Abstract
We have developed a sensitive and quantitative reverse-transcription polymerase chain reaction (RT-PCR) assay for detection of PML-RARα, the fusion oncogene present as a specific marker in >99% of cases of acute promyelocytic leukemia (APL). The assay is linear over at least 5 orders of magnitude of input DNA or RNA, and detects as few as 4 copies of PML-RARα plasmid DNA. PML-RARα transcripts could be detected in mixtures containing 2 to 5 pg of RNA from fusion-containing cells in a background of 1 μg of RNA from PML-RARα-negative cells. Using 1.0 to 2.5 μg of input RNA, the sensitivity of the assay was between 10−5 and 10−6. Furthermore, determination of GAPDH copy number in each reaction allowed an accurate assessment of sample-to-sample variation in RNA quality and reaction efficiency, with consequent definition of a detection limit for each sample assayed. Using an internal calibrator, assay precision was high, with coefficients of variation between 10 and 20%. An interlaboratory study using coded samples demonstrated excellent reproducibility and high concordance between laboratories. This assay will be used to test the hypothesis that sensitive and quantitative measurement of leukemic burden, during or after therapy of APL, can stratify patients into discrete risk groups, and thereby serve as a basis for risk-adapted therapy in APL.
Many cases of leukemia are caused by translocations that result in the formation and expression of chimeric fusion genes, 1 and it is now routinely possible to detect extremely small numbers of transcripts from these leukemia-associated fusion genes in the blood or bone marrow of affected patients. Clinical application of these exquisitely sensitive reverse-transcription polymerase chain reaction (RT-PCR) assays has been slow due to a variety of factors, including a lack of interlaboratory standardization and the non-quantitative nature of most such assays. Interpretation of negative results is complicated by issues of RNA integrity and assay sensitivity that can vary dramatically from one center to another and even from day to day in the same laboratory. Positive results are more readily accepted, but the mere detection of transcripts of leukemia-associated fusion genes has not proven to be a reliable marker for impending relapse. 2, 3, 4 The accumulated evidence suggests that there exists a threshold of leukemic burden above which relapse is highly likely, but below which continued remission is the expected outcome. Definition of such a threshold is an important goal in APL and other leukemias, and will require a standardized, sensitive, and quantitative assay that can expeditiously analyze large numbers of samples from uniformly treated patients enrolled on collaborative clinical trials.
Themajority of APL patients harbor a translocation between chromosomes 15 and 17, with resultant fusion of the PML gene, at 15q12, with the retinoic acid receptor α (RARα) gene, at 17q22. 5 This gene fusion results in production of a leukemia-specific chimeric mRNA, PML-RARα. In the present study, we present the technical details, pre-clinical validation, and initial clinical application of a sensitive, quantitative RT-PCR assay that can detect and measure the PML-RARα fusion transcript. The assay utilizes the 5′-nuclease technique, in which nuclease degradation of a dual-labeled fluorogenic probe results in a measurable fluorescence signal that is detected during the PCR process (“real-time” PCR, or TaqMan assay). 6, 7 Our data indicate that the sensitivity of this quantitative method is at least equal to that of a published manual RT-PCR assay for detection of PML-RARα, and that it is 100% specific. Furthermore, the precision of this method is good, and it is amenable to interlaboratory standardization; these features suggest that it is ideally suited for high throughput analysis of large numbers of clinical specimens.
Materials and Methods
Cells and Cell Culture
NB4 8 and UF1 cells 9 were cultured in RPMI 1640 medium supplemented with 10% fetal calf serum and antibiotics. HL-60 cells were cultured in Iscove’s modified Dulbecco’s medium + 20% fetal calf serum. The NB4 cell line contains the PML-RARα type L (long) isoform, while UF1 cells express the S (short) isoform (Figure 1) . The HL-60 promyelocytic cell line does not express PML-RARα. For RNA dilution experiments, RNA was extracted from exponentially growing cells, quantified by spectrophotometry, and mixed at dilutions indicated in the text or figure legends. After informed consent, blood or bone marrow was collected from normal donors or from patients with leukemia into Vacutainer tubes containing either disodium heparin or EDTA anticoagulant. Mononuclear cells were prepared from blood or bone marrow by Ficoll-Hypaque density-gradient centrifugation.
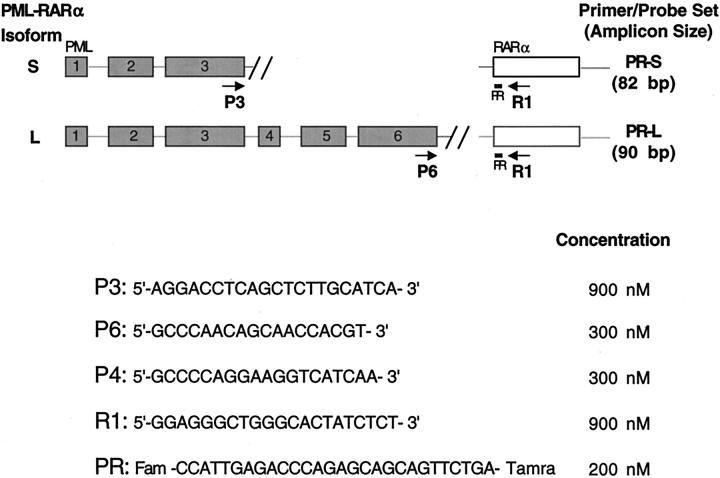
Figure 1.
Primers and probe used in real-time PML-RARα PCR. The primer and probe names, locations, and sequences are shown, along with the final concentrations used in PCR, and the S- and L-form amplicon sizes. PML exons 1–6 are shown as shaded rectangles, while RARα exon 3 is represented by an open rectangle. The S and L genomic breakpoints are designated by double slanted lines.
PCR Precautions
All pre-PCR manipulations (RNA isolation, RT and PCR set-up) were performed in a clean room that was physically isolated from the PCR machine and the post-PCR processing area. 10 Dedicated pipettes and reagents were used. Negative controls were run with all assays and were uniformly negative. Plasmids used for standard curve generation were added to 96-well plates in a separate room, and after experimental wells had been capped. PML-RARα and GAPDH plasmid standards were never taken into the RT-PCR set-up room.
Isolation of RNA and Reverse Transcription into cDNA
Total cellular RNA was isolated using either a modification of the guanidinium acid-phenol extraction method 11 (STAT-60 reagent; Tel-Test, Inc., Friendswood, TX), or one of two proprietary column affinity procedures (RNeasy kits, Qiagen, Inc., Valencia, CA; or RNAqueous kits, Ambion Inc., Austin, TX), in each case according to the respective manufacturers’ instructions. RNA quality was assessed visually by confirmation of intact 28S and 18S ribosomal bands following agarose gel electrophoresis and ethidium bromide staining. Before cDNA synthesis, RNA (1.0–2.5 μg) was heated at 75°C for 10 minutes and snap-cooled on ice. Two different RT protocols were used, which differed only by the inclusion of either gene-specific primers or random hexamers to prime first strand cDNA synthesis. Both protocols included 1X Taq polymerase buffer II (Applied Biosystems, Foster City, CA; final concentrations: 50 mmol/L KCl, 10 mmol/L Tris-HCl, pH 8.3), 5 units/μl Moloney murine leukemia virus (Applied Biosystems) or Superscript II RNase H–(Life Technologies, Grand Island, NY) reverse transcriptase, 250 μmol/L each dGTP, dCTP, dATP, dTTP, 3.0 mmol/L MgCl2, and 1.6 units/μl RNase inhibitor (Applied Biosystems or Promega, Madison, WI). Random hexamers were used at a final concentration of 5 μmol/L, and gene-specific primers at a final concentration of 0.125 μmol/L each. Sequences for the gene-specific primers were as follows: RARα, 5′-TGTTCTTCTGGATGCTG-3′; GAPDH, 5′-ACTTGATTTTGGAGGGA-3′. The final reaction volume was either 20 or 25 μl. cDNA synthesis was performed at 48°C for 60 minutes, followed by an incubation at 95°C for 10 minutes to inactivate the RT enzyme; in reactions including random hexamers, an initial 10-minute incubation at 25°C was included before the 48°C cDNA synthesis reaction. In the experiment shown in Figure 4 , gene-specific primers were used for RT, dNTP concentrations were 1 mmol/L each, and MgCl2 concentration was 7.5 mmol/L. These variations did not materially affect the sensitivity or specificity of the assay.
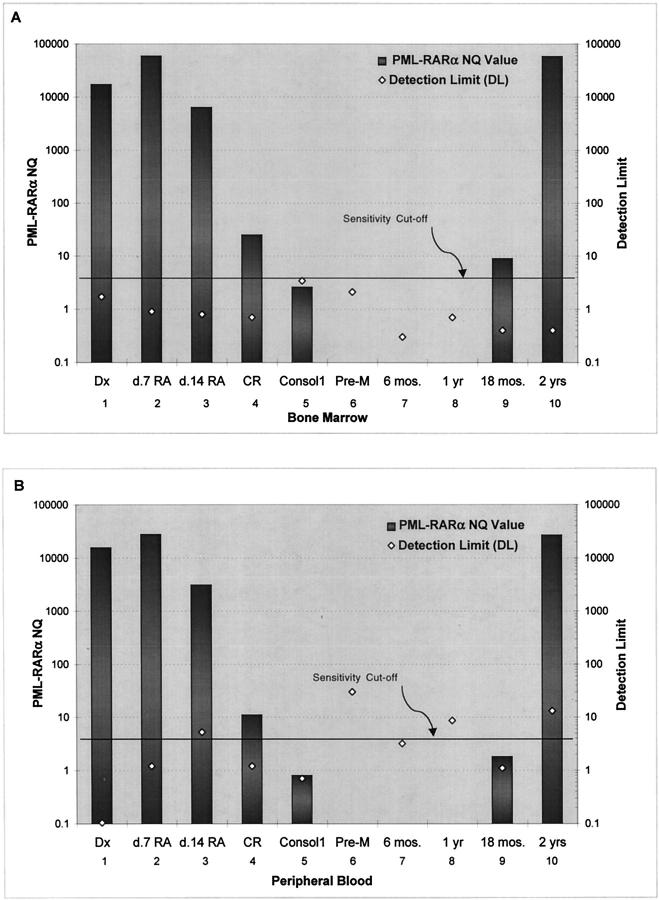
Figure 4.
Application of real-time PML-RARα RT-PCR to analysis of clinical specimens. Total RNA was isolated from bone marrow (A) or blood (B) mononuclear cells from a single patient with APL (L-isoform) who was treated with all-trans retinoic acid (RA) until complete remission (CR), and then with 2 cycles of daunorubicin and cytosine arabinoside consolidation chemotherapy. Real-time RT-PCR was performed as described in Materials and Methods using both L-form and GAPDH primers and probes. Dx, diagnosis; Consol1, sample obtained after one cycle of consolidation; Pre-M, sample obtained after two cycles of consolidation and before randomization to maintenance therapy. Additional samples were taken at 6-month intervals during follow-up. Pathological analysis of the sample taken at 2 years revealed relapsed APL. The definitions of NQ value and detection limit are given in the text.
Primer and Probe Design
Primers and probes were designed using Primer Express software, version 1.0 (Applied Biosystems). The names, sequences, and locations of the PML and RARα primers and probe used in this work are indicated in Figure 1 . All probes contained a 3′ TAMRA (6-carboxy-tetramethyl-rhodamine) quencher dye and were synthesized at Perkin Elmer. The P4 primer was synthesized at the Biopolymer facility of Roswell Park Cancer Institute. The RARα probe (R1) was labeled at the 5′ end with a FAM (6-carboxyfluorescein) reporter fluorescent dye, while the GAPDH probe contained a 5′ JOE label. GAPDH primers and probe were purchased from or provided by Applied Biosystems. Final concentrations of probes and primers (Figure 1) were chosen based on preliminary optimization experiments (data not shown).
Standard Curve Generation
Plasmids containing the PML-RARα and GAPDH cDNAs were constructed using standard recombinant DNA methods. Plasmid DNA was prepared and its concentration determined using the PicoGreen assay. 12 The DNA was 10-fold serially diluted in water and aliquots were stored at −70°C until use. Standard curves were generated using from 4 to 4 × 107 copies of plasmid DNA. It is emphasized that the copy numbers/μg RNA, as cited in the text for test samples, were derived by reference to these standard curves and do not precisely indicate the number of mRNA molecules, since the efficiency of RT was not directly determined.
Real-Time PCR
For PML-RARα PCR, the 50 μl reaction contained 25 μl 2X TaqMan Universal PCR Master Mix (Applied Biosystems), 2.5–5 μl of RT product, and appropriate PML-RARα primers and probe at concentrations listed in Figure 1 . For GAPDH PCR, the final reaction volume was either 25 or 50 μl, and generally 2.5 μl of RT product was used. Amplifications were carried out at 50°C for 2 minutes, 95°C for 10 minutes, followed by 40 PCR cycles at 95°C (15 seconds) and 60°C (1 minute). All reactions were carried out in MicroAmp optical tubes or MicroAmp optical 96-well reaction plates (Applied Biosystems) using an ABI PRISM 7700 sequence detection system. Real-time PCR data were collected using the ABI Prism 7700 sequence detection system (Applied Biosystems) as previously published. 7 Briefly, the normalized fluorescence intensity of the reporter dye (ΔRn) is plotted against cycle number to derive a graphical representation of the PCR reaction. The threshold cycle CT is defined as the cycle number at which the ΔRn crosses a software-generated threshold defined as 10 standard deviations above baseline (during cycles 3–15). The CT is linearly proportional to the logarithm of the input copy number. By construction of an appropriate standard curve, the starting copy number of an unknown sample relative to the standard can be derived.
Manual RT-PCR
Reverse transcription from random hexamers and first-round PCR amplification of PML-RARα were performed essentially as described previously, 13 making adjustments to ensure that 1 μg RNA equivalent of cDNA was used in the PCR reaction. After 40 PCR cycles, 2 μl of a 1:20 dilution of the first-round PML-RARα L-form product was amplified in a standard 50 μl second-round, nested reaction for 37 PCR cycles, using the following primers: upstream, anchored in the proximal portion of PML exon 6 (5′-ACAACGACAGCCCAGAAGAGGAAGT-3′); downstream, crossing the PML-RARα junction (5′-GCTGCTCTGGGTCTCAATGGCTG-3′). PCR amplifications were performed in a PTC-100 programmable thermal controller (MJ Research, Inc., Watertown, MA). We found that utilization of the PTC-100 “Hot Bonnet” attachment, performance under a layer of mineral oil, and thorough denaturation of both rounds of PCR amplification, using heat-activatable Taq polymerase (Amplitaq Gold, Perkin Elmer, Inc., Norwalk, CT) were important elements in achieving high sensitivity. The effectiveness of RNA transcription was evaluated by using 0.1 μg RNA equivalent of cDNA from each RT reaction for PCR amplification of GAPDH mRNA in the presence of a fixed amount of a Mimic DNA competitor, as previously described. 14 Gel electrophoretic analysis of the second-round products was as previously described. 13
Results
Pre-Clinical Assay Validation
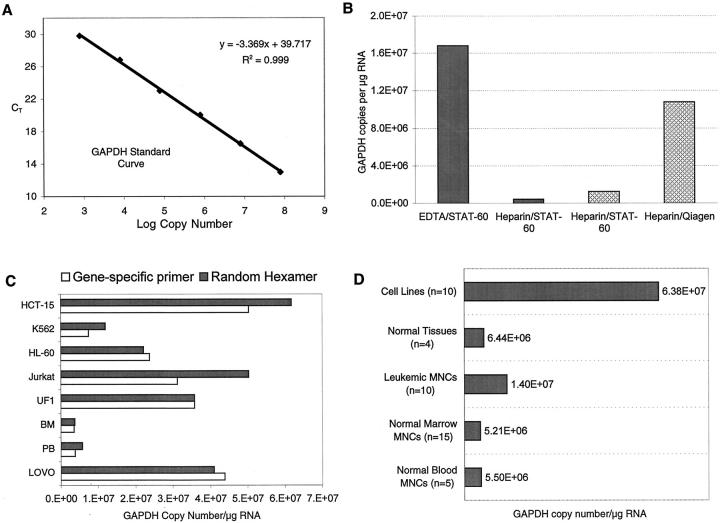
The PR-S and -L primer and probe sets each include a distinct, exon-specific PML forward primer, a common RARα reverse primer, and a probe that hybridizes to RARα sequence at the junction of the fusion transcript (Figure 1) . To optimize protocols for efficient cDNA synthesis and real-time PCR, GAPDH mRNA levels were measured in clinical samples or cell lines by reference to standard curves generated with GAPDH plasmid DNA (Figure 2A) . As previously reported, 15 RNA isolated from blood or marrow samples anti-coagulated with heparin appeared to contain an inhibitor of RT or PCR (Figure 2B) . This problem was overcome by isolation (or “clean-up”) of RNA using either of two column-based purification protocols (Rneasy, Qiagen; or RNAqueous, Ambion) (Figure 2B) . cDNA yield was also influenced, albeit modestly, by the type of primer used to initiate cDNA synthesis (Figure 2C and data not shown). Using optimal RNA isolation procedures and plasmid-based standard curves, the calculated number of GAPDH copies/μg RNA was approximately 5 × 106 (±3 × 106) using RNA from Ficoll-purified normal blood or marrow mononuclear cells (Figure 2D) , and approximately 1.5 × 107 (±5 × 106) using RNA from leukemic blood or marrow mononuclear cells. The number of calculated GAPDH copies per μg of RNA from cultured hematopoietic cell lines (HL-60, Jurkat, etc.) was significantly higher (Figure 2D) .
Figure 2.
GAPDH copy number related to sample processing technique, type of primer used to initiate cDNA synthesis, and tissue source. A: GAPDH standard curve generated with 10-fold serial dilutions of GAPDH plasmid standards. B: Effects of sample processing and RNA extraction. Solid columns: bone marrow was collected in EDTA or heparin and RNA was isolated using STAT-60. Cross-hatched columns: bone marrow was collected in heparin and cryopreserved. Vials were thawed, divided into two aliquots, and RNA was extracted using either STAT-60 or Qiagen RNeasy columns. In all cases, equivalent amounts of RNA (measured by spectrophotometry) were reverse-transcribed into cDNA and equal amounts of cDNA were amplified using GAPDH primers and probe. The number of GAPDH copies was determined by reference to the GAPDH standard curve shown in A. C: Comparison of random hexamers to gene-specific primers in cDNA synthesis. Equal amounts of the RNA from the cells shown were reverse-transcribed into cDNA using either gene-specific primers or random hexamers, and GAPDH copy number was calculated from the same amount of cDNA equivalent using real-time PCR. D: GAPDH copy number/μg of RNA from different tissue sources. The cell lines were a mixture of hematopoietic and epithelial cells. RNA from normal tissue (spleen, placenta, colon, and lung, all pooled) was purchased from Clontech. All RNAs, except those purchased from Clontech, were isolated using Qiagen RNeasy columns.
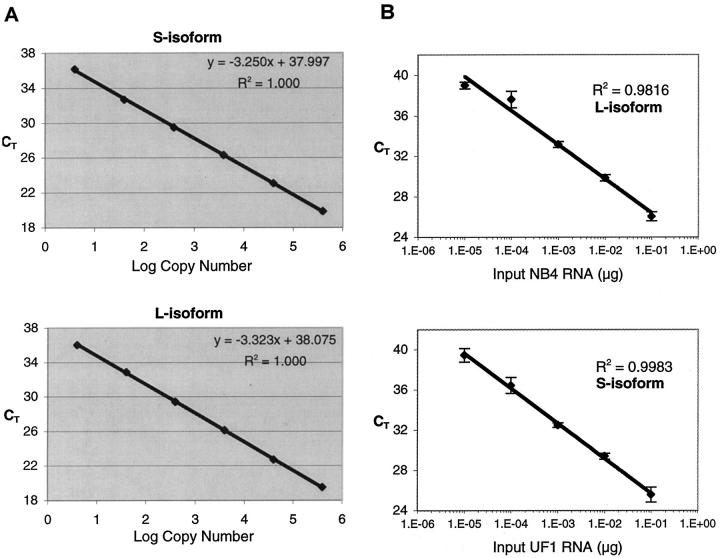
Using plasmid DNA standards, as few as 4 copies of L- or S-form DNA could be reproducibly detected (Figure 3A) , and the CT was linearly proportional to the logarithm of the input copy number over at least 5 orders of magnitude (R2 > 0.99). The dynamic range and detection limit was similar for GAPDH (Figure 2A and data not shown). The Y intercept reflects the theoretical CT value at one copy of input DNA, while the slope (S) of the linear regression curve correlates with the efficiency (E) of the PCR reaction, according to the following formula: E = 10−1/S −1. A PCR efficiency of 1 (slope = −3.322) indicates that the copy number doubles in each cycle. Using plasmid DNA as a template, the reaction efficiencies for the PR-S, PR-L, and GAPDH probe and primer sets were generally 0.95 or higher, indicating near optimum PCR amplification. A similar analysis was performed using NB4 or UF1 RNA as starting material. The results (Figure 3B) indicate that PML-RARα transcripts could be detected using as little as 10 pg of either NB4 or UF1 RNA, and confirm that the assay is linear over at least 4 orders of magnitude, from 10 pg to 100 ng of input NB4 or UF1 RNA (R2 = 0.982 and 0.998, respectively). The respective efficiencies, calculated from the slopes of the linear regression equations, were 98% and 94%, suggesting little to no decrease in PCR efficiency despite the more complex nature of the cDNA starting material. Additional experiments confirmed an even lower detection limit of PML-RARα in RNA dilutions, in the range of 3 to 5 pg of NB4 or UF1 RNA admixed with control RNA (data not shown).
Figure 3.
A: Detection limit and dynamic range of detection of PML-RARα using plasmid DNA standards. Ten-fold serial dilutions of PML-RARα standards (L and S isoform) were amplified in TaqMan PCR using conditions outlined in Materials and Methods. The threshold cycle CT was computed using the 7700 Sequence Detector software and is plotted against the logarithm of the input copy number. The linear regression equations and correlation coefficients are shown. B: Detection limit and dynamic range of detection of PML-RARα using RNA from APL cell lines. NB4 or UF1 total RNA was mixed with HL-60 total RNA at dilutions ranging from 10−1 to 10−6. Five μg of RNA from each dilution was reverse-transcribed into cDNA and 20% (1 μg RNA equivalent) was amplified in TaqMan PCR reactions using the PR-L or -S primer/probe set. The amount of NB4 or UF1 RNA equivalent in each TaqMan reaction ranged from 1 pg (10−6 dilution) to 100 ng (10−1 dilution).
The specificity of the assay was evaluated by testing 28 non-APL samples, including RNA from blood and/or marrow from 6 normal donors, RNA from 5 different pooled human tissues (bone marrow, colon, spleen, thymus, and placenta), RNA from 8 cases of various hematological malignancies (5 non-M3 AML, 2 chronic myeloid leukemia, one myelodysplastic syndrome), and RNA from 9 hematopoietic and non-hematopoietic cell lines. For all 28 specimens, from 1.0 to 2.5 μg of RNA was reverse-transcribed into cDNA, and 20% of the RT product was analyzed in duplicate in TaqMan PCR reactions with both the PR-L and PR-S primer and probe sets (112 total data points). All 112 CT values were 40, indicating no aberrant amplification, and no detection of false products, with either L or S primer/probe set (data not shown). The integrity of the RNA from each sample was confirmed by simultaneous GAPDH amplification (data not shown). To evaluate interlaboratory reproducibility, coded RNA samples were assayed blindly and independently at separate sites (University of New Mexico and Roswell Park). As shown in Table 1 , there was excellent concordance between the two laboratories, and the sensitivities achieved (10−5 for UF1 [S isoform] dilutions, and 10−4 or 10−5 for NB4 [L isoform] dilutions) were similar. Finally, the PML-RARα normalized quantities (NQ, number of PML-RARα copies per 1 × 106 GAPDH copies), were also similar between the two laboratories (Table 1) .
Table 1.
Blinded Inter-Laboratory Comparison of Real-Time RT-PCR for Detection of PML-RARα
| L isoform sample (dilution) | PML-RARα CT-L (UNM) | PML-RARα CT-L (RPCI) | NQ1 (UNM) | NQ (RPCI) |
|---|---|---|---|---|
| NB4 (10−3) | 33.0 | 34.3 | 30.3 | 22.8 |
| NB4 (10−4) | 36.2 | 36.8 | 3.31 | 3.90 |
| NB4 (10−5) | 39.0 | 40.0 | 0.76 | 0.00 |
| NB4 (5× 10−6) | 40.0 | 40.0 | 0.00 | 0.00 |
| NB4 (10−6) | 40.0 | 40.0 | 0.00 | 0.00 |
| Blood MNCs | 40.0 | 40.0 | 0.00 | 0.00 |
| Bone Marrow MNCs | 40.0 | 40.0 | 0.00 | 0.00 |
| S isoform sample (dilution) | PML-RARα CT-S (UNM) | PML-RARα CT-S (RPCI) | NQ (UNM) | NQ (RPCI) |
| UF1 (10−3) | 32.6 | 32.5 | 47.0 | 38.7 |
| UF1 (10−4) | 37.4 | 36.7 | 2.70 | 3.77 |
| UF1 (10−5) | 38.7 | 37.5 | 0.83 | 1.12 |
| UF1 (5× 10−6) | 40.0 | 39.5 | 0.00 | 0.42 |
| UF1 (10−6) | 39.9 | 40.0 | 0.12 | 0.00 |
| Blood MNCs | 40.0 | 40.0 | 0.00 | 0.00 |
| Bone Marrow MNCs | 40.0 | 40.0 | 0.00 | 0.00 |
One laboratory (University of New Mexico) prepared coded samples of UF1 or NB4 RNA mixed with RNA from normal blood leukocytes at dilutions ranging from 10−3 to 10−6 (or RNA from normal bone marrow and blood mononuclear cells). Samples were assayed independently in both laboratories using the same amount (1 μg) of RNA for RT, and the same amounts of cDNA for TaqMan PCR with PR-L, PR-S, and GAPDH probe and primer sets (225 ng cDNA equivalent for PR-L and PR-S, and 100 ng for GAPDH).
NQ, normalized quantity (number of PML-RARα copies per 1 × 106 GAPDH copies).
MNCs, mononuclear cells. UNM, University of New Mexico.
Assay precision was evaluated by serial testing, over 4 working days, of the same RNA from both a high and low copy number clinical specimen. Three replicates for the high and low copy number PML-RARα samples were tested on each day, as well as an internal calibrator (NB4 RNA diluted 1:100 in HL-60 RNA), and a negative control (HL-60 RNA). All 12 HL-60 assays were negative, as expected. On each day, separate standard curves were generated for PML-RARα and GAPDH. The results (Table 2) are reported as 1) the number of PML-RARα copies per μg of starting RNA; 2) as a GAPDH normalized quantity, NQ; and 3) as a normalized, calibrated quantity (NQC), in which the NQ for the experimental sample is divided by the NQ for the internal calibrator. The coefficients of variation (CVs) were lowest (12–16%) when data were reported with reference to an internal calibrator (Table 2) .
Table 2.
Precision of PML-RARα TaqMan Assay
| Input PML-RARα copy number* | PML-RARα copies/μg† | PML-RARα NQ‡ | PML-RARα NQC§ | ||||||
|---|---|---|---|---|---|---|---|---|---|
| Mean (range) | SD | CV (%) | Mean (range) | SD | CV (%) | Mean (range) | SD | CV (%) | |
| Low | 37 (23–59) | 16 | 43 | 24 (16–27) | 5.2 | 22 | 0.87 (0.82–1.02) | 0.10 | 12 |
| High | 4768 (3335–6490) | 1485 | 31 | 1979 (1519–2402) | 398 | 20 | 73 (59–86) | 11 | 16 |
Assays were performed four times on 4 successive days. Results shown are the means (ranges) of the four independent determinations, with standard deviation (SD) and coefficient of variation (CV) shown.
RNA from L-isoform blasts at diagnosis of APL. High, undiluted RNA; low, RNA diluted 1:100 in RNA from normal peripheral blood mononuclear cells.
Number of PML-RARα copies per μg of input RNA, calculated by reference to a standard curve.
NQ, normalized quantity: number of PML-RARα copies per 106 GAPDH copies.
NQC, normalized, calibrated quantity: number of PML-RARα copies per 106 GAPDH copies divided by an internal calibrator.
Comparison of the Sensitivity of Real-Time and Manual RT-PCR
Table 3presents the data from a direct comparison, using the same RNA preparation, between real-time and manual RT-PCR procedures following serial dilution of NB4 cells in HL-60 cells (or NB4 RNA in HL-60 RNA). Although the amounts of RNA differed between the qualitative and quantitative assays, it can be concluded that the real-time procedure is at least as sensitive, and perhaps more sensitive, than the optimized manual procedure in detecting PML-RARα, and that the dilution range over which the real-time procedure becomes variably positive is more narrow than for the manual assay. The results in Table 3 also suggest that the practical sensitivity limit of the real-time assay is between 1 in 10−5 and 1 in 10−6, assuming a more standard input RNA equivalent of 500 ng to 1 μg, commonly achieved with clinical samples.
Table 3.
Comparison of the Sensitivity of Real-Time RT-PCR and Manual RT-PCR for the Detection of PML-RARα in NB4 Cells or RNA Serially Diluted in HL-60 Cells or RNA
| RT-PCR type | cDNA equivalent assayed (range) | Dilution method | NB4 cell/RNA dilution (no. positive/no. tested) | Control | |||
|---|---|---|---|---|---|---|---|
| 10−4 | 10−5 | 10−6 | 10−7 | ||||
| Real-time | 4.52 μg (3.08–6.08) | Cell | 12 /12 | 12 /12 | 12 /12 | 4 /12 | 0 /12 |
| Manual | 1 μg | Cell | 10 /10 | 9 /10 | 3 /10 | NT | 0 /1 |
| 1 μg | RNA | 12 /12 | 16 /21 | 5 /21 | 0 /1 | 0 /3 | |
For the manual assay, 1 μg of RNA equivalent cDNA was used in each PCR reaction, and samples were interpreted as positive or negative based on the presence or absence of strong ethidium-bromide staining bands of appropriate size after double nested PCR and gel analysis. A corresponding reaction for GAPDH was performed for each sample in the presence of a fixed amount of competitor DNA using 0.1 μg RNA equivalent to ensure that negative reactions were not false negatives due to RNA degradation or failure of RNA transcription. 14 For real-time RT-PCR, performed in a separate laboratory, the amount of input RNA equivalent cDNA varied for each dilution, as indicated (average 4.52 μg). For each real-time PCR assay, 1/20 of the RT reaction product was amplified using GAPDH primers and probe to confirm RNA integrity.
Analysis of Clinical Samples: Application of the Detection Limit Concept
A representative case of APL was analyzed (Figure 4) to demonstrate the clinical utility of the assay and introduce several concepts related to analysis of clinical samples. The detection limit (DL) is defined as [(1/GAPDH copy number) × 106], and thus is the NQ value that would result if one copy of PML-RARα were present in the PCR reaction (the arbitrary 106 multiplier is the same multiplier used to calculate the NQ values). Note that any sample with an NQ value above the DL is considered positive (>1 copy). By definition, the DL depends on the number of GAPDH molecules in each tested sample, and thus is a quantitative measurement of the sensitivity achieved with each specimen. In a small percentage of cases (ie, sample 5 in Figure 4A ), the PML-RARα NQ value, while measurable, is nevertheless below the theoretical DL of 1 copy. These samples generally have very high CT values (≥39), and reference to the standard curve generates a calculated starting PML-RARα copy number of <1, which is a theoretical impossibility. While the appropriate way to handle such data points is not clear, it seems likely that most such samples are true positives, and that the calculated copy number of < 1 reflects quantitative imprecision at very low input copy numbers.
Proper interpretation of negative results (samples 6–8 in Figure 4 , A and B) requires definition of a sensitivity threshold, which is based on the number of GAPDH copies in each sample. This threshold is by definition user-selected but should be consistently applied across all samples (in Figure 4 , this threshold is set at 2.5 × 105 GAPDH copies, or an approximately 10−4 sensitivity). If the DL falls below this threshold in a given specimen (samples 6–8 in Figure 4A , and sample 7 in Figure 4B ), then the assay was theoretically capable of detecting 1 PML-RARα copy at that sensitivity threshold, and such a sample is considered to be a true negative. Specimens with NQs of 0 but with DLs above the sensitivity threshold (Figure 4B , samples 6 and 8) are considered to be false negatives. The sensitivity threshold can be applied consistently across a large clinical dataset to ensure exclusion of samples that do not meet minimum requirements for RNA input and assay performance. Figure 4 also graphically demonstrates the kinetics of clearance of PML-RARα in this patient who received all-trans retinoic acid for induction and chemotherapy consolidation. The eventual relapse at 2 years was clearly anticipated by TaqMan analysis of both blood and bone marrow samples analyzed 6 months earlier, when the PML-RARα quantity was 4 orders of magnitude less than the value observed at frank relapse.
Discussion
The management of APL may be facilitated by use of highly sensitive and specific molecular assays that measure the amount of residual leukemia burden at various times during and after therapy. 16 Since manual end-point RT-PCR assays can detect fusion transcripts in bone marrow from APL patients in long-term CR, 3 there is a clear mandate to develop quantitative assays and to relate the level (not just presence) of fusion PML-RARα mRNA with relapse risk. From a technical standpoint, cumbersome competitive quantitative RT-PCR protocols are being replaced by assays that use dual-labeled fluorescent probes and continuous detection of PCR amplicons in “real time.” 7, 17 These TaqMan, or real-time, RT-PCR assays are high-throughput, quantitative, and, assuming proper probe and primer design and adequate RNA input, highly sensitive. The assays can be used to quantify serial changes in RNA expression relative to a baseline sample or, by reference to RNA- or DNA-based standard curves, to determine absolute starting copy number of a specific RNA in all samples. The purpose of the current report is to present the technical details and pre-clinical validation of a newly developed TaqMan protocol for detection of PML-RARα in bone marrow or blood specimens from APL patients. The assay described here is sufficiently sensitive and specific to quantify low levels of minimal residual disease in APL, and will be useful to test the hypothesis that a threshold of leukemic burden can be defined that will correlate with outcome. Indeed, we have recently applied this assay to a large clinical dataset of over 800 APL samples, and our preliminary results show a statistically significant correlation between the level of PML-RARα at the end of consolidation and relapse risk. 18 (Gallagher RE, Yeap B, Bi W, Slack JL, Harrington DP, Livak K, Appelbaum FR, Bloomfield CD, Tallman MS, Willman CW, manuscript submitted for publication).
A major criticism of end-point RT-PCR assays is the lack of a quantitative measure of sensitivity on a per-sample basis. Sensitivities are generally inferred from serial dilutions of cell line RNA, but extrapolation from such ideal mixtures to clinical specimens is not necessarily valid, given issues such as PCR inhibitors, poor RNA integrity, etc. Using TaqMan, the success (and by extension sensitivity) of each individual RT-PCR reaction can be quantified by measurement of GAPDH or other control transcript. A minimum sensitivity level can be established that is then used for an entire data set, and the detection limit concept can be applied to each sample to determine whether one copy of PML-RARα was theoretically detectable in that sample. The critical point is that sensitivity, as generally and previously defined, has not accounted in a quantifiable way for day-to-day differences in specimen quality and reaction efficiency that can clearly be significant, particularly using clinical samples. TaqMan RT-PCR can measure these differences and include them in the read-out, removing one of the largest variables, and sources of inaccuracy, in studies using manual or end-point RT-PCR. Thus, the power of the TaqMan assay is that it not only provides quantitative results that allow comparison among positive samples, but it also permits direct assessment of the sensitivity achieved for each negative sample. For the TaqMan assay to be accurate in determination of sensitivity for a specific sample, the degradation rates of the target (PML-RARα) and control (GAPDH or other) transcripts must be relatively similar. This issue is of paramount importance to clinical application of the assay, since blood or marrow samples are often shipped long distances, with unavoidable delays in processing of cells. If the target mRNA undergoes significant degradation on storage or shipment, while the control transcript is stable, then the sensitivity of some samples would likely be overestimated. Experiments to address this theoretical concern are clearly required.
Additional issues that are critical to application of this assay to large clinical datasets include assay precision and potential for interlaboratory standardization. The precision of an assay can be estimated by calculation of a coefficient of variation (CV), which reflects the day-to-day differences in results of testing the same sample. Clearly, the CV needs to be acceptably low to propose application of a given assay to a large dataset. The CVs reported in Table 3 (approximately 20%) compare favorably with clinically accepted and commercially used tests, eg, for measurement of HIV RNA levels in plasma. 19 The use of an internal PML-RARα calibrator, in addition to the use of GAPDH as a measure of RNA integrity and reverse transcription efficiency, gave the lowest CVs (12 to 16%), and may be a useful protocol addition if the assay is to be performed by multiple independent laboratories. 20 The results of a blinded, interlaboratory comparison (Table 1) indicated an acceptable degree of agreement, both in CT values and in calculated copy numbers for PML-RARα, suggesting that this assay can be standardized for use by different laboratories, which is a prerequisite for its application to a rare disease like APL.
The TaqMan, or 5′-nuclease assay, has previously been used to quantify disease-specific transcripts from a variety of hematological malignancies. 21, 22, 23, 24, 25, 26, 27 Several groups 21, 22, 26 have published data regarding detection of BCR-ABL (the leukemia-specific fusion transcript found in chronic myeloid leukemia) using real-time RT-PCR. Using a one-step protocol, Preudhomme et al 22 could detect 20 pg of positive control RNA from the BCR-ABL-positive cell line K562, which translated into a sensitivity of approximately 10−4. Mensink et al, 21 using a different primer and probe set and using a two-step procedure similar to that reported in the current study, could detect at least 10 copies of BCR-ABL plasmid DNA and 10 pg of K562 RNA, for a calculated sensitivity of 1 in 105. The TaqMan probe used to detect the BCR-ABL amplicon was, in both cases, derived from ABL cDNA sequence and did not cross the BCR-ABL fusion junction. Similar to our data using a probe entirely from RARα, this approach did not lead to any appreciable incidence of false positive results, suggesting that non-junction-specific probes provide acceptable specificity in the TaqMan assay. These studies and others suggest that real-time RT-PCR, assuming properly designed primers and probes, will have sufficient sensitivity to provide valuable prognostic information in human leukemias.
While these experiments were in progress, Cassinat et al 28 published a slightly different protocol to detect PML-RARα using the TaqMan method. Although those authors used a different PML-RARα primer and probe set, and a different internal control mRNA (porphobilinogen deaminase), the sensitivity, specificity, and precision of their assay was similar to that reported here. Given the rarity of APL, it may be desirable to agree on a “universal” primer and probe set that can be used world-wide to quantify PML-RARα. Such an approach would allow more ready comparison of quantitative RT-PCR results among different APL molecular diagnostic centers, and perhaps lead to more universal agreement about the level of PML-RARα that correlates with poor clinical outcome. Our results, combined with those of Cassinat et al, 28 strongly suggest that “real-time” RT-PCR methodology is sufficiently robust to accurately quantify small amounts of PML-RARα mRNA in clinical specimens, and it is expected that clinical application of this assay will provide important information for the management of APL patients. However, while this elegant technology can clearly provide a precise measurement of the amount of fusion RNA present in a given clinical specimen, extensive further study is necessary to formally define transcript levels that correlate with relapse risk at specific time points.
Acknowledgments
We thank Drs. M. Lanotte and M. Kizaki for NB4 and UF1 cells, respectively, and Dr. Srinivas Rao for assistance with the development of the high-sensitivity manual RT-PCR assay.
Footnotes
Supported by National Institutes of Health grants CA75049 (to J.L.S.) and CA56771 (to R.E.G.).
Address requests for reprints to James L. Slack, MD, Department of Medicine, Roswell Park Cancer Institute, Elm & Carlton Streets, Buffalo, NY 14263. E-mail: james.slack@roswellpark.org.
References
- 1.Look AT: Oncogenic transcription factors in the human acute leukemias. Science 1997, 278:1059-1064 [DOI] [PubMed] [Google Scholar]
- 2.Nucifora G, Larson RA, Rowley JD: Persistence of the 8;21 translocation in patients with acute myeloid leukemia type M2 in long-term remission. Blood 1993, 82:712-715 [PubMed] [Google Scholar]
- 3.Tobal K, Liu Yin JA: RT-PCR method with increased sensitivity shows persistence of PML-RARα fusion transcripts in patients in long-term remission of APL. Leukemia 1998, 12:1349-1354 [DOI] [PubMed] [Google Scholar]
- 4.Jurlander J, Caligiuri MA, Ruutu T, Baer MR, Strout MP, Oberkircher AR, Hoffmann L, E.D B, Frei-Lahr DA, Christiansen NP, Block AW, Knuutila S, Herzig GP, Bloomfield CD: Persistence of the AML1/ETO fusion transcript in patients treated with allogeneic bone marrow transplantation for t(8;21) leukemia. Blood 1996, 88:2183-2191 [PubMed] [Google Scholar]
- 5.Slack JL, Gallagher RE: The molecular biology of acute promyelocytic leukemia. Cancer Treat Res 1999, 99:75-124 [DOI] [PubMed] [Google Scholar]
- 6.Holland PM, Abramson RD, Watson R, Gelfand DH: Detection of specific polymerase chain reaction product by utilizing the 5′-3′ exonuclease activity of Thermus aquaticus DNA polymerase. Proc Natl Acad Sci USA 1991, 88:7276-7280 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Heid CA, Stevens J, Livak KJ, Williams PM: Real time quantitative PCR. Genome Res 1996, 6:986-994 [DOI] [PubMed] [Google Scholar]
- 8.Lanotte M, Martin-Thouvenin V, Najman S, Balerini P, Valensi F, Berger R: NB4, a maturation inducible cell line with t(15;17) marker isolated from a human acute promyelocytic leukemia (M3). Blood 1991, 77:1080-1086 [PubMed] [Google Scholar]
- 9.Kizaki M, Matsushita H, Takayama N, Muto A, Ueno H, Awaya N, Kawai Y, Asou H, Kamada N, Ikeda Y: Establishment and characterization of a novel acute promyelocytic leukemia cell line (UF-1) with retinoic acid-resistant features. Blood 1996, 88:1824-1833 [PubMed] [Google Scholar]
- 10.Kwok S, Higuchi R: Avoiding false positives with PCR. Nature 1989, 339:237-239 [DOI] [PubMed] [Google Scholar]
- 11.Chomczynski P, Sacchi N: Single-step method of RNA isolation by acid guanidium thiocyanate-phenol-chloroform extraction. Anal Biochem 1987, 162:156-159 [DOI] [PubMed] [Google Scholar]
- 12.Singer VL, Jones LJ, Yue ST, Haugland RP: Characterization of PicoGreen reagent and development of a fluorescence-based solution assay for double-stranded DNA quantitation. Anal Biochem 1997, 249:228-238 [DOI] [PubMed] [Google Scholar]
- 13.Gallagher RE, Willman CL, Slack JL, Andersen J, Li Y, Viswanatha D, Bloomfield CD, Applebaum FR, Schiffer CA, Tallman MS, Wiernik PH: Association of PML-RARa fusion mRNA type with pre-treatment hematologic characteristics but not treatment outcome in acute promyelocytic leukemia: an intergroup molecular study. Blood 1997, 90:1656-1663 [PubMed] [Google Scholar]
- 14.Zhou DC, Hallam SJ, Lee SJ, Klein RS, Wiernik PH, Tallman MS, Gallagher RE: Constitutive expression of cellular retinoic acid binding protein II and lack of correlation with sensitivity to all-trans retinoic acid in acute promyelocytic leukemia cells. Cancer Res 1998, 58:5770-5776 [PubMed] [Google Scholar]
- 15.Beutler E, Gelbart T, Kuhl W: Interference of heparin with the polymerase chain reaction. Biotechniques 1990, 9:166. [PubMed] [Google Scholar]
- 16.Diverio D, Rossi V, Avvisati G, DeSantis S, Pistilli A, Pane F, Saglio G, Martinelli G, Petti MC, Santoro A, Pelicci PG, Mandelli F, Biondi A, Lo Coco F: Early detection of relapse by prospective reverse transcriptase-polymerase chain reaction analysis of the PML/RARalpha fusion gene in patients with acute promyelocytic leukemia enrolled in the GIMEMA-AIEOP multicenter “AIDA” trial. Blood 1998, 92:784-789 [PubMed] [Google Scholar]
- 17.Wattjes MP, Krauter J, Nagel S, Heidenreich O, Ganser A, Heil G: Comparison of nested competitive RT-PCR and real-time RT-PCR for the detection and quantification of AML1/MTG8 fusion transcripts in t(8;21) positive acute myelogenous leukemia. Leukemia 2000, 14:329-335 [DOI] [PubMed] [Google Scholar]
- 18.Willman C, L., Yeap B, Bi W, Slack JL, Harrington DP, Livak KJ, Appelbaum FR, Bloomfield CD, Tallman MS, Gallagher RE: Predictive value of automated, quantitative real-time RT-PCR performed at the end of consolidation of acute promyelocytic leukemia (APL): A SWOG, CALGB, and ECOG Intergroup Study (INT-0129). Proc Am Soc Clin Oncol 2000, 19:5a (Abstract 9)
- 19.Sun R, Ku J, Jayakar H, Kuo JC, Brambilla D, Herman S, Rosenstraus M, Spadoro J: Ultrasensitive reverse transcription-PCR assay for quantitation of human immunodeficiency virus type 1 RNA in plasma. J Clin Microbiol 1998, 36:2964-2969 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Meijerink J, Mandigers C, van De Locht L, Tonnissen E, Goodsaid F, Raemaekers J: A novel method to compensate for different amplification efficiencies between patient DNA samples in quantitative real-time PCR. J Mol Diagn 2001, 3:55-61 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Mensink E, van de Locht A, Schattenberg A, Linders E, Schaap N, Geurts van Kessel A, De Witte T: Quantitation of minimal residual disease in Philadelphia chromosome positive chronic myeloid leukaemia patients using real-time quantitative RT-PCR. Br J Haematol 1998, 102:768-774 [DOI] [PubMed] [Google Scholar]
- 22.Preudhomme C, Revillion F, Merlat A, Hornez L, Roumier C, Duflos-Grardel N, Jouet JP, Cosson A, Peyrat JP, Fenaux P: Detection of BCR-ABL transcripts in chronic myeloid leukemia (CML) using a “real time” quantitative RT-PCR assay. Leukemia 1999, 13:957-964 [DOI] [PubMed] [Google Scholar]
- 23.Pongers-Willemse MJ, Verhagen OJ, Tibbe GJ, Wijkhuijs AJ, de Haas V, Roovers E, van der Schoot CE, van Dongen JJ: Real-time quantitative PCR for the detection of minimal residual disease in acute lymphoblastic leukemia using junctional region specific TaqMan probes. Leukemia 1998, 12:2006-2014 [DOI] [PubMed] [Google Scholar]
- 24.Marcucci G, Livak KJ, Bi W, Strout MP, Bloomfield CD, Caligiuri MA: Detection of minimal residual disease in patients with AML1/ETO-associated acute myeloid leukemia using a novel quantitative reverse transcription polymerase chain reaction assay. Leukemia 1998, 12:1482-1489 [DOI] [PubMed] [Google Scholar]
- 25.Gerard CJ, Olsson K, Ramanathan R, Reading C, Hanania EG: Improved quantitation of minimal residual disease in multiple myeloma using real-time polymerase chain reaction and plasmid-DNA complementarity determining region III standards. Cancer Res 1998, 58:3957-3964 [PubMed] [Google Scholar]
- 26.Branford S, Hughes TP, Rudzki Z: Monitoring chronic myeloid leukaemia therapy by real-time quantitative PCR in blood is a reliable alternative to bone marrow cytogenetics. Br J Haematol 1999, 107:587-599 [DOI] [PubMed] [Google Scholar]
- 27.Pallisgaard N, Clausen N, Schroder H, Hokland P: Rapid and sensitive minimal residual disease detection in acute leukemia by quantitative real-time RT-PCR exemplified by t(12;21) TEL-AML1 fusion transcript. Genes Chromosomes Cancer 1999, 26:355-365 [DOI] [PubMed] [Google Scholar]
- 28.Cassinat B, Zassadowski F, Balitrand N, Barbey C, Rain JD, Fenaux P, Degos L, Vidaud M, Chomienne C: Quantitation of minimal residual disease in acute promyelocytic leukemia patients with t(15;17) translocation using real-time RT-PCR. Leukemia 2000, 14:324–328 [DOI] [PubMed]