19 |
| 13 | M | MD | FOM | 2.2 | III | Yes | Yes | 1.71 | 27 |
| 14 | M | MD | Tongue | 4.5 | II | Yes | Yes | 1 | 22 |
| 15 | M | MD | Tongue | 1.5 | U | Unknown | Social | 1.35 | 17 |
| 16 | F | WD | FOM | 4.5 | II | No | No | 1 | 6 |
| 18 | M | MD | Tongue | 2 | II | Yes | Yes | 1.72 | 10 |
| 19 | M | PD | Tongue | 4.5 | III | Yes | No | 1 | 8 |
| 20 | M | MD | Tongue | n/a | III | Yes | Quit | 1.58 | 16 |
| 22 | M | MD | FOM | 4 | U | Yes | Quit | n/a | n/a |
| 23 | M | PD | Tongue | 1.7 | U | Quit | Yes | 1.73 | 24 |
| 24 | M | PD | Tongue | 0.7 | I | Quit | Yes | n/a | n/a |
| 25 | M | PD | FOM | 8 | III | No | No | 1 | n/a |
| 26 | M | MD | Tongue | 2 | IV | No | Social | 1 | 9 |
| 27 | M | PD | FOM | n/a | III | Quit | Quit | n/a | n/a |
| 28 | M | MD | FOM | n/a | II | Yes | Yes | n/a | n/a |
| 29 | F | MD | FOM | n/a | III | Quit | Yes | n/a | n/a |
| 30 | F | MD | Tongue | n/a | II | No | Social | n/a | n/a |
| 31 | M | MD | Tongue | 1.5 | II | Yes | Social | n/a | n/a |
| 32 | M | MD | Tongue | 1.8 | III | Quit | Social | 1 | 5 |
| 33 | M | MD | Tongue | 2.2 | II | No | Social | 1 | 14 |
| 34 | F | MD | Tongue | n/a | II | No | No | n/a | n/a |
| 35 | M | PD | Gingiva | 2.7 | III | Quit | Unknown | 1 | 4 |
| 36 | F | MD | Tongue | 1.5 | II | Yes | Social | n/a | n/a |
| 37 | M | PD | Tongue | 3.5 | III | Yes | Quit | 1.66 | 6 |
| 38 | F | PD | Tongue | 2 | IV | Yes | Social | 2.33 | 12 |
| 39 | F | MD | Tongue | n/a | II | No | No | n/a | n/a |
| 40 | M | WD | FOM | n/a | I | Quit | Social | 1 | 5 |
Cases 10, 17, and 21 were dropped during the study for lack of sufficient DNA to complete the study.
Quit: ceased smoking for at least one year and no current drinking; Yes: alcohol >7 drinks/week; smoking, at least 1 pack/day.
M, male; F, female; WD, well differentiated; MD, moderately differentiated; PD, poorly differentiated; FOM, floor of mouth; U, unknown; DI, DNA index; PI, proliferative index; Sm, smoking; n/a, not available.
Table 2.
Incidence of LOH in Heterozygous Cases (%) of Saliva and Tumor Specimens of Patients with HNSC
| Marker | Heterozygous (informative) | Tumor only | Saliva only | Tumor and Saliva |
|---|---|---|---|---|
| D3S656 | 24 | 10 | 0 | 2 |
| D3S1293 | 32 | 5 | 2 | 2 |
| D3S1234 | 23 | 7 | 3 | 5 |
| D3S1217 | 33 | 11 | 2 | 2 |
| D3S1261 | 33 | 12 | 2 | 1 |
| D8S254 | 26 | 11 | 1 | 0 |
| D8S261 | 23 | 9 | 0 | 0 |
| LPL-tet | 28 | 9 | 0 | 1 |
| D8S298 | 26 | 7 | 0 | 0 |
| D8S283 | 30 | 7 | 1 | 0 |
| D9S104 | 31 | 12 | 1 | 1 |
| D9S156 | 28 | 16 | 3 | 1 |
| D9S168 | 23 | 11 | 2 | 1 |
| D9S171 | 29 | 16 | 2 | 1 |
| D9S199 | 32 | 13 | 0 | 1 |
| D17S513 | 27 | 10 | 0 | 2 |
| TP53 | 33 | 9 | 1 | 1 |
| D17S799 | 29 | 9 | 1 | 2 |
| CHRNB1 | 29 | 9 | 1 | 1 |
| D17S122 | 26 | 7 | 0 | 1 |
| D18S46 | 31 | 11 | 1 | 1 |
| D18S363 | 20 | 7 | 0 | 0 |
| D18S35 | 28 | 9 | 0 | 1 |
| D18S39 | 22 | 10 | 0 | 0 |
| D18S41 | 23 | 6 | 0 | 0 |
Figure 1.
Frequency of LOH by chromosomal arm in tumor and saliva samples.
Saliva
Of all 37 saliva specimens, 18 samples (49%) manifested LOH, and 19 samples (51%) lacked any abnormalities. The most frequent LOH (21.6%) was found at marker D3S1234 (8 cases), but adding markers D17S799, and D9S156 (13 35%) of the 37 cases analyzed showed LOH which led to 100% specificity and 44% sensitivity of LOH detection. Eleven instances of instability were identified in saliva from three cases. None of the 10 informative saliva samples from healthy individuals showed any LOH at the markers used.
Tumor
Thirty-two (86%) tumors showed LOH in at least one marker, and five (14%) had no LOH. The two markers that exhibited LOH most frequently in tumor tissue were D9S171 and D9S156, which identified 17 (53%) incidences of LOH each. The combination of markers D9S171 + D3S1234 identified 24 (75%) of the 32 tumors with LOH (P = 0.051). Addition of marker D8S254 to the above combination identified three more instances of LOH for a total of 27 cases (P = 0.009). This marker combination gave 84% sensitivity and 100% specificity of LOH detection. Various other combinations of markers, especially those of D9S171, D3S1234, D8S254, and D9S156, identified similar incidence of tumors with LOH. Four incidences of instability were also noted in tumors from four different patients.
Saliva versus Tumor
Sixteen tumors from the 18 patients with LOH in saliva samples (in one or more of the 25 markers) had LOH in corresponding tumors. Also, 16 tumors from the 19 patients with no LOH in saliva samples had LOH (P = 1.00, Fisher’s exact test); the occurrence of LOH in saliva did not predict the LOH in corresponding tumor samples. One pair of markers (D3S1234 + D9S199) showed a statistically significant correlation in LOH between saliva and tumor (P = 0.038), as did many sets of three markers. The majority of these sets of three markers showed high agreement between saliva and tumor (LOH in both saliva and tumor or lack of LOH in both saliva and tumor). However, there was no significant correlation between LOH in saliva and in tumor for individual markers. In three instances, reciprocal LOH was noted between saliva and matching tumor specimens (Figure 2A) . In these instances, the finding was considered discordant. No concomitant instability was found in any saliva and tumor samples manifesting this feature.
Figure 2.
Representative illustrations of LOH in specimens. A: LOH in both tumor and saliva; B: LOH in tumor alone; C: LOH in saliva alone. N, normal; T, tumor; S, saliva.
Cytologic Atypia and LOH in Saliva
No malignant cells were identified in any of the saliva samples by cytologic examination of giemsa-stained slide preparation. Twenty (54%) of the 37 saliva specimens had cytologic atypia. Nine (45%) of the 20 cases with cytologic atypia had no alterations, and 11 of the remaining (55%) had LOH; 7 (41%) of the 17 samples with cytologically normal epithelial cells had LOH in at least one marker (P = 0.79). Although no correlation between LOH at single marker and atypia was found, two markers showed the highest association with cytological atypia (D9S156 and D9S168, P = 0.132 and P = 0.217, respectively). Combining either of these markers with D3S1261 showed statistical correlation between LOH and atypia (P = 0.02) with 60% and 100% specificity (P = 0.020). The inclusion of a third marker did not improve the correlation.
LOH and Clinicopathological Factors
A correlation between certain marker combinations and smoking history at four markers alone or in various combinations (D3S1293, CHRNB1, D8S298, and D9S104) showed correlation with this feature. This and other quadruple combinations of markers identified LOH in 15 of the 18 smokers (P = 0.0001). Interestingly, patients who quit smoking showed an LOH frequency that was between that of smokers and nonsmokers: five of the 10 patients who quit smoking had LOH in at least one of these markers in their tumors. Multiple markers in tumors correlated with stage (e.g., D3S1261, D8S283, D9S156, D9S168, and TP53); the combination with the highest correlation was D3S1261, D9S156, and TP53 (P = 0.002). Similarly, the combination D18S363 and D8S283 was significantly correlated with the DNA index (P = 0.002). The only factor significantly correlated with LOH at marker D3S1248 in saliva was the DNA index (P = 0.014).
Saliva from Smoking and Nonsmoking Normal Volunteers
All of the 10 normal saliva samples were informative for at least one of the markers used. None of the DNA extracted from the 10 saliva specimens from normal nonsmoking and smoking individuals manifested any LOH at the markers analyzed. Only one saliva specimen from a normal smoker showed instability at one marker on the short arm of chromosome 3.
Discussion
Molecular genetic studies of HNSC have demonstrated frequent genetic alterations at certain chromosomal regions in premalignant lesions and invasive tumors. 1, 2, 3, 4, 5, 6, 7, 8, 9 The studies also showed that certain regions on chromosomes 3p, 9p, 8p, and 17p are frequently altered in dysplastic lesions 1, 6, 7, 8 and may constitute an early event in lesion development. Analysis of these markers in oral secretions and other accessible specimens may allow for rapid, inexpensive, and objective assessment of the genetic abnormalities at these sites for early detection, screening of individuals at high risk, and follow-up of patients with cancer. 2, 3, 26
In this study, approximately 50% of the saliva samples and 86% of the tumor specimens manifested microsatellite LOH. The incidence of LOH in saliva, however, could have been higher had a method to enrich the epithelial cells in specimens been used. 27 The results, however, support those of previous studies and further underscore the early association of these markers with HNSC tumorigenesis. 1, 2, 6, 7 The genetic heterogeneity between saliva negative for malignant cells and corresponding tumor specimens in certain cases, highlight the presence of genetic alterations in the squamous epithelial cells lining the oral cavity exclusive of the cancer site. 28, 29, 30, 31, 32, 33, 34
We, however, found no LOH in any of the histologically normal squamous mucosas used as control and we attribute this to the relatively small contribution of the squamous epithelium relative to the subepithelial elements in the tissues used. Microdissection of epithelial cells may have led to the identification of LOH in these histologically nondysplastic squamous mucosa as reported in at high risk patients 7, 34 studied by our group. Nonetheless, the finding of LOH in saliva samples lacking malignant cells on cytologic evaluation, lends further credence to the field cancerization hypothesis and the increased risk of a second primary cancer developing in these patients 7, 26, 35, 36, 37 . Although none of the patients in this study manifested evidence of recurrence or secondary tumors thus far, a longer follow-up period is required to substantiate this notion. Other studies, however, have shown a high concordance in matched secretion and tumor specimens, indicating common clonal derivation 16, 17, 20, 23, 24, 38, 39 . In these studies, however, malignant cells were identified and analyzed in the cellular sources used.
In our study, although no cytological evidence of malignancy in any of the saliva samples was identified, cytologic atypia was found in more than 50% of the specimens. This feature correlated significantly with LOH at two markers on the short arm of chromosomes 3 and 9. Our findings, along with those from previous studies of head and neck, lung, and bladder tumors indicate that combining molecular and cytologic analyses in secretions may provide additional information for better identification of high risk patients 5, 7, 8, 9, 20, 39, 40 . Our results also show that alterations at microsatellite markers in tumors correlated significantly with aggressive clinicopathologic factors in these patients. A similar correlation has been reported in other studies, indicating that alterations in certain chromosomal loci are associated with the pathobiologic characteristics of these tumors and may be used for the biological assessment of these tumors. 10, 11 Of particular interest in our study is the finding of a significant correlation between LOH at certain chromosomal regions in tumor and smoking and alcohol consumption. Additional investigations of these regions may lead to identifying the markers or genes that are associated with carcinogenesis. Previous studies have also reported an association between LOH in these same chromosomal loci and tumors in the upper respiratory tract. 41, 42
In conclusion, the results of our study indicate that, in patients with HNSC, saliva is a readily available source of DNA for genetic analysis. 42, 43 Enriching for epithelial cells may increase the correlation obtained between saliva and tumor specimens. Alternatively, cells obtained by buccal brushing of multiple sites in the oral cavity may be used as a noninvasive substitute for molecular analysis. 26, 44 Our results lend molecular evidence for field cancerization 45, 46 and the use of selective genetic markers in early detection of HNSC in people at high risk. 24, 43, 47
Acknowledgments
The authors thank Sue M. Martinez for typing the manuscript.
Address reprint requests to Adel K. El-Naggar, M.D., Ph.D., Department of Pathology, The University of Texas M. D. Anderson Cancer Center, 1515 Holcombe Blvd., Box 85, Houston, TX 77030. E-mail: anaggar@notes.mdacc.tmc.edu.
Footnotes
Supported in part by Oral Cancer Center of Excellence Grant 1P5011960601, the M. D. Anderson Tobacco Settlement Research Initiatives Program, and The Kenneth D. Muller Professorship (A.E.-N.).
References
- 1.El-Naggar AK, Hurr K, Batsakis JG, Luna MA, Goepfert H, Huff V: Sequential loss of heterozygosity at microsatellite motifs in preinvasive and invasive head and neck squamous carcinoma. Cancer Res 1995, 55:2656-2659 [PubMed] [Google Scholar]
- 2.Califano J, Ahrendt SA, Meininger G, Westra WH, Joch WM, Sidransky D: Detection of telomerase activity in oral rinses from head and neck squamous cell carcinoma patients. Cancer Res 1996, 56:5720-5722 [PubMed] [Google Scholar]
- 3.Huang MF, Chang YC, Liao PS, Huang TH, Tsay CH, Chou MY: Loss of heterozygosity of p53 gene of oral cancer detected by exfoliative cytology. Oral Oncol 1999, 35:296-301 [DOI] [PubMed] [Google Scholar]
- 4.The American Cancer Society: Cancer Facts and Figures. New York, The American Cancer Society, 1998
- 5.Mao L, Lee DJ, Tockman MS, Erozan YS, Askin F, Sidransky D: Microsatellite alterations as clonal markers for the detection of human cancer. Proc Natl Acad Sci USA 1994, 91:9871-9875 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Califano J, Riet VDP, Westra W, Nawroz H, Clayman G, Piantadosi S, Corio R, Lee D, Greenberg B, Koch W, Sidransky D: Genetic progression model for head and neck cancer: implications for field cancerization. Cancer Res 1996, 56:2488-2492 [PubMed] [Google Scholar]
- 7.Mao L, El-Naggar AK, Papadimitrakopoulou V, Shin DM, Shin HC, Fan Y, Zhou X, Clayman G, Lee JJ, Lee JS, Hittelman WN, Lippman SM, Hong WK: Phenotype and genotype of advanced premalignant head and neck lesions after chemopreventive therapy. J Natl Cancer Inst 1998, 90:1545-1551 [DOI] [PubMed] [Google Scholar]
- 8.El-Naggar AK, Lai S, Clayman G, Lee JK, Luna MA, Goepfert H, Batsakis JG: Methylation, a major mechanism of p16/CDKN2 gene inactivation in head and neck squamous carcinoma. Am J Pathol 1997, 151:1767-1774 [PMC free article] [PubMed] [Google Scholar]
- 9.Riet VDP, Nawroz H, Hruban RH, Corio R, Tokino K, Koch W, Sidransky D: Frequent loss of chromosome 9p21–22 early in head and neck cancer progression. Cancer Res 1994, 54:1156-1158 [PubMed] [Google Scholar]
- 10.Field JK, Kiaris H, Risk JM, Tsiriyotis C, Adamson R, Zoumpourlis V, Rowley H, Taylor K, Whittaker J, Howard P, Beirne JC, Gosney JR, Woolgar J, Voughan ED: Allotype of squamous cell carcinoma of the head and neck: fractional allelic loss correlates with survival. Br J Cancer 1995, 72:1180-1188 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Frank CJ, McClatchey KD, Devaney DO, Carey TE: Evidence that loss of chromosome 18q is associated with tumor progression. Cancer Res 1997, 57:824-827 [PubMed] [Google Scholar]
- 12.Buchhagen DL, Worsham MJ, VanDyke DL, Carey TE: Two regions of homozygosity on chromosome 3p in squamous cell carcinoma of the head and neck: comparison with cytogenetic analysis. Head Neck 1996, 18:529-537 [DOI] [PubMed] [Google Scholar]
- 13.Oshita F, Nomura I, Yamada K, Kato Y, Tanaka G, Noda K: Detection of K-ras mutations of bronchoalveolar lavage fluid cells aids the diagnosis of lung cancer in small pulmonary lesions. Clin Cancer Res 1999, 5:617-620 [PubMed] [Google Scholar]
- 14.Mao L, Hruban RH, Boyle JO, Tockman M, Sidransky D: Detection of oncogene mutations in sputum precedes diagnosis of lung cancer. Cancer Res 1994, 54:1634-1637 [PubMed] [Google Scholar]
- 15.Mills NE, Fishman CL, Scholes J, Anderson SE, Rom WN, Jacobson DR: Detection of K-ras oncogene mutations in bronchoalveolar lavage fluid for lung cancer diagnosis. J Natl Cancer Inst 1995, 87:1056-1060 [DOI] [PubMed] [Google Scholar]
- 16.Sidransky D, von Eschenbach A, Tsai YC, Jones P, Summerhayes I, Marshalla F, Paul M, Green P, Hamilton SR, Frost P, Vogelstein B: Identification of p53 gene mutations in bladder cancers and urine samples. Science 1991, 252:706-709 [DOI] [PubMed] [Google Scholar]
- 17.Sidransky D, Tokino T, Hamilton SR, Kinzler KW, Levin B, Frost P, Vogelstein B: Identification of ras oncogene mutations in the stool of patient with curable colorectal tumors. Science 1992, 256:102-105 [DOI] [PubMed] [Google Scholar]
- 18.Ahrendt SA, Chow JT, Xu LH, Yang SC, Eisenberger CF, Esteller M, Herman JG, Wu L, Decker PA, Jen J, Sidransky D: Molecular detection of tumor cells in bronchoalveolar lavage fluid from patients with early stage lung cancer. J Natl Cancer Inst 1999, 91:332-339 [DOI] [PubMed] [Google Scholar]
- 19.Miozzo M, Sozzi G, Musso K, Pilotti S, Incarbone M, Pastorino U, Pierotti MA: Microsatellite alterations in bronchial and sputum specimens of lung cancer patients. Cancer Res 1996, 56:2285-2288 [PubMed] [Google Scholar]
- 20.Mao L, Schoenberg MP, Scicchitano M, Erozan YS, Merlo A, Schwab D, Sidransky D: Molecular detection of primary bladder cancer by microsatellite analysis. Science 1996, 271:659-662 [DOI] [PubMed] [Google Scholar]
- 21.Itoi T, Takei K, Shinohara Y, Takeda K, Nakamura K, Horibe T, Sanada A, Ohno H, Matsubayashi H, Saito T, Watanabe H: K-ras codon 12 and p53 mutations in biopsy specimens and bile from biliary tract cancer. Pathol Int 1999, 49:30-37 [DOI] [PubMed] [Google Scholar]
- 22.Marchetti A, Buttitta F, Carnicelli V, Pellegrini S, Bertacca G, Merlo G, Bevilacqua G: Enriched SSCP: a highly sensitive method for the detection of unknown mutations. Application to the molecular diagnosis of lung cancer in sputum samples. Diagn Mol Pathol 1997, 6:185-191 [DOI] [PubMed] [Google Scholar]
- 23.Linn JF, Lango M, Halachmi S, Schoenberg MP, Sidransky D: Microsatellite analysis and telomerase activity in archived tissue and urine sample of bladder cancer patients. Int J Cancer 1997, 74:625-629 [DOI] [PubMed] [Google Scholar]
- 24.Rosin MP, Epstein JB, Berean K, Durham S, Hay J, Cheng X, Zeng T, Huang Y, Zhang L: The use of exfoliative cell samples to map clonal genetic alterations in the oral epithelium of high-risk patients. Cancer Res 1997, 57:5258-5260 [PubMed] [Google Scholar]
- 25.Traganos F, Darzynkiewicz Z, Sharpless T, Melamed MR: Simultaneous staining of ribonucleic and deoxyribonucleic acids in unfixed cells using acridine orange in a flow cytofluorometric system. J Histochem Cytochem 1997, 25:46-56 [DOI] [PubMed] [Google Scholar]
- 26.Richards B, Skoletsky J, Shuber AP, Balfour R, Stern RC, Dorkin HL, Parad RB, Witt D, Klinger KW: Multiplex PCR amplification from the CFTR gene using DNA prepared from buccal brushes/swabs. Hum Mol Genet 1993, 2:159-163 [DOI] [PubMed] [Google Scholar]
- 27.Coombes MM, Mao L, Steck KD, Luna MA, El-Naggar AK: Genotypic analysis of flow-sorted and microdissected head and neck squamous lesions by whole genome amplification. Diagn Mol Pathol 1998, 7:197-201 [DOI] [PubMed] [Google Scholar]
- 28.Scholes AG, Woolgar JA, Boyle MA, Brown JS, Vaughan ED, Hart CA, Jones AS, Field JK: Synchronous oral carcinomas: independent or common clonal origin? Cancer Res 1998, 58:2003-2006 [PubMed] [Google Scholar]
- 29.Takes RP, Baatenburg de Jong RJ, Schuuring E, Litvinov SV, Hermans J, Van Krieken JH: Differences in expression of oncogenes and tumor suppressor genes in different sites of head and neck squamous cell. Anticancer Res 1998, 18:4793-4800 [PubMed] [Google Scholar]
- 30.Califano J, Westra WH, Joch W, Meininger G, Reed A, Yip L, Boyle JO, Lonardo F, Sidransky D: Unknown primary head and neck squamous cell carcinoma: molecular identification of the site of origin. J Natl Cancer Inst 1999, 91:599-604 [DOI] [PubMed] [Google Scholar]
- 31.Chung KY, Mukhopadhyay T, Kim J, Casson A, Ro JY, Goepfert H, Hong WK, Roth JA: Discordant p53 gene mutations in primary head and neck cancers and corresponding second primary cancers of the upper aerodigestive tract. Cancer Res 1993, 53:1676-1683 [PubMed] [Google Scholar]
- 32.Worsham MJ, Wolman SR, Carey TE, Zarbo RJ, Benninger MS, Van Dyke DL: Common clonal origin of synchronous primary head and neck squamous cell carcinomas: analysis by tumor karyotypes and fluorescence in situ hybridization. Hum Pathol 1995, 26:251-261 [DOI] [PubMed] [Google Scholar]
- 33.Bedi GC, Westra WH, Gabrielson E, Koch W, Sidransky D: Multiple head and neck tumors: evidence for a common clonal origin. Cancer Res 1996, 56:2484-2487 [PubMed] [Google Scholar]
- 34.Mao L, Lee JS, Fan YH, Ro J, Batsakis JG, Lippman S, Hittleman W, Hong WK: Frequent microsatellite alterations at chromosome 9p21 and 3p14 in oral pre-malignant lesions and their value in cancer risk assessment. Nat Med 1996, 2:682-685 [DOI] [PubMed] [Google Scholar]
- 35.Leonog PP, Rezai B, Koch WM, Reed A, Eisele D, Lee DJ, Sidransky D, Jen J, Westra WH: Distinguishing second primary tumors from lung metastases in patients with head and neck squamous cell carcinoma. J Natl Cancer Inst 1998, 90:972-977 [DOI] [PubMed] [Google Scholar]
- 36.Sozzi G, Miozzo M, Pastorino U, Pilotti S, Donghi R, Giarola M, DeGregorio L, Manenti G, Radice P, Minoletti F, Porta GD, Pierotti MA: Genetic evidence for an independent origin of multiple preneoplastic and neoplastic lung lesions. Cancer Res 1995, 55:135-140 [PubMed] [Google Scholar]
- 37.Van Oijen MG, Slootweg PJ: Oral field cancerization: carcinogen-induced independent events or micrometastatic deposits? Cancer Epidemiol Biomarkers Prev 2000, 9:249-256 [PubMed] [Google Scholar]
- 38.Fey MF, Tobler A: Tumour heterogeneity and clonality: an old theme revisited. Ann Oncol 1996, 7:121-128 [DOI] [PubMed] [Google Scholar]
- 39.Boyle JO, Mao L, Brennan JA, Koch WM, Eisele DW, Saunders JR, Sidransky D: Gene mutations in saliva as molecular markers for head and neck squamous cell carcinoma. Am J Surg 1994, 168:429-432 [DOI] [PubMed] [Google Scholar]
- 40.Kennedy TC, Proudfoot SP, Franklin WA, Merrick TA, Saccomanno G, Corkill ME, Mumma DL, Sirgi KE, Miller YE, Archer PG, Prochazka A: Cytopathological analysis of sputum in patients with airflow obstruction and significant smoking histories. Cancer Res 1996, 56:4673-4678 [PubMed] [Google Scholar]
- 41.Liloglou T, Scholes AG, Spandidos DA, Vaughan ED, Jones AS, Field JK: p53 mutations in squamous cell carcinoma of the head and neck predominate in a subgroup of former and present smokers with a low frequency of genetic instability. Cancer Res 1997, 57:4070-4074 [PubMed] [Google Scholar]
- 42.Mao L, Lee JS, Kurie JM, Fan YH, Lippman SM, Lee JJ, Ro JY, Broxson A, Yu R, Morice RC, Kemp BL, Khuri FR, Walsh GL, Hittelman WN, Hong WK: Clonal genetic alterations in the lungs of current and former smokers. J Natl Cancer Inst 1997, 89:857-862 [DOI] [PubMed] [Google Scholar]
- 43.Liao P-H, Chang Y-C, Huang M-F, Tai K-W, Chou M-Y: Mutation of p53 gene codon 63 in saliva as a molecular marker for oral squamous cell carcinoma. Oral Oncol 2000, 36:272-276 [DOI] [PubMed] [Google Scholar]
- 44.Powell CA, Klares S, O’Connor G, Brody JS: Loss of heterozygosity in epithelial cells obtained by bronchial brushing: clinical utility in lung cancer. Clin Cancer Res 1999, 5:2025-2034 [PubMed] [Google Scholar]
- 45.Slaughter DP, Southwick HW, Smejkal W: Field cancerization in oral stratified squamous epithelium: clinical implications of multicentric origin. Cancer 1953, 6:963-968 [DOI] [PubMed] [Google Scholar]
- 46.Garcia SB, Park HS, Novelli M, Wright NA: Field cancerization, clonality, and epithelial stem cells: the spread of mutated clones in epithelial sheets. J Pathol 1999, 187:61-81 [DOI] [PubMed] [Google Scholar]
- 47.Sidransky D: Nucleic acid-based methods for the detection of cancer. Science 1997, 278:1054-1058 [DOI] [PubMed] [Google Scholar]