Preimplantation genetic diagnosis (PGD) is a procedure that allows embryos to be tested for genetic disorders before they enter the uterus and before pregnancy has begun. Embryos obtained by in vitro fertilization undergo a biopsy procedure in which one or two cells are removed and tested for a specific disorder. If the cell is unaffected, the embryo from which it was taken is judged to be free of the disorder. The embryo can then be transferred to the uterus to initiate pregnancy. Couples whose children are at increased risk for a specific genetic disorder can benefit from PGD. Some of these couples may have affected family members or family ancestry that puts them at high risk for transmitting a particular disorder to their offspring. PGD is an alternative to prenatal tests such as amniocentesis or chorionic villus sampling and since it is performed before a pregnancy has begun, it may be more acceptable to couples who have either had an affected child, previous termination of pregnancy, or who have objections to termination of pregnancy.
PGD tests have largely focused on two methodologies: fluorescent in situ hybridization (FISH) and polymerase chain reaction (PCR). This review will focus on the use of PCR-based methodologies to diagnose single gene disorders in single cells; specifically describing the characteristics and limitations of single cell PCR and mutation detection strategies which have been developed for use in clinical PGD.
The hundreds of cycles of preimplantation diagnosis performed to date have resulted in the birth of several hundred healthy children. 1 As shown in Table 1 , the genetic conditions for which PGD has been applied are numerous and the various methods used for diagnosis reflect the heterogeneity of causative mutations.
Table 1.
Strategies for PCR-Based Tests Used for Clinical Preimplantation Genetic Diagnosis
| Method | Disorder to be diagnosed | Mutation type |
|---|---|---|
| Single PCR, agarose gel (+/− Y band) | X-linked disorders 2 | Various (gender determination to exclude hemizygotes) |
| Nested PCR, agarose gel (+/− X/Y) | X-linked disorders 6 | Various (gender determination to exclude hemizygotes) |
| Nested PCR, heteroduplexing | Cystic fibrosis 22, 38, 44, 75 Tay-Sachs disease 111 | 3 bp deletion (ΔF508) 4 bp insertion |
| Nested PCR, allele-specific amplification | RhD blood typing 3 Myotonic dystrophy 127 | +/− RhD gene determines Rh status Expansion of (CTG)n trinucleotide repeat |
| Nested PCR, restriction enzyme | Cystic fibrosis, 23 Beta thalassemia, 83 Marfan syndrome, 107 Epidermolysis Bullosa, 100 Lesch-Nyhan syndrome, 101 Sickle cell anemia, 102 Fanconi’s anemia, 103 Ornithine transcarbamylase deficiency, 104 Spinal muscular atrophy 108, 109, 110 | Various point mutations Deletion. Distinguish between gene and pseudogene |
| Nested PCR, restriction enzyme (2 mutations in 1 fragment) | Skin fragility ectodermal dysplasia syndrome 66 | Allows detection of ADO |
| Whole genome amplification and comparative genome hybridization | Aneuploidy screening 96 | NA |
| Whole genome amplification (PEP) | Familial adenomatous polyposis coli 60 | Multiple analyses from each sample |
| Nested PCR, linked markers | Duchenne muscular dystrophy 10, 125 Ornithine transcarbamylase deficiency 104 | Exon deletions Point mutation (linked marker for ADO detection) |
| Nested PCR, SSCP | Familial Adenomatous Polyposis Coli 60 | Point mutation |
| Nested PCR, direct cycle sequencing | Skin fragility ectodermal dysplasia syndrome 66 | Point mutations (cycle sequencing to confirm restriction digest) |
| Nested PCR, DGGE | Beta thalassemia 115 | Point mutations |
| Heminested PCR, site specific mutagenesis | Retinitis pigmentosa 99 Ornithine transcarbamylase deficiency 104 | Point mutation Point mutation |
| Heminested PCR, allele dependent length polymorphism | Retinitis pigmentosa 99 | Point mutation |
| Nested multiplex PCR (including linked markers) | Marfan syndrome 119 Epidermolysis Bullosa 100 Beta thalassemia 83 | Unknown mutation Monitor allele dropout |
| Nested multiplex PCR (including linked and non-linked markers) | Sickle cell anemia, 41 hemophilia B, 41 cystic fibrosis, 41 Gaucher disease, 61 Long chain 3-hydroxyacyl-CoA dehydrogenase deficiency 61 | Monitor allele dropout and contamination |
| Fluorescent PCR, allele size (fragment analysis) | Huntington disease 32 Cystic fibrosis 106 Myotonic dystrophy 55 Fragile X syndrome 128 | Expansion of (CAG)n trinucleotide repeat 3 bp deletion Expansion of (CTG)n trinucleotide repeat Expansion of (CGG)n trinucleotide repeat |
| Fluorescent PCR, SSCP | Medium chain acyl CoA dehydrogenase deficiency 76 | Point mutation |
| Fluorescent PCR, ARMS | Spinal muscular atrophy 77 | Exon deletion in gene but not pseudogene |
| Fluorescent PCR, restriction analysis | Congenital adrenal hyperplasia, 78 osteogenesis imperfecta, 105 medium chain acyl CoA dehydrogenase deficiency, 33 Sickle cell anemia 34 | Point mutations |
| Fluorescent PCR, restriction analysis (2 mutations in 1 fragment) | Beta thalassemia 34 | Point mutations, small deletion |
| Multiplex Fluorescent PCR | Beta thalassemia 34 | Point mutations, small deletion |
| Multiplex Fluorescent PCR (including unlinked marker) | Myotonic dystrophy 130 | Expansion of (CTG)n trinucleotide repeat/contamination control |
| Multiplex Fluorescent PCR (including linked marker) | Medium chain acyl CoA dehydrogenase deficiency 76 | Maternal mutation unknown |
| Fluorescent PCR, linked markers only | Fragile X syndrome 120 Marfan syndrome 121 Charcot Marie Tooth disease 45 Cystic fibrosis 123 | Expanded (CGG)n repeat (refractory to PCR) Unknown mutation Gene duplication Heterogeneous mutations |
The first clinical application of PGD used a generic PCR protocol for gender determination to avoid the transfer of male embryos which have a 50% probability of being affected by an X-linked recessive disorder. Gender was determined in a single blastomere by a single round of PCR using primers for Y-chromosome specific repetitive DNA sequences. The presence of Y-specific PCR amplification products was indicative of a male embryo and the absence of products was scored as female. 2 Although this approach had some success, a misdiagnosis, presumably due to amplification failure, did occur and emphasized the challenges inherent in single cell analysis and, more specifically, the danger in relying on the absence of amplification to diagnose genotype. 3 Subsequently, PCR protocols for preimplantation gender determination were refined to include primer sets which simultaneously amplify sequences common to both sex chromosomes (for example single copy genes such as ZFX/ZFY, 4 AMELX/AMELY, 5 ) and repetitive sequences such as DXZ1 and DYZ1. 6, 7 Sequences common to the sex chromosomes are identical at the site of primer annealing but differ internally in terms of size or include minor polymorphisms. Despite these technical improvements, the majority of embryo sexing is now accomplished using fluorescent in situ hybridization (FISH) which is less prone to contamination and can also provide the copy number for each chromosome tested thereby potentially avoiding the transfer of common chromosome abnormalities such as triploidy or X-monosomy. 8, 9
Although FISH has largely superseded PCR for sex determination, the specific diagnosis of single-gene defects remains dependent on DNA amplification with PCR. In the case of X-linked disorders, testing of the specific gene has the added advantage of ensuring that all embryos free of the mutant gene can be selected for transfer, irrespective of gender. 10, 11, 12 The list of disorders and the particular mutation detection strategies used for PCR-based clinical PGD application are given in Table 1 .
Materials and Methods
Essentially there are two laboratory components involved in PGD. The first involves the collection of diagnostic material for testing. This is usually performed in a clinical in vitro fertilization (IVF) laboratory under sterile conditions. A set of micromanipulators linked to an inverted microscope with contrast optics and facilities for extended embryo culture are the minimum essential requirements to carry out diagnostic biopsy procedures. The second step involves the diagnostic test itself, which can be performed in a region of the IVF laboratory, an adjacent laboratory equipped to perform molecular analyses or in a completely separate dedicated molecular genetics laboratory equipped to process single cell samples. Minimum requirements include a PCR preparation area (usually a small, dedicated flow hood), dedicated PGD reagent storage facilities, thermal cycler, and access to the necessary post-PCR mutation detection apparatus. The critical component of the diagnostic step is to minimize the level of contamination and a number of possible laboratory designs and procedures may fulfill this requirement.
Theoretically, diagnostic material can be collected at any developmental stage between the mature oocyte and blastocyst. To date, four distinct stages have been targeted; metaphase II oocyte, zygote, cleavage stage embryo, and blastocyst. The four stages dictate different diagnostic strategies, each with its own limitations. The different technical approaches required to obtain the material and the material itself can affect the success rate of the procedure. The strengths and limitations of each approach are summarized in Table 2 .
Table 2.
Strategic Considerations for PCR Analysis of Diagnostic Material Biopsied at Different Developmental Stages for Preimplantation Genetic Diagnosis
| Stage | Advantages | Disadvantages |
|---|---|---|
| Oocyte (1st polar body) | Removal has no effect on embryo development Increased time to perform PCR analysis prior to transfer | Only 1 cell available for analysis Increased risk of diagnostic error Maternally inherited disease only Fewer embryos for transfer (recombination) |
| Zygote (1st and 2nd polar body) | 2 cells for analysis (greater accuracy/reliability) Removal has no effect on embryo development Increased time to perform PCR analysis prior to transfer | Maternally inherited disease only Narrow time window to complete biopsy |
| Cleavage stage (blastomeres) | Diagnosis of maternally/paternally inherited disorders Large body of clinical data available 2 cells available for analysis (greater accuracy/reliability) | Chromosomal mosaicism present Selection of nucleated blastomere is critical |
| Blastocyst (trophectoderm) | Sample multiple cells (eliminate PCR failure/ADO) Trophectoderm sampled rather than inner cell mass Embryo quality preselected Higher implantation rate/lower multiple gestation rate | Time for PCR analysis may be limited Cells may not be representative of embryo Fewer embryos for analysis Limited clinical data available |
Each of the biopsy methods involves at least two steps; the first step being to breach the zona pellucida while the second involves the removal of cellular material (be that polar body, blastomere, or trophectodermal cells). Zona breaching can be achieved mechanically (by means of a sharp microneedle), chemically (using acidified Tyrodes solution, pH 2.2), or by thermal ablation (using a non-contact laser). Removal of cellular material is generally carried out using a glass micropipette attached to a pneumatic or hydraulic based suction system. 13
At present, polar body biopsy in combination with PCR based assays is performed almost exclusively by one group 14, 15 while the majority of PGD centers 16 obtain genetic material for PGD by cleavage stage biopsy on the third day following insemination when the embryo has between 6 and 10 cells. At this stage, blastomeres are believed to be totipotent and embryo survival and metabolism appears to be unaffected by biopsy. 17 While blastocyst biopsy appears to be a promising approach 18, 19, 20 its clinical utility for PGD has yet to be demonstrated in clinical practice.
Diagnostic Methods
The success of PCR in amplifying small quantities of DNA to a level at which they can be visualized and subjected to further genetic analysis has made the technique one of the most important diagnostic techniques in the modern molecular laboratory. Application of PCR protocols to single cell analyses has proved to be challenging but ultimately highly successful, and remains the only means of detecting specific mutant alleles in human preimplantation embryos. The limited amount of template DNA (approximately 7 pg) available in a single diploid cell leads to a number of problems which are rarely, if ever, observed in routine diagnostic PCR (in which a starting amount of DNA template of at least 10 ng is usually available). Problems frequently encountered include an increased incidence of detectable contamination, amplification failure, and extreme preferential amplification of one allele or complete absence of one allele (allele dropout) in heterozygous samples.
Characteristics of Single Cell PCR (SCPCR)
Amplification Efficiency
Amplification efficiencies at the single cell level are generally lower than those encountered during the routine PCR of DNA samples in which the amount of starting template may be larger by several orders of magnitude. Reduced amplification efficiency can be the result of many problems encountered between sample collection and the PCR procedure itself. Operator problems such as cell loss during the delicate process of cell transfer to the tube or spontaneous cell lysis before the cell entering the tube contribute to amplification failure or reduced amplification efficiency. Intrinsic factors such as anucleate or degenerating cells with concomitant absence or degradation of DNA respectively are more difficult to control. Indeed, blastomeres from poor quality embryos yield lower amplification efficiencies than their high quality counterparts 21, 22 underlining the importance of blastomere selection during embryo biopsy. Following successful transfer of a high quality nucleated cell, the cell lysis protocol used also influences amplification success. Consecutive rounds of freezing and thawing in distilled water or boiling do achieve cell lysis 23 but the use of either alkaline or proteinase based lysis buffers has proved more effective. 24, 25, 26, 27, 28, 29 Nevertheless, there is no consensus as to which lysis buffer is the most effective. 16
Contamination
With only one or two DNA molecules present per haploid (second polar body, oocyte, or sperm) or diploid (blastomere or first polar body) cell respectively, extraneous DNA can easily lead to a misdiagnosis in clinical PGD. Contamination is an omnipresent threat in any molecular diagnostics laboratory but the large number of PCR cycles required for detectable amplification in combination with a single genome starting template exacerbates this threat. A series of stringent experimental practices can be implemented to counter contamination but there is no guaranteed method of eliminating sporadic contamination. Sources of contamination are numerous since DNA (particularly in the form of previously amplified PCR products 30 ) can exist in aerosol form and, as such, is likely to be present on all exposed laboratory surfaces. Such “carry-over” contamination caused by the inadvertent amplification of PCR products generated in previous experiments is a cumulative problem and probably the most significant contamination threat in single cell PCR. To address this problem single cell reactions should be set up in a room designated for this purpose (pre-PCR area) and physically separated from the area in which PCR product analysis occurs (post-PCR area). Pre-PCR areas (including the cell preparation area, the reagent preparation area, and the PCR set-up area) kept under constant positive pressure can prevent the entry of contaminants but much of the cellular and PCR product contamination is introduced by human traffic. For this reason, dedicated gowns, gloves, overshoes, caps, and masks should be worn and remain in this room, together with dedicated equipment such as tubes, racks, and pipettes. Ideally, a unidirectional work flow prevents the re-introduction of items from a post-PCR area into a pre-PCR area. Filtration and autoclaving of reagents, incubation of component reagents of the reaction mix with restriction enzymes to destroy any PCR product (for example exonuclease III, Alu I, Hae III, and Hinf I) 31, 32, 33, 34 or the use of a mineral oil overlay to provide a physical barrier against contamination may be of some value but the introduction of additional components into any clean system may be counterproductive and could potentially reintroduce contaminants. Routine decontamination of work surfaces and equipment using 10% bleach 35 or exposure to ultraviolet light to destroy DNA is also recommended. Unfortunately, no single strategy can be considered to be 100% effective or render the continuous monitoring of contamination levels obsolete. For this reason all PCR reagents, cell washing, and lysis solutions should be rigorously tested for contamination before any clinical diagnostic application.
Another measure for contamination control which has been used extensively in infectious disease screening by sensitive PCR but not yet in PGD is post-PCR sterilization. One method uses uracil DNA glycosylase (UDG) to cleave uracil bases from PCR products in which dUTP is substituted for dTTP in the PCR mix. In this way the action of DNA polymerase is blocked exclusively with carry-over contamination products but not native DNA. 36 A different technique uses isopsoralen which binds to PCR products such that photoactivation following amplification damages the DNA strand preventing it from functioning as a template in subsequent PCRs. 37
Cellular DNA from sperm or maternal cumulus cells (both of which may be present on the zona pellucida of the human embryo) is another potential source of contamination but can be largely eliminated by means of precautions in the IVF laboratory. All cumulus cells must be carefully removed before biopsy and the embryo checked under an inverted microscope. Moreover, the use of intracytoplasmic sperm injection (ICSI) a technique used to introduce a single sperm into the cytoplasm of the oocyte has circumvented the problems caused by supernumerary sperm which frequently bind to the zona pellucida in large numbers following standard insemination techniques. 38 The biopsied blastomeres themselves should be washed through a series of fresh drops of holding medium known to be contamination-free before transfer to the PCR tube. The commonly used precautions against contamination are listed in Table 3 .
Table 3.
Precautions against Contamination in Single Cell PCR
| Type of contamination | Precautions |
|---|---|
| Operator contamination (can be cellular or product contamination; see below) | Elimination or reduction Protective clothing: gloves (close fitting), cap, overshoes, gown Frequent changes of gloves Operator technique Detection DNA Fingerprinting (incorporate informative polymorphic markers) |
| Product contamination (PCR products from previous reactions also known as “carry-over” contamination) | Elimination or reduction Dedicated equipment (PCR machine, pipettes, tubes) Dedicated reagents (all solutions) Filtration of reagents Filtered pipette tips Positive displacement pipettes Aliquot all reagents One-time use of tips and reagent aliquots UV irradiation of preparation area/equipment/reagents Autoclaving equipment and reagents Restriction enzyme digestion of PCR master mix (component reagents) Switch from nested PCR to FPCR Geographical separation of pre-PCR/PCR and post-PCR activities Preparation of PCR reagents in laminar flow Decontamination of surfaces/equipment with 10% bleach Post-PCR sterilization (dUTP and UDG/isopsoralen) Purchase reagents as ready-made ‘molecular biology grade’ solutions Reduce number of tube-opening events Mineral oil overlay Detection Switch from nested PCR to FPCR Use of multiple negative controls (cell wash blanks and reagent blanks) Test all component reagents before clinical case |
| Genomic DNA (gDNA) contamination (eg, DNA used for positive controls/assay development) | Elimination or reduction Isolate procedures involving gDNA (eg, no gDNA in reagent prep room) |
| Cellular contamination (eg, Maternal cells (cumulus), paternal cells (sperm), embryonic material (from different embryos) or operator cells (epithelial)) | Elimination or reduction Rinse embryo thoroughly (to remove cumulus cells) Exclusive use of Intracytoplasmic sperm injection for fertilization (to prevent supernumerary sperm exposure) Wash blastomere thoroughly Change micropipettes if a cell lyses during biopsy or dish-to-tube transfer Detection DNA “fingerprinting” (incorporate informative polymorphic markers) Wash blank controls |
Allele Dropout
Another problem unique to single cell PCR is that of allele dropout (ADO), a phenomenon whereby only one of the two alleles present is successfully amplified. 39, 40, 41 ADO is only detectable when heterozygous alleles are present but appears to be indiscriminate, in that the allele successfully amplified is random (even when only differing by a single nucleotide). 42 ADO remains the biggest obstacle to accurate and efficient PGD for single gene disorders and the severity of its consequences is closely linked to the mode of inheritance of the disorder under test. For autosomal recessive conditions when both partners are carrying the same mutation, ADO should not, in the absence of contamination, result in the transfer of an affected embryo. However, the number of embryos available for transfer will decrease as the ADO rate increases, potentially reducing the likelihood of pregnancy. In such cases, there is some reassurance in the calculation that a 10% allele dropout rate would only result in the exclusion of, on average, 2.5% of embryos for which a diagnosis was successfully made (based on a 90% amplification rate). In contrast, for compound heterozygous or autosomal dominant conditions, the consequences of ADO can be catastrophic, as misdiagnosis and subsequent transfer of affected embryos can occur. 43 Indeed ADO is the most likely cause of reported errors in PGD of cystic fibrosis in which affected compound heterozygote embryos were misdiagnosed as carrier embryos because the analysis used could only detect one of the inherited mutations. 9, 44
The frequency of allele dropout reported in the literature varies widely and has been reported to be as high as 25% in a clinical PGD case. 45 This figure could be considered unacceptably high, but the concept of an acceptable ADO rate is meaningful only when parameters relevant to the PGD case have been assessed. For example, a more accurate ADO rate can be established as more cells are analyzed and a higher ADO rate tolerated with contamination rates close to zero in combination with diagnosis of a homozygous recessive mutation.
Reports suggest that blastomeres generally exhibit a greater ADO rate than polar bodies, lymphocytes, or fibroblasts 41, 46 although such differences have not been unanimously reported. 47 Observations that amplification rates are generally lower for blastomeres than other cell types even when a nucleus is present 48, 49 and the detection of haploidy in an estimated 7 to 15% of blastomeres provide further evidence for a cell-specific effect on the observed frequency of ADO. 50, 51
The origins of ADO remain elusive but experimental data supports the causative factors being suboptimal PCR conditions and/or incomplete cell lysis. Adequate denaturation is essential for amplification of both alleles as demonstrated by a reduction in ADO when the denaturation temperature is increased in the first cycles of PCR. 27
ADO could also arise from DNA deterioration or damage such as strand breaks caused by endogenous nucleases. As with reduced amplification efficiencies, increased ADO is noted in degenerating cells presumably the result of strand-specific DNA degradation. 52 Additionally, access to the target genomic sequence by the primers and Taq polymerase may be restricted by, for example, adjacent G/C rich regions reducing denaturation efficiency or differing degrees of folding perhaps related to the stage of the cell cycle. 27 Whatever the exact cause, ADO likely arises in the initial cycles of the primary PCR before the number of target molecules is increased by the process of amplification. Evidence for this suggestion comes from experiments in which different proportions of two separate populations of single cells (each homozygous for a different sized triplet repeat sequence) are mixed, demonstrating that the minority allele is undetectable when the starting template ratio is less than one in four cells. 53
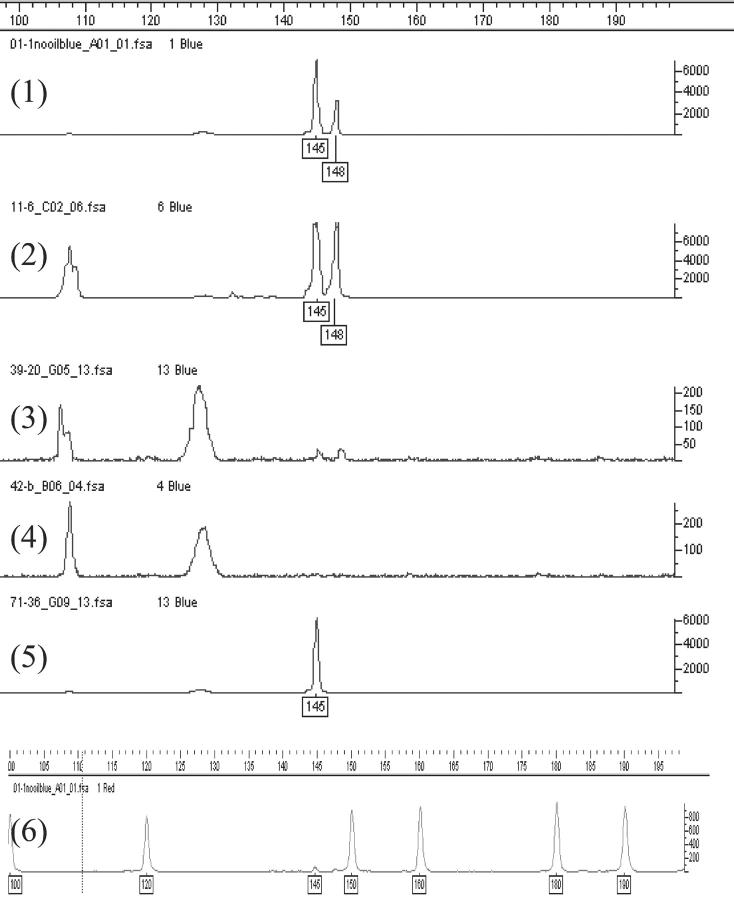
Allele dropout observed during conventional nested PCR with ethidium bromide detection comprises both extreme preferential amplification, in which the PCR product from one allele is present but at extremely low levels, and true allele dropout (in which one allele is either absent or has totally failed to amplify). Enhanced detection methods such as the use of fluorescent primers 40 and SYBR green I staining 54 have shown that a proportion of observed ADO is due to extreme preferential amplification. A fourfold reduction in ADO for both lymphoblasts and blastomeres has been reported after switching from an ethidium based protocol to one using fluorescence primers. 55 However, a significant proportion of true ADO exists even using fluorescent PCR. 33, 42 Such observations from single cell fluorescent PCR reinforce the need for cut-off values to distinguish background noise, contamination, extremely low amplification, preferential amplification, and allele dropout. Examples of preferential amplification and allele dropout from samples of single heterozygous cells are shown in Figure 1 .
Figure 1.
Genotyping of single heterozygous cells after fluorescent PCR. Lanes 1, 2, 3, 5: Single lymphoblast cells heterozygous for deltaF508 mutation (3-bp deletion) in cystic fibrosis. Lane 1 demonstrates preferential amplification of the deleted allele (145 bp). Lane 2 shows equivalent amplification from both alleles. Lane 3: Scored as amplification failure. Note the extremely low peaks in lane 3 (corresponding to peaks at 145 and 148 bp) considered technical artifacts in view of the extremely low signal amplitude and proximity to a strong positive lane. Lane 4: Negative control (wash drop blanks). No amplification observed. Lane 5: Allele dropout in which the wild-type allele (148 bp) has failed to amplify to detectable levels. Lane 6: ROX-labeled size standard with peaks at 100, 120, 150,160, 180, and 190 bp. This size standard is labeled with a red fluorescent dye and is added to all samples to allow accurate sizing in each lane. Lanes 1–5 are shown with the size standard trace removed for clarity. The y axis for each trace represents units of fluorescence and the x axis represents sizing (in bp) according to the internal size standard. All PCR products were generated using FAM-labeled primer and identified using an ABI3100 DNA analyzer with Genescan software.
The use of alkaline lysis buffer or lysis buffer containing proteinase and detergent also seems to be beneficial in reducing ADO 24, 25, 26, 27, 28, 29 although there is no consensus as to which lysis buffer to use. 16 Indeed, two reports offered dramatically different conclusions with one favoring proteinase K 28 and the other favoring alkaline lysis buffer. 29
Protocols that rely on reverse transcription of abundant mRNA molecules followed by PCR (RT-PCR) and subsequent mutation analysis have been proposed as a means of reducing amplification failures and ADO since multiple targets should not be subject to allele specific amplification failure. Single cell expression assays have been developed for the diagnosis of Marfan 56 and Lesch-Nyhan 57 syndromes. Such assays could prove valuable for genes that are expressed at the cleavage stage, provided that they are not subject to genomic imprinting and that residual maternally-derived transcripts from the oocyte or alternatively spliced products 57 do not confuse the diagnosis.
Several other methods of decreasing ADO include the use of restriction enzymes before PCR to shorten genomic template strands (presumably making them more accessible to the polymerase enzyme during the first few cycles of PCR) 58 and the use of Taq/Pwo polymerase mixture (perhaps because of the proof-reading ability of Pwo polymerase). 59
In addition to reducing ADO, strategies have been proposed to increase the detection of ADO. One such strategy is the use of linked markers 46, 60, 61 which simultaneously controls for contamination. 62 Use of one or two linked markers reduces undetected ADO by approximately 50% and 75% respectively and with three linked markers ADO is virtually always detected. 46 The use of linked markers carries considerable advantages not only from the point of view of reducing the possibility of misdiagnosis, but also by potentially increasing the number of embryos available for transfer. 63 For example, in a homozygous recessive condition, carrier embryos could still be transferred even when the normal allele appears to be absent due to ADO, but a linked marker (or markers) is present. However, the identification and work-up of reliable informative linked markers can be labor intensive and may not always be cost effective for all diseases, particularly when the patient population is very small.
Another strategy used to increase ADO detection is special design of the PCR assay itself. For disorders in which a triplet repeat expansion (which is refractory to PCR) is the disease causing DNA sequence change, an assay based on detection of two normal sized triplet repeat alleles will prevent transfer of affected embryos when the parental alleles are informative. In addition, assays in which a single amplified fragment encompasses both mutations of a compound heterozygous condition should always allow the detection of allele dropout in an affected blastomere 24, 64, 65, 66 (Figure 2) . In such cases, ADO of the wildtype allele in carrier embryos will result in a restriction pattern suggesting homozygosity for one particular mutation. This result is not possible when the parents carry different mutations. Such a pattern in a clinical diagnosis would result in rejection of that particular embryo for transfer since it would be impossible to distinguish between an unaffected carrier and an affected compound heterozygous embryo.
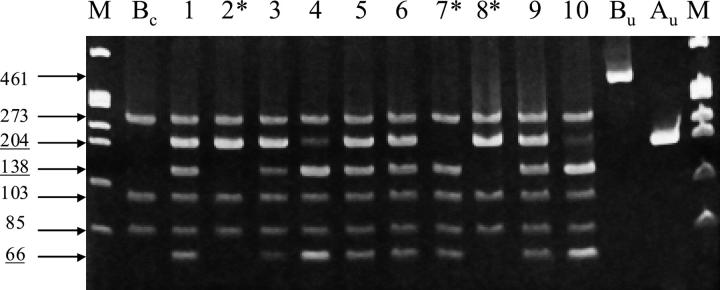
Figure 2.
Use of internal restriction digestion control to avoid misdiagnosis. The PCR reaction generates a 204-bp product from the LAMA3 gene and a 461-bp product from the LAMB3 gene (which is used as an internal control). Digestion with Dde I cleaves the LAMB3 product into fragment sizes of 273, 103, and 85 bp (Bc) and cleaves only the LAMA3 allele that contains the R650X mutation (into fragment sizes 138 bp and 66 bp). M, Marker 1-kb DNA ladder (φX174 DNA/Hae III marker, Promega Corporation). Lanes 1–10: Single lymphocytes heterozygous for R650X mutation in LAMA3 gene. Au and Bu, uncut LAMA3 and LAMB3 PCR products, respectively. Allele dropout (*) is apparent in lanes 2, 7 and 8. The internal digestion control prevents an affected cell being misdiagnosed as unaffected (as a consequence of failed Dde I digestion) since the LAMB3 product remains undigested (Bu) at 461 bp.
The existence of ADO and contamination (which may have been responsible for a serious misdiagnosis of myotonic dystrophy) prompted the directive to test two cells from each embryo regardless of the mode of inheritance. 67 Certainly, the probability of ADO affecting the same allele in both cells in independent reactions is low. 63 In a mouse embryo model, dual blastomere biopsy combined with independent blastomere analysis improved preimplantation diagnostic reliability dramatically for a dominant 68 condition but only slightly when the inheritance was recessive. 69 Whereas increasing the DNA template threefold is not effective in reducing ADO, 42 the risk of error due to ADO can be virtually eliminated if more than four cells are taken and independently analyzed. 70 Although blastocyst biopsy could make this approach feasible, the routine removal of four cells from cleavage stage embryos would be unacceptable in terms of the negative impact on subsequent embryo development. If stringently applied, even a two-cell policy would be dramatically affected by suboptimal embryo development on day 3, instantly reducing the cohort for biopsy and ultimately the number of potentially unaffected embryos available for transfer. A recent retrospective analysis showed that implantation rates of biopsied embryos were equivalent regardless of whether one or two cells had been removed 67 but so far no prospective randomized studies have been performed to test this hypothesis. A summary of the methods used for the reduction and detection of ADO is shown in Table 4 .
Table 4.
Strategies for the Reduction and Detection of Allele Dropout (ADO)
| Action/measure | Reduce/detect ADO | Mechanism | Potential problems/disadvantages |
|---|---|---|---|
| Use of any lysis buffer | Reduce | Protein removal/DNA accessibility/destroys endogenous nucleases (fewer DNA strand breaks) | Quality control of additional reagents |
| Choice of lysis buffer | Reduce | As above | Quality control of additional reagents |
| Increase denaturation temperature in first ten cycles of PCR | Reduce | Accessibility of DNA, complete denaturation of DNA strands | Taq polymerase failure due to prolonged exposure to high temperature |
| ≥2 cells taken from cleavage stage embryo (analyzed together) | Reduce | Increase starting template reduces probability of ADO | Possible detrimental effects of 2 cell biopsy |
| Reverse-transcriptase PCR | Reduce | Increased starting template | Prone to contamination/ presence of maternal transcripts/imprinted genes will exhibit ADO |
| Restriction enzyme digestion prior to PCR | Reduce | Shortens genomic DNA template strands—facilitating primer-template annealing | Limited data available |
| Use of Taq/Pwo-polymerase mixture | Reduce | Proof-reading activity? | No data available for single cells |
| Blastocyst biopsy (>2 cells) | Reduce/detect | Increase starting template reduces probability of ADO | Reduced embryo cohort at blastocyst stage |
| ≥2 cells taken from cleavage stage embryo (analyzed independently) | Detect | Low probability of two independent analyses both exhibiting ADO | Possible detrimental effects of 2 cell biopsy |
| Fluorescent PCR | Detect | ∼1000 times more sensitive than ethidium. High sensitivity can identify preferential amplification | Equipment and reagent cost |
| SYBR green I fluorescent stain | Detect | ∼25 times more sensitive than ethidium bromide. High sensitivity can identify preferential amplification | Reagent cost |
| Same fragment PCR | Detect | Impossible to have ADO if fragment contains both mutations of interest. Either both alleles are detected or amplification failure is observed | Only a small proportion of compound heterozygous conditions have mutations within several hundred base pairs of each other |
| Diagnosis of two normal alleles | Detect | In the absence of contamination, the presence of two normal alleles indicates that a mutant (expanded) allele cannot be present | Maternal and paternal alleles must be informative |
| Use of linked markers | Detect | Low probability of ADO occurring at a series of different adjacent loci | Requires design of single cell duplex/multiplex PCR |
PCR Strategies
Nested PCR
Using a conventional ethidium bromide-based detection system, around 50 to 60 cycles of PCR are required to obtain detectable products from unique sequences. Since the enzyme Taq polymerase incorporates mistakes after approximately 40 cycles, aspecific products appear with such large numbers of cycles. This problem led to the development of nested PCR in which two sequential rounds of PCR are used to improve sensitivity and specificity when amplifying unique sequences from single cells. 71, 72 The primary PCR generates DNA fragments encompassing the mutation site but which are insufficient in number to be visualized. PCR products from the first reaction are transferred to a new PCR tube and are amplified to detectable levels using a different set of primers situated internally to the first. This strategy enhances the specificity of PCR, as well as reducing the risk of carry-over contamination for subsequent primary amplifications, as the secondary product cannot be amplified by the outer set of primers. However, the threat of contamination from primary PCR products in the first round PCR and secondary products in the second round PCR remains.
Fluorescent PCR
Fluorescent PCR (FPCR) is fast becoming the method of choice for laboratories performing single cell PCR for a number of reasons. Compared with nested PCR, FPCR combines increased sensitivity and throughput, shorter turnaround time, 73 and superior precision in fragment sizing. 62 The use of a laser system to perform automated fragment analysis with various fluorescent molecules, each with their own unique wavelength of emitted light, allows simultaneous discrimination of unrelated, similarly sized products. Furthermore, the thousandfold increase in sensitivity 74 compared with ethidium bromide allows a single round of PCR, potentially avoiding the contamination which can result from multiple tube openings.
The accuracy of automated fragment analysis enables, for example, a deletion of 3 bp in the ΔF508 mutation causing cystic fibrosis, to be clearly differentiated from the normal allele after fluorescent PCR (Figure 1) removing the need for either heteroduplex analysis 75 or lengthy conventional electrophoresis using a high resolution gel. Several different instruments are available for such analyses and the technique has been successfully applied to PGD development and clinical cases in many laboratories. 55, 62, 73 Fluorescent PCR is also compatible with many established forms of mutation analysis such as SSCP, 76 ARMS, 77 and restriction enzyme digestion. 78
Multiplex PCR
By using combinations of unrelated primer sets in one PCR assay (multiplex PCR) it is possible to amplify multiple loci simultaneously and attempt to overcome the limitations of the single cell. 62, 79, 80, 81 Providing there is no interaction between unrelated primers or PCR products, the various loci should be amplified simultaneously within a single reaction. Each multiplex PCR need only be optimized for the combination of primers involved. Successful multiplex reactions enable the simultaneous assessment of numerous loci, with as many as 15 analyzed from larger DNA samples. 82 It may also be possible to assess similar numbers of loci in single cells but to date the maximum number of sequences amplified simultaneously from a single cell is seven using either conventional ethidium detection 12 or fluorescent PCR. 62 Unfortunately the problems of allele dropout and preferential amplification persist even with the FPCR approach. 40, 80
Multiplex PCR can also be used to detect ADO by simultaneous amplification of a disease causing mutation and an informative “linked” polymorphism. This is a particularly useful strategy when diagnosing dominant disorders, but has also been reported for a number of recessive disorders including cystic fibrosis, 41 β-thalassemia, 83 and medium chain acyl CoA dehydrogenase deficiency. 76 The probability of ADO affecting both mutation site and linked polymorphism is very low and consequently the mutant allele can almost always be detected.
Whole Genome Amplification
One of the most exciting developments in single cell analysis has been the evolution of protocols designed to amplify the entire genome from a single cell. Depending on the particular whole genome amplification (WGA) method used, a starting template of approximately 7 pg of DNA can be amplified up to 1000 times apparently overcoming the limitation of a single cell. 84 The technical difficulties sometimes associated with multiplex PCR, such as incompatibility of primer sets and problems distinguishing the various amplified products, are not encountered using WGA. Moreover WGA provides a supply of sample DNA that can be reassessed, allowing confirmation of diagnosis using the same or different methods or the analysis of other genes. The most commonly used method has been primer extension preamplification (PEP) which utilizes 15 base oligonucleotide primers of random sequence to initiate DNA synthesis throughout the genome. 85 Reports estimate that between 70% and 96% of the genome is amplified between 30 and 1000 times. 84, 85, 86
PEP has been used to develop PGD protocols for single cell analysis of Tay-Sachs disease, 87 cystic fibrosis, 88, 89 hemophilia A, 90 and Duchenne muscular dystrophy, 91 but its clinical application has been limited. One problem is the length of time necessary, since PEP mandates an embryo transfer on day 4 post-fertilization at the earliest; however, a modified protocol has been reported that reduces the time required from >14 hours to 5 hours 30 minutes. 92 Nevertheless, PEP was successfully used for PGD in the dominant cancer syndrome familial adenomatous polyposis coli (FAP) allowing the subsequent amplification of two different fragments, one containing a mutation and the other an informative polymorphism. 60
Another form of WGA is degenerate oligonucleotide primed PCR (DOP-PCR) which was designed to give general amplification of target DNAs at frequently occurring priming sites, without restrictions due to the complexity of the DNA or the species from which it was derived. It rests on the principle of priming from short sequences specified by the 3′ end of the oligonucleotides used, during the initial low temperature cycles of the PCR protocol. Since these short sequences occur frequently and are evenly distributed throughout the genome, amplification of target DNA proceeds at multiple loci simultaneously. Annealing of the specified 3′-most primer sequence is stabilized by the adjacent six bases of degenerate sequence which create a pool of 4096 primers of different sequence, as opposed to the single sequence of a nondegenerate primer. At the 5′ end of the primer is a further specified sequence which allows efficient annealing of primers to previously amplified DNA, enabling a higher annealing temperature to be used in later PCR cycles. 93 DOP-PCR amplifies a similar proportion of the genome to PEP, but to a much more significant level. A single cell subjected to DOP-PCR can provide enough DNA for over 100 subsequent PCR amplifications. 86 Furthermore sufficient DNA is produced to allow additional experimental procedures such as comparative genomic hybridization (CGH) for the detection of chromosome copy number in embryos 94, 95 —an approach recently applied in clinical PGD. 96
ADO rates after PEP and DOP-PCR are comparable to those obtained by direct amplification of single cell loci. 86 A significant drawback of WGA techniques is that amplification of repetitive DNA sequences, such as short tandem repeats, is error prone if performed on WGA products. 86, 97 In some studies over 50% of fragments amplified differed from their expected size presumably due to the uniformly low temperatures needed for WGA which could allow slippage of the DNA chain during product generation. 86 Such errors would currently rule out the use of WGA for the clinical diagnosis of triplet repeat expansion diseases or diagnoses based on linkage analysis with STRs. The current difficulties associated with the WGA approach will no doubt be overcome because of the enormous potential of the technique to be combined with repeated simplex and multiplex PCR analysis, CGH, 86 and microarray analysis. 84
Detection Methods
Once DNA from a single cell has been amplified to a detectable level, most of the mutation detection techniques currently available in diagnostic laboratories can be used for its analysis (Table 1) . Mutation analyses can be broadly divided into three categories: those that are tailor made for the detection of one specific mutation, those that detect a variety of different mutations with a single protocol, and those that do not attempt to detect the mutation but infer the presence of the mutation. Techniques that fall into the first category are generally used in a diagnostic context and usually provide a rapid means of detecting common mutations. Methods in the second category are known as “scanning” methods and are usually applied to searches for mutations that have not been characterized. Scanning methodologies are particularly useful for the diagnosis of inherited disorders caused by a heterogeneous spectrum of mutations, as a single methodology can usually be applied for detection of most of the DNA sequence alterations. The third category, linkage analysis, is frequently used in the presence of a suitable pedigree, when pathological mutations are uncharacterized or when known mutations are refractory to PCR. Although such indications are encountered in couples requesting PGD, the detection of contamination and allele dropout are becoming powerful indications in their own right for the inclusion of linked markers.
Amplification Refractory Mutation System
The annealing of allele specific oligonucleotides is the basis of the amplification refractory mutation system (ARMS) technique. ARMS employs one oligonucleotide to anneal upstream of the mutation site while two other oligonucleotides each anneal exclusively to either the mutant or normal alleles. These allele-specific oligonucleotides merely serve as primers for PCR and are not detected directly. The presence or absence of a specific allele is inferred from the presence of PCR product which is only seen when primer annealing occurs. If nested PCR is used, the outer set of primers is designed to produce an amplicon containing the mutation site. Two different inner amplifications are set up from separate aliquots of the outer reaction, one containing a primer specific to the normal allele and the other a primer for the mutant allele. Amplification from only the mutant allele specific primer would result in the embryo being diagnosed as affected and excluded from transfer. Heterozygous samples would show positive amplification for both normal and mutant primer sets. As with restriction enzyme digestion-based diagnoses, the detection of both mutant and normal alleles is a safer and more informative test than the detection of the mutant allele alone. This methodology has been used for the analysis of the five most prevalent cystic fibrosis mutations in single cells 98 using a nested PCR approach.
A slight modification of this technique which has been applied clinically for the diagnosis of spinal muscular atrophy 77 allows different primers specific for mutant or normal alleles to be included in the same PCR mixture. This provides rapid analysis using a single round of PCR. Normal and mutant alleles are distinguishable using automated fragment analysis following fluorescent PCR 77 or conventional electrophoresis. The latter detection method requires the design of allele specific primers with different lengths, so called allele-dependent length polymorphism (ADLP), and has been clinically applied for the preimplantation diagnosis of retinitis pigmentosa. 99
Restriction Endonuclease Digestion
Amplification of DNA followed by restriction digestion is a common form of mutation detection in preimplantation diagnosis and has allowed single cell diagnosis of a wide variety of disorders, generally involving point mutations. 100, 101, 102, 103, 104, 105, 106 With knowledge of the DNA sequence and the exact mutation, a restriction enzyme may be selected which will cleave a normal DNA strand while a mutant strand remains undigested and the products of digestion can distinguished by electrophoresis. This is the ideal design for a clinical PGD PCR protocol. Conversely, enzymes which digest the mutant but not the normal allele can usually be found, but in such cases incomplete or failed digestion could lead to an embryo being incorrectly diagnosed as normal. 33, 107, 108, 109 For such suboptimal assays the inclusion of an internal digestion control 29 could help to prevent a misdiagnosis (Figure 3) .
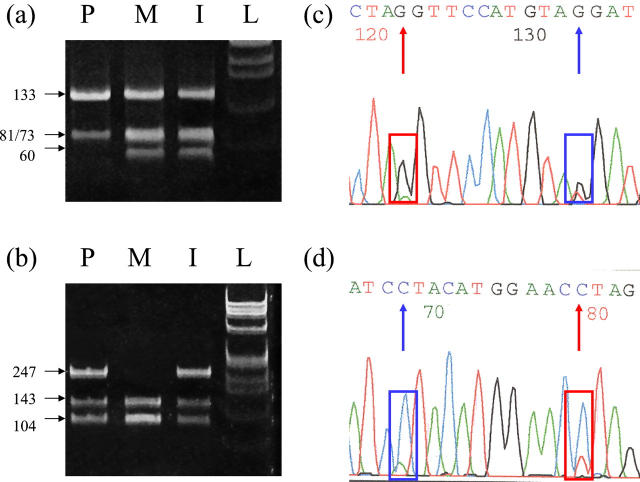
Figure 3.
Restriction enzyme analysis and parallel direct sequencing of PCR products from single cells to detect two different mutations in the same PCR fragment. a and b:. Electrophoretic analysis following Fok I (a) or Bfa I (b) restriction digestion of PCR products from single lymphocytes obtained from carrier parents (each with a separate mutation in the Plakophilin 1 gene) and their affected child who is a compound heterozygote for both mutations. [M, maternal; P, paternal; I, index case (child); L, 1 kb ladder]. a: Maternal mutation is a T-to-G transversion which creates an additional Fok I cut site (GGATG) to generate additional digestion products (133 bp cleaved to 60 and 73 bp) in heterozygous cells (M, I). b: Paternal mutation is a G-to-A transition which removes an existing Bfa I cut site (CTAG) preventing complete digestion of the 247-bp product (P, I). c and d: Direct sequencing of a purified PCR product from a compound heterozygous cell (I) using big dye terminators on an ABI 310 genetic analyzer. Use of both forward (c) and reverse (d) primers clearly shows the presence of two different alleles at each mutation site (red and blue boxes/arrows). Boxes/arrows correspond to the location of the paternal (red) and maternal (blue) mutations respectively. Note the peak size difference between the guanine base (black) and the corresponding cytosine base (blue) at the maternal mutation site in the forward and reverse panels respectively. This observation underlines the importance of sequencing in both forward and reverse directions for single cell analysis as a precaution against preferential incorporation of different bases.
Enzyme digestion has also been an essential component of the preimplantation diagnosis of spinal muscular atrophy (SMA), in which a causative deletion in the survival motor neuron gene (SMN1) prevents PCR amplification of the mutant allele, but product from a highly homologous copy gene (SMN2) is specifically cut by the restriction enzyme Dra I. 108, 109 In these assays, no naturally occurring restriction site exists and an artificial cut site specific for the SMN2 is introduced during PCR using a primer mismatch, known as site specific mutagenesis (SSM), a strategy which has also been used for clinical PGD of retinitis pigmentosa. 99 An improved diagnosis for SMA uses an alternative restriction site Hinf I which is contained in both SMN1 and SMN2. SSM was used to introduce an additional cut site in SMN1 only to allow differentiation of the two sequences. 110 Restriction digestion is a straightforward and generally reliable method for mutation detection, but the digestion time required (between 3 and 6 hours) and the requirement for purification in some reported cases 78 can make this approach cumbersome for clinical PGD.
Heteroduplex Analysis
Heteroduplex analysis can identify a wide variety of mutations (particularly small deletions or insertions) and has been used extensively for identification of the ΔF508 mutation (a 3-bp deletion) causing cystic fibrosis. Since homozygous samples do not produce heteroduplexes, ΔF508/ΔF508 affected samples can be identified through heteroduplex formation following the addition of equivalent wild-type PCR product and absence of heteroduplex formation following addition of mutant product. 75 As well as extensive use in PGD of cystic fibrosis 22 heteroduplex analysis has also allowed diagnosis of Tay-Sachs disease 111 and was one of a series of methods used in parallel for PGD of familial adenomatous polyposis coli. 60 One potential problem with this method is the requirement for DNA of known genotype for mutation detection which could provide an opportunity for sample mix-up errors.
Single Strand Conformational Polymorphism Analysis
Single strand conformational polymorphism (SSCP) analysis is a “scanning” assay which is capable of detecting small DNA deletions and insertions and even single bp substitutions. 112 SSCP has become one of the most frequently used strategies for mutation detection and, in its simplest form, is uncomplicated and inexpensive requiring only a minimal amount of equipment. Single strands of DNA, generated by denaturing a PCR amplified sample, take on sequence specific conformations that are stabilized by intrastrand interactions. Allele-specific DNA strands frequently adopt distinct conformations which migrate at distinct rates when subjected to nondenaturing polyacrylamide gel electrophoresis. A single protocol can detect a number of genotypes so long as both mutations lie within the same amplified fragment. This may simplify the diagnosis of compound heterozygotes as such samples usually give a unique pattern of bands easily distinguished from other genotypes. Furthermore, SSCP has been performed using ethidium bromide (to detect the causative mutation in PGD of the dominant cancer syndrome familial adenomatous polyposis coli, 60 sensitive silver staining (to diagnose β-thalassemia at the single cell level), 113 or highly sensitive fluorescent PCR (to diagnose medium chain acyl CoA dehydrogenase deficiency). 76 A disadvantage of SSCP is that experimental conditions need to be carefully controlled to ensure reproducible assay sensitivity. This challenge is exacerbated by the single cell specific problems such as preferential amplification and ADO.
Denaturing Gradient Gel Electrophoresis
Denaturing gradient gel electrophoresis (DGGE) is another popular scanning method which, like SSCP, relies on physical properties of the DNA strand determined by base sequence. Mutations are detected indirectly by virtue of altered melting characteristics which affect the migration of the DNA strand as it passes through a polyacrylamide gel with increasing concentration of denaturant. The primers usually used for DNA amplification before DGGE are modified to include a stretch of approximately 40 guanine or cytosine residues (GC-clamp). These additional nucleotides significantly increase the proportion of sequence variants that can be detected in a given DNA fragment. However, under some circumstances the GC-clamp can reduce the efficiency of PCR and may actually be refractory to amplification if used at the single cell level. 114 The use of nested PCR with GC-clamped primers used only in the secondary amplification may overcome this difficulty. An advantage of DGGE over some other techniques is its ability to detect multiple mutations within the same PCR fragment. This has led to its use in clinical PGD for the detection of mutations causing β-thalassemia. 115 Like SSCP, DGGE can give banding patterns which are difficult to interpret at the single cell level.
Despite limitations in applying mutation SSCP or DGGE analysis to single cells, these methodologies can be very useful in identifying mutations in the couple before initiation of a PGD cycle. Other mutation-specific techniques or sequencing then can be used to specifically test for the parental mutations in PGD.
Sequencing
Direct sequencing is accurate, reliable and the time required to obtain results can be markedly reduced by confining the sequence analysis (post-PCR) to a smaller region of interest containing the mutation. Sequencing could be applied as a generic approach for PGD of any disease involving point mutations, small deletions, or insertions particularly when a series of mutations lie fairly close together within the same gene (as is the case for mutations in the β-globin gene resulting in thalassemia). Amplification of both parental mutation sites in the same fragment allows most ADO to be detected and prevent the transfer of affected compound heterozygous embryos.
Recently, direct sequencing was used in a PGD case involving a novel skin fragility ectodermal dysplasia syndrome to confirm restriction enzyme digestion results. 66 To attempt PGD, it was necessary to identify reliably and accurately the presence of the two different parental mutations (which lead to a functional knockout of the plakophilin 1 gene) in a single cell assay. Fortunately, the mutations lay within 11 bp of each other making a nested PCR approach feasible for the restriction assay, whereby both mutation sites could be amplified in the same fragment during the first round of PCR. Restriction analysis was carried out using two separate digests (one for each mutation). In parallel, cycle sequencing using big dye terminators on an ABI 310 genetic analyzer (Applied Biosystems, Foster City, CA) was performed in both forward and reverse primers for each purified sample (Figure 3) .
Linkage Analysis
Even when the exact mutation causing a disorder is unknown, the particular disorder may still be avoided by the detection of linked markers. Any informative polymorphism, which lies in close proximity to the disease locus, can be used as a tool to indicate the presence or absence of the mutation without its direct detection. Markers that are intragenic or situated close to the gene are preferred for this approach, as they are unlikely to be separated from the mutation by recombination during meiosis. To perform linkage analysis, a family pedigree must be obtained and DNA from family members tested to determine which polymorphic variant is inherited along with the disease phenotype. Many types of polymorphism are used for this purpose, the most commonly used are microsatellites (eg, Simple Tandem Repeats or STRs). These are highly polymorphic and consequently have the greatest probability of being informative for a given family.
Prior knowledge of the STR allele sizes of couples undergoing PGD allows the calculation of all possible zygote genotypes. Any deviation from these possibilities indicates the presence of contaminating DNA. 62, 80, 116, 117 The polymorphic nature of STR markers also permits the detection of haploidy and uniparental disomy. For these reasons many groups involved in PGD are now attempting to incorporate polymorphic markers into their molecular diagnoses. 41, 60, 104 The use of tetranucleotide repeats in preference to dinucleotide repeats and the application of commercially available optimized reaction buffers should reduce the frequency of artifacts known as “stutter bands” that complicate analysis of results. 118 The preferred future method for linkage analysis may use Single Nucleotide Polymorphisms (SNPs) which are DNA alterations that occur at a frequency of approximately 1 per 1000 bp throughout the genome. Since variability at a particular locus is limited to the four deoxynucleotides, a large number of SNPs is required for reliable linkage analysis. Microarray analysis (following whole genome amplification of single cells) will be a prerequisite to using SNPs as an alternative to STRs for linkage analysis.
Linkage analysis for PGD has been used for a number of different reasons. The causative mutation may be unknown, 119 the sequence containing the mutation may be refractory to PCR, 120 or heterogeneity of the causative mutations may make linkage analysis a more cost-effective way to provide a generic test for a disorder. 121, 122, 123 In addition, detection of allele dropout (particularly relevant for dominant conditions) and contamination make linkage analysis a powerful tool in clinical PGD. Finally, linkage analysis has allowed non-disclosure testing of embryos for Huntington disease. 124 For PGD by linkage analysis, many laboratories rely on informative markers from a two-generation pedigree—frequently available in couples with previous affected pregnancies or children.
The first clinical application of linkage analysis for PGD was to identify the autosomal dominant disorder, Marfan syndrome, in which the specific mutation was unknown. Affected embryos were identified by tracing the inheritance of a dinucleotide repeat polymorphism linked to the causative fibrillin gene. 119 Since this application, linkage analysis has also been used to detect embryos carrying mutant alleles of the dystrophin gene 125 and has been combined with mutation analysis using multiplex PCR 126 or whole genome amplification. 60
Diseases caused by the inheritance of large trinucleotide repeat expansions, such as fragile X syndrome and myotonic dystrophy, pose an additional problem for single cell analysis. In these cases the expanded allele is frequently too large to be amplified using PCR or may be subject to in vitro expansion producing erroneous results. 53 Consequently, inheritance of the disease allele in a tested blastomere would be inferred by the failure of PCR amplification across the expansion and the absence of the normal allele from the carrying parent. Indeed, conventional electrophoresis and later automated fragment analysis to detect non-expanded alleles was the basis for clinical preimplantation diagnosis of myotonic dystrophy 55, 127 and fragile X syndrome. 128 Alternatively, linkage analysis may be used with informative markers flanking the expansion. Strategies of this kind have been successfully developed for fragile X syndrome 120, 129 and myotonic dystrophy. 130 The inclusion of linked markers for the detection of allele dropout has become a standard in some laboratories and such a strategy should have a positive impact on pregnancy rates following PGD since the number of correctly diagnosed embryos available for transfer should increase as ADO is detected. 46, 63
Finally, linkage analysis can be of use in exclusion testing as a means by asymptomatic individuals who are at high risk of carrying HD can obtain antenatal genetic testing without incurring the emotional, social, and financial burdens that might result from the presymptomatic disclosure of their own carrier status. 124
Methodologies for Future Application to Clinical PGD
Cell Recycling
Another technique, which provides cytogenetic and also molecular genetic information, is known as cell recycling. 131 Fixed single cells are subjected to sequential PCR and FISH analysis allowing the investigation of specific gene sequences as well as chromosomal copy number. This combination of information would be particularly useful for PGD cases in which patients of advanced maternal age present with risk for having a child with a single gene disorder. Two clinical PGD cases have been reported, in which embryos free from the particular single gene disorder under test, resulted in pregnancies which miscarried and were found to be trisomy 16 132 and trisomy 22 66 respectively. In either case, an additional FISH test to rule out common chromosomal abnormalities would have been beneficial. Despite its potential, clinical application of cell recycling is not recommended at present since ADO rates are significantly higher with fixed template DNA than those observed using routine single cell protocols. 133
Quantitative Fluorescent PCR
Quantitative fluorescent PCR (QF-PCR) assays are based on the amplification of DNA sequences unique for each chromosome pair and have been developed to establish the number of specific chromosomes present in a cell. 79 These tests amplify STR or microsatellite markers with quantitation of products. Although QF-PCR is robust and reliable and can be completed within one working day, its application at the single cell level is hampered by an unacceptably high (25%) rate of preferential amplification which results in artificially skewed ratios of PCR products and the potential for misdiagnosis of chromosomal copy number in PGD. 80 STR markers can confidently identify aneuploidy with tri-allelic trisomies in single cells but this potential has yet to be fully realized owing to a lack of highly polymorphic chromosomal markers.
Real-Time PCR
Real-time PCR allows the rate of amplicon accumulation to be measured by detection of fluorescently tagged probes at each cycle of the reaction. The use of probes directed to either wild-type or mutant sequence also allows genotyping to be performed. The technique is rapid and has the added convenience that the amplification and detection procedures are carried out in the same tube (ie, as a homogeneous assay), thereby greatly reducing the chances of laboratory contamination. For example, addition of wild-type or mutant hairpin probes (which contain a fluorophore and quencher molecule at opposite ends of the probe) allows accurate mutation analysis as PCR products accumulate in the reaction tube. As the probes anneal to target sequence, the fluorophore and quencher are separated and fluorescence can be measured. The degree of homology between probe and target determines the particular annealing temperature at which the fluorescence can be measured. Real-time PCR assays have been used very effectively to detect multiple copy Y chromosomal sequences 49 (using molecular beacon technology) and BRCA1 sequences 134 (using LightCycler technology, Roche Diagnostics Corporation, Indianapolis, IN) in single cells and shows considerable promise for application to clinical PGD.
Denaturing High Performance Liquid Chromatography
This technique provides an efficient and inexpensive method for the rapid detection of single nucleotide mismatches and small deletions or insertions within an amplified DNA fragment without the need for fluorescence. Denaturing high performance liquid chromatography (DHPLC) exploits the differential retention of double stranded heteroduplex and homoduplex molecules, allowing the automatic comparison of PCR amplicons for variation. 135 A recent study 136 analyzing the CAG repeat region of the Huntington gene in single fibroblasts and blastomeres using this technology showed promising results in terms of amplification efficiency and ADO rates. However, aside from the markedly lower cost when compared with fluorescent PCR technology, it is difficult to see the advantages this technique can provide as fluorescent PCR becomes more readily available for routine molecular diagnostics in laboratories.
Microarrays
DNA microarrays (chips) are one of the latest and most promising tools for genetic analysis. These chips offer the possibility of simultaneously analyzing thousands of predefined DNA sequences and can be applied to DNA diagnostics, gene expression analysis, and aneuploidy detection. The most significant application has been in monitoring expression profiles to deduce genes relevant to particular disease pathologies (by comparing cDNA extracts from tissues derived from normal or disease states). Detection of aneuploidy using chip technology would work in a similar fashion to that of expression analysis. 137 Pieces of genomic DNA from specific chromosomes act as probes on the slide and a competitive hybridization process between samples from known normal karyotype and unknown occurs. Aneuploidy detection using microarrays is proving to be more difficult than expression analysis because copy number changes seen in aneuploidies are more subtle than gene expression changes which can vary by orders of magnitude. 138
Several methodologies for mutation analysis using microarrays have also been described. One of the more common examples of this is minisequencing in which an oligonucleotide is attached to the chip by its 5′ end. Each spot on the surface of the chip can contain several million of these oligonucleotides. The oligonucleotide is complimentary to the sequence of a disease causing gene and its 3′ end terminates at the base before a known mutation site. When the surface of the chip is exposed to sample DNA with DNA polymerase and di-deoxynucleotides triphosphates, the sample DNA acts as the template for the extension. By labeling each ddNTP with a different color it is possible to determine which nucleotide was added indicating the presence or absence of the mutation. Solid-phase minisequencing following whole genome amplification by PEP correctly genotyped single cells at 96% of the nucleotide positions analzyed. 84 Current drawbacks to using microarray technology in PGD include high cost, poor reproducibility, complex and lengthy data analysis, and the absolute requirement for some form of whole genome amplification.
Quality Control and Quality Assurance for Single Cell PCR
Reliability and accuracy in any molecular genetics diagnostics laboratory rely on stringent quality control (QC) and quality assurance (QA) measures, many of which are specific to PCR. 139 Such routine QC and QA measures would include appropriate assay validation, participation in proficiency testing surveys, testing of reagents before a clinical case, incorporation of measures to prevent and detect sample mix-up or contamination, routine equipment maintenance, and laboratory accreditation. The costs of these standard measures are generally absorbed within a general quality assurance plan in larger reference diagnostic laboratories. To ensure the highest standards of analytic reliability and accuracy for single cell analyses, additional measures are required (Table 5) . The combination of general and single cell specific QC/QA costs could be prohibitive in IVF laboratories providing single cell diagnostics.
Table 5.
Quality Control/Quality Assurance for Single Cell PCR
| Process | Measures |
|---|---|
| Routine/general QC | Comprehensive training/protocols (esp. contamination control) |
| Avoid specimen mix-up (multiple samples/embryos per patient) | |
| Overlap batches of tested and untested reagents | |
| Test all reagents prior to a clinical case | |
| Check temperatures of water-baths/thermal cyclers, etc | |
| Pipette calibration | |
| External quality assessment (unavailable at present) | |
| Ensure appropriate testing | Medical genetics consultation recommended |
| Verification of diagnosis (documentation or laboratory re-test) | |
| Apply PGD test offered to DNA/single cells from the couple | |
| Karyotype couple to exclude chromosomal abnormality? | |
| Assay development | Minimum number of single cells analyzed for assay development |
| Use heterozygous single cells to establish ADO/amplification rates | |
| Blastomere analysis for assay development | |
| Analyze DNA from−/−,+/− and +/+ sources | |
| Optimize primer design (particularly first round of nested PCR) | |
| Perform “dummy-runs” in simulated case conditions | |
| Clinical assay | Selection of mononucleate blastomeres only for analysis |
| Observe cell transfer to reaction tube | |
| Use of check gel to avoid post-PCR processing of failed samples | |
| Use of positive and multiple negative controls per clinical assay | |
| Contamination (observe precautions described in Table 3 ) | |
| Allele dropout (observe precautions described in Table 4 ) | |
| Minimize turn-around-time (for timely embryo selection/transfer) | |
| Mutation detection | Design PCR such that normal allele is cut into new product sizes |
| Use of internal controls for restriction enzyme digestion | |
| Purify PCR product prior to restriction digestion (if necessary) | |
| Establish cut-off values for failed amplifications/contamination in fluorescent PCR | |
| Sequence using forward and reverse primers | |
| Documentation | Labeling± color coding of tubes |
| Worksheet to contain all tube labels, gel loading sequence, etc. | |
| Diagnostic laboratory supervisor/director sign off for all cases | |
| Witness in IVF laboratory for embryo selection and transfer | |
| Misdiagnosis rates/ADO rates | Assess single blastomeres from non-transferred embryos |
| Confirm PGD result by amniocentesis/CVS/cord blood |
For example, primer design is critical when working with only 7 pg of DNA. In nested PCR, the outer primers are more critical than the inner primers and will have a significant effect on ADO rates if suboptimal. In view of the time-constraints for clinical PGD, smaller PCR products can help to reduce timings for electrophoretic separation and fragment analysis and even accentuate small differences in allele sizes on conventional gels. Optimization of the reaction with respect to magnesium and primer concentration, annealing temperatures and so on can be achieved using small amounts of informative DNA (20 to 50 ng) before using readily available single cells (such as lymphocytes, buccals cells, or fibroblasts). It is imperative to optimize the assay using such single cells since amplification rates in blastomeres are frequently lower and more variable. The minimum number of single cells for such validation studies has yet to be standardized. However, it is not unreasonable to conduct a series of between 5 and 10 consecutive experiments with at least 10 to 20 single cells per experiment to establish amplification efficiency and contamination rates with any degree of confidence. Furthermore, ADO rates can be established using heterozygous cells which can be obtained from commercial sources (as lymphoblast or fibroblast cell lines) or fresh from known carriers and prospective PGD patients (lymphocytes or buccal cells). The use of embryonic blastomeres as part of the assay development before clinical test implementation is controversial since a given mutation will be absent in most embryos generated by routine IVF (donated to research in excess of clinical need). Indeed, the majority of such results will be uninformative with respect to allele dropout rates. However, the use of non-transferred embryos (surplus in a clinical PGD case) is recommended to obtain misdiagnosis rates on embryos informative for the particular assay. Although a minimum number of single cells for assay validation has yet to be determined, development studies can be expensive, particularly in view of the need for large numbers of contamination controls. With a shortage of generic tests applicable at the single cell level for single gene disorders exhibiting molecular heterogeneity, the high cost of custom assay development has, so far, been a limiting factor in the uptake of PGD. Furthermore, a single PGD treatment involving 10 embryos could create up to 45 samples (depending on the number of blastomeres and appropriate controls analyzed) making the cost of reagents and personnel time significantly different compared with analysis of a single blood sample. As high-throughput technologies become widespread, however, the cost of molecular testing to the patient should not be prohibitive. Indeed, in the United States, the diagnostic test itself probably accounts for less than 10% of the total cost of the PGD treatment, the IVF procedures accounting for most of the cost.
In view of the cost of PGD treatment and the unique nature and origin of the test material, the additional cost and inconvenience to the patient of pre-cycle screening to ensure appropriateness of testing is justified. Before commencing a PGD cycle, it is vital to verify the DNA diagnosis using peripheral blood from the couple. Furthermore, it is prudent to apply the specific PGD test to DNA or single cells from the particular couple to discover any unexpected test results which could render future blastomere results questionable (for example, a polymorphism which may exist under a primer used in the single cell assay but not in the routine laboratory assay).
A number of QC processes apply to the single cell PCR procedure itself relating to amplification failure, contamination (Table 3) , and allele dropout (Table 4) . With respect to amplification failure, intrinsic problems relating to the biopsied material can be reduced by selecting only mononucleate blastomeres for analysis and using a check gel (when appropriate) to avoid time-consuming and costly post-PCR processing of failed samples. This latter measure is particularly important in view of the high number of samples expected to have no amplification (ie, wash blank controls). The number of blanks to include in assay development and clinical cases presents something of a dilemma in view of the calculation that 300 negative blanks are required to ensure that the contamination rate is less than 1%. A two-stage testing procedure has been suggested to maintain this low contamination rate. Before clinical implementation, a large series of blanks (eg, 100) should be run. After this, smaller series should be run periodically. 63 Specific QC and QA measures taken for mutation detection procedures have been discussed in previous sections of this review and are listed in Table 5 .
Finally, the documentation and hand-over procedures in clinical PGD must be stringent to avoid sample mix-up at any stage of the process. Such procedures are critical for the reporting of any genetic test result. Fortunately, some of the more recent technologies (such as real-time PCR) should make it possible to avoid transfer of material between tubes since amplification and mutation detection takes place in the same tube. Regardless of this possibility, it is recommended that critical procedures (eg, transfer of embryos between dishes) be witnessed by a second person and a rigorous protocol for labeling tubes and loading gels be implemented.
One QA measure, conspicuous by its absence in clinical PGD at present, is external quality assessment (EQA) or proficiency testing. If satellite PGD (in which IVF laboratories collect embryonic material for analysis at distant diagnostic laboratories) is to become more accepted, EQA is essential to maintain the highest standards of patient care. The different mutation detection strategies used to diagnose the same disorders (as shown in Table 1 ) demonstrate the lack of consensus and standardization in PGD. An analogous area of genetic testing is PCR screening for Y chromosomal microdeletions in the work-up for male infertility in which an EQA project is providing laboratories worldwide with overall misdiagnosis rates and an individual performance rating. 140 Organizing a similar scheme for PGD is essential but represents an enormous challenge which may ultimately only be met under the auspices of such organizations as the ESHRE PGD consortium 1, 16 or the International Working Group on Preimplantation Genetics. 141
Ethical, Legal and Social Issues Relating to PGD
Considerable differences in the regulatory oversight of PGD exists among countries, ranging from total bans on any embryo manipulation to the almost complete absence of any regulations or authority. 142 The high cost of practice, low pregnancy rate, 143 problems with patient access, and insurance coverage appear to be the biggest drawbacks to universal acceptance in societal terms. Ethical discussions considering the moral status of the human embryo 144 and what constitutes severe genetic disease have been debated elsewhere 145, 146 but such discussions are clearly outside of the purview of this methodological review. Somewhat reassuring for those centers currently offering PGD is the acknowledgment from professional organizations that PGD can now be considered a “standard of care” rather than an experimental treatment. 147
Conclusions
Robust and reliable single cell PCR diagnoses require optimization of reaction conditions and appropriate mutation detection strategies. For this to be achieved, one must fully appreciate the difficulties of amplifying DNA from a single cell. Once the limitations of a single cell have been overcome, using some form of DNA amplification, the mutation detection methods available in the molecular genetics armory are all applicable with only minor modifications.
The use of informative polymorphisms, which can provide confirmatory results to mutation analysis, identify contamination and increase the detection rate of ADO, will increase the reliability and accuracy of many of the diagnostic strategies already reported. For single cell applications, fluorescent PCR will likely replace conventional PCR strategies in view of its speed, throughput, and sensitivity, all of which could help to reduce the cost of each diagnostic test. Increasing amounts of chromosomal information from single cells will also be obtained using PCR-based techniques such as improved methods of quantitative fluorescent PCR and whole genome amplification in combination with comparative genomic hybridization or microarray technology. Whatever the difficulties faced by single cell diagnosis, the growing patient demand for PGD will continue to drive research into the application of further strategies for the diagnosis of an increasing variety of inherited diseases.
As can be seen from Table 1 , a large number of assays have been developed over the past decade to detect a variety of disorders. Development of any single cell assay can be costly and time-consuming and the development of assays for couples with unique mutations is a tribute to the dedication of researchers in the field of preimplantation genetics. However, the focus of the next decade should be to develop robust single cell assays with an emphasis on making such tests generic (for example, using linkage analysis with STRs or SNPs) to use limited resources cost-effectively to help more couples. Each of the various mutation detection methods and PCR strategies described above is associated with a different turn-around time (for example, real-time PCR requires less time than nested PCR followed by restriction digestion). The improvement of embryo culture medium supporting embryo development to the blastocyst stage now provides up to 3 days for analysis, more than enough time for any of the methods described above. Expansion of the analytic window has also made possible the geographical separation of IVF center and diagnostic laboratory, although significant logistics issues remain.
In conclusion, PGD testing is largely unregulated by any accrediting agency at present. The introduction of standardization, proficiency testing, and external quality assessment procedures among centers offering PGD (whether on-site or at a satellite laboratory) is in accordance with other forms of molecular testing and would ensure the highest quality of care for all patients.
Address reprint requests to Dr. Alan R. Thornhill, Mayo Clinic, Department of Laboratory Medicine and Pathology, Division of Laboratory Genetics, 200 First Street SW, Rochester, MN 55905. E-mail: thornhill.alan@mayo.edu.
References
- 1.: ESHRE preimplantation genetic diagnosis (PGD) consortium: data collection II. Hum Reprod 2000, 15:2673-2683 [DOI] [PubMed] [Google Scholar]
- 2.Handyside AH, Kontogianni EH, Hardy K, Winston RM: Pregnancies from biopsied human preimplantation embryos sexed by Y-specific DNA amplification. Nature 1990, 344:768-770 [DOI] [PubMed] [Google Scholar]
- 3.Avner R, Reubinoff BE, Simon A, Zentner BS, Friedmann A, Mitrani Rosenbaum S, Laufer N: Management of rhesus isoimmunization by preimplantation genetic diagnosis. Mol Hum Reprod 1996, 2:60-62 [DOI] [PubMed] [Google Scholar]
- 4.Chong SS, Kristjansson K, Cota J, Handyside AH, Hughes MR: Preimplantation prevention of X-linked disease: reliable and rapid sex determination of single human cells by restriction analysis of simultaneously amplified ZFX and ZFY sequences. Hum Mol Genet 1993, 2:1187-1191 [DOI] [PubMed] [Google Scholar]
- 5.Nakahori Y, Hamano K, Iwaya M, Nakagome Y: Sex identification by polymerase chain reaction using X-Y homologous primer. Am J Med Genet 1991, 39:472-473 [DOI] [PubMed] [Google Scholar]
- 6.Levinson G, Keyvanfar K, Wu JC, Fugger EF, Fields RA, Harton GL, Palmer FT, Sisson ME, Starr KM, Dennison-Lagos L, Calvo L, Sherins RJ, Bick D, Schulman JD, Black SH: DNA-based X-enriched sperm separation as an adjunct to preimplantation genetic testing for the prevention of X-linked disease. Hum Reprod 1995, 10:979-982 [DOI] [PubMed] [Google Scholar]
- 7.Hashiba T, Sueoka K, Kuroshima M, Asada H, Kuji N, Yoshimura Y: Accurate multiplex polymerase chain reaction assay for gender determination from a single cell. Gynecol Obstet Invest 2000, 49:217-220 [DOI] [PubMed] [Google Scholar]
- 8.Griffin DK, Wilton LJ, Handyside AH, Atkinson GH, Winston RM, Delhanty JD: Diagnosis of sex in preimplantation embryos by fluorescent in situ hybridisation. Br Med J 1993, 306:1382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Grifo JA, Tang YX, Munne S, Alikani M, Cohen J, Rosenwaks Z: Healthy deliveries from biopsied human embryos. Hum Reprod 1994, 9:912-916 [DOI] [PubMed] [Google Scholar]
- 10.Liu J, Lissens W, Van Broeckhoven C, Lofgren A, Camus M, Liebaers I, Van Steirteghem A: Normal pregnancy after preimplantation DNA diagnosis of a dystrophin gene deletion. Prenat Diagn 1995, 15:351-358 [DOI] [PubMed] [Google Scholar]
- 11.Hussey ND, Donggui H, Froiland DA, Hussey DJ, Haan EA, Matthews CD, Craig JE: Analysis of five Duchenne muscular dystrophy exons and gender determination using conventional duplex polymerase chain reaction on single cells. Mol Hum Reprod 1999, 5:1089-1094 [DOI] [PubMed] [Google Scholar]
- 12.Ray PF, Vekemans M, Munnich A: Single cell multiplex PCR amplification of five dystrophin gene exons combined with gender determination. Mol Hum Reprod 2001, 7:489-494 [DOI] [PubMed] [Google Scholar]
- 13.Harper JC, Thornhill AR: Embryo biopsy. Preimplantation Genetic Diagnosis. Edited by Harper JC, Delahanty JDA, Handyside AH. Chicester UK, John Wiley and Sons, 2001, pp 141–163
- 14.Verlinsky Y, Ginsberg N, Lifchez A, Valle J, Moise J, Strom CM: Analysis of the first polar body: preconception genetic diagnosis. Hum Reprod 1990, 5:826-829 [DOI] [PubMed] [Google Scholar]
- 15.Verlinsky Y, Rechitsky S, Cieslak J, Ivakhnenko V, Wolf G, Lifchez A, Kaplan B, Moise J, Walle J, White M, Ginsberg N, Strom C, Kuliev A: Preimplantation diagnosis of single gene disorders by two-step oocyte genetic analysis using first and second polar body. Biochem Mol Med 1997, 62:182-187 [DOI] [PubMed] [Google Scholar]
- 16.: ESHRE PGD Consortium Steering Committee: ESHRE Preimplantation Genetic Diagnosis (PGD) Consortium: preliminary assessment of data from January 1997 to September 1998. Hum Reprod 1999, 14:3138-3148 [DOI] [PubMed] [Google Scholar]
- 17.Hardy K, Martin KL, Leese HJ, Winston RML, Handyside AH: Human preimplantation development in vitro is not adversely affected by biopsy at the 8-cell stage. Hum Reprod 1990, 5:708-714 [DOI] [PubMed] [Google Scholar]
- 18.Dokras A, Sargent IL, Ross C, Gardner RL, Barlow DH: Trophectoderm biopsy in human blastocysts. Hum Reprod 1990, 5:821-825 [DOI] [PubMed] [Google Scholar]
- 19.Pickering SJ, Muggleton-Harris AL: Reliability and accuracy of polymerase chain reaction amplification of two unique target sequences from biopsies of cleavage-stage and blastocyst-stage human embryos. Hum Reprod 1995, 10:1021-1029 [DOI] [PubMed] [Google Scholar]
- 20.Veiga A, Sandalinas M, Benkhalifa M, Boada M, Carrera M, Santalo J, Barri PN, Menezo Y: Laser blastocyst biopsy for preimplantation genetic diagnosis in the human. Zygote 1997, 5:351-354 [DOI] [PubMed] [Google Scholar]
- 21.Cui KH, Matthews CD: Nuclear structural conditions and PCR amplification in human preimplantation diagnosis. Mol Hum Reprod 1996, 2:63-71 [DOI] [PubMed] [Google Scholar]
- 22.Ray PF, Ao A, Taylor DM, Winston RML, Handyside AH: Assessment of the reliability of single blastomere analysis for preimplantation diagnosis of the ΔF508 deletion causing cystic fibrosis in clinical practice. Prenat Diagn 1998, 18:1402-1412 [DOI] [PubMed] [Google Scholar]
- 23.Avner R, Laufer N, Safran A, Kerem BS, Friedmann A, Mitrani Rosenbaum S: Preimplantation diagnosis of cystic fibrosis by simultaneous detection of the W1282X and ΔF508 mutations. Hum Reprod 1994, 9:1676-1680 [DOI] [PubMed] [Google Scholar]
- 24.Sermon K, Lissens W, Nagy ZP, Van Steirteghem A, Liebaers I: Simultaneous amplification of the two most frequent mutations of infantile Tay-Sachs disease in single blastomeres. Hum Reprod 1995, 10:2214-2217 [DOI] [PubMed] [Google Scholar]
- 25.Gitlin SA, Lanzendorf SE, Gibbons WE: Polymerase chain reaction amplification specificity: incidence of allele dropout using different DNA preparation methods for heterozygous single cells. J Assist Reprod Genet 1996, 13:107-111 [DOI] [PubMed] [Google Scholar]
- 26.Kontogianni EH, Griffin DK, Handyside AH: Identifying the sex of human preimplantation embryos in X-linked disease: amplification efficiency of a Y-specific alphoid repeat from single blastomeres with two lysis protocols. J Assist Reprod Genet 1996, 13:125-132 [DOI] [PubMed] [Google Scholar]
- 27.Ray PF, Handyside AH: Increasing the denaturation temperature during the first cycles of amplification reduces allele dropout from single cells for preimplantation genetic diagnosis. Mol Hum Reprod 1996, 2:213-218 [DOI] [PubMed] [Google Scholar]
- 28.El-Hashemite N, Delhanty JDA: A technique for eliminating allele specific amplification failure during DNA amplification of heterozygous cells for preimplantation diagnosis. Mol Hum Reprod 1997, 3:975-978 [DOI] [PubMed] [Google Scholar]
- 29.Thornhill AR, McGrath JA, Braude P, Eady R, Handyside AH: A comparison of different lysis buffers to assess allele dropout from single cells for preimplantation genetic diagnosis. Prenat Diagn 2001, 21:490-497 [DOI] [PubMed] [Google Scholar]
- 30.Kwok S, Higuchi R: Avoiding false positives with PCR. Nature 1989, 339:237-238 [DOI] [PubMed] [Google Scholar]
- 31.Zhu YS, Isaacs ST, Cimino CD, Hearst JE: The use of exonuclease III for polymerase chain reaction sterilization. Nucleic Acids Res 1991, 19:2511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sermon K, Goossens V, Seneca S, Lissens W, De Vos A, Vandervorst M, Van Steirteghem A, Liebaers I: Preimplantation diagnosis for Huntington’s disease (HD): clinical application and analysis of the HD expansion in affected embryos. Prenat Diagn 1998, 18:1427-1436 [DOI] [PubMed] [Google Scholar]
- 33.Sermon K, Henderix P, Lissens W, De Vos A, Vandervorst M, Vanderfaeillie A, Vamos E, Van Steirteghem A, Liebaers I: Preimplantation genetic diagnosis for medium-chain acyl-CoA dehydrogenase (MCAD) deficiency. Mol Hum Reprod 2000, 6:1165-1168 [DOI] [PubMed] [Google Scholar]
- 34.De Rycke M, Van de Velde H, Sermon K, Lissens W, De Vos A, Vandervorst M, Vanderfaeillie A, Van Steirteghem A, Liebaers I: Preimplantation genetic diagnosis for sickle-cell anemia and for beta-thalassemia. Prenat Diagn 2001, 21:214-222 [DOI] [PubMed] [Google Scholar]
- 35.Prince AM, Andrus L: PCR: how to kill unwanted DNA. Biotechniques 1992, 12:358-360 [PubMed] [Google Scholar]
- 36.Longo MC, Berninger MS, Hartley JL: Use of uracil DNA glycosylase to control carry-over contamination in polymerase chain reactions. Gene 1990, 93:125-128 [DOI] [PubMed] [Google Scholar]
- 37.Cimino GD, Metchette KC, Tessman JW, Hearst JE, Isaacs ST: Post-PCR sterilization: a method to control carryover contamination for the polymerase chain reaction. Nucleic Acids Res 1991, 19:99-107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Liu J, Lissens W, Silber SJ, Devroey P, Liebaers I, Van Steirteghem A: Birth after preimplantation diagnosis of the cystic fibrosis delta F508 mutation by polymerase chain reaction in human embryos resulting from intracytoplasmic sperm injection with epididymal sperm. JAMA 1994, 272:1858-1860 [PubMed] [Google Scholar]
- 39.Ray PF, Winston RML, Handyside AH: Single cell analysis for diagnosis of cystic fibrosis and Lesch-Nyhan syndrome in human embryos before implantation. Miami Bio/Technol 1994, 5:46 (Abstract)
- 40.Findlay I, Ray P, Quirke P, Rutherford A, Lilford R: Allelic drop-out and preferential amplification in single cells and human blastomeres: implications for preimplantation diagnosis of sex and cystic fibrosis. Hum Reprod 1995, 10:1609-1618 [DOI] [PubMed] [Google Scholar]
- 41.Rechitsky S, Strom C, Verlinsky O, Amet T, Ivakhnenko V, Kukharenko V, Kuliev A, Verlinsky Y: Allele dropout in polar bodies and blastomeres. J Assist Reprod Genet 1998, 15:253-257 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Hahn S, Garvin AM, Di Naro E, Holzgreve W: Allele drop-out can occur in alleles differing by a single nucleotide and is not alleviated by preamplification or minor template increments. Genet Test 1998, 2:351-355 [DOI] [PubMed] [Google Scholar]
- 43.Navidi W, Arnheim N: Using PCR in preimplantation genetic disease diagnosis. Hum Reprod 1991, 6:836-849 [DOI] [PubMed] [Google Scholar]
- 44.Ao A, Ray P, Harper J, Lesko J, Paraschos T, Atkinson G, Soussis I, Taylor D, Handyside A, Hughes M, Winston RM: Clinical experience with preimplantation genetic diagnosis of cystic fibrosis (delta F508). Prenat Diagn 1996, 16:137-142 [DOI] [PubMed] [Google Scholar]
- 45.De Vos A, Sermon K, Van de Velde H, Joris H, Vandervorst M, Lissens W, Mortier G, DeSutter P, Lofgren A, Van Broeckhoven C, Liebaers I, Van Steirteghem A: Pregnancy after preimplantation genetic diagnosis for Charcot-Marie-Tooth disease type 1A. Mol Hum Reprod 1998, 4:978-984 [DOI] [PubMed] [Google Scholar]
- 46.Verlinsky Y, Kuliev A: An Atlas of Preimplantation Genetic Diagnosis. New York, Parthenon, 2000
- 47.Findlay I, Matthews P, Quirke P: Multiple genetic diagnoses from single cells using multiplex PCR: reliability and allele dropout. Prenat Diagn 1998, 18:1413-1421 [DOI] [PubMed] [Google Scholar]
- 48.Pickering SJ, McConnell JM, Johnson MH, Braude PR: Reliability of detection by polymerase chain reaction of the sickle cell-containing region of the beta-globin gene in single human blastomeres. Hum Reprod 1992, 7:630-636 [DOI] [PubMed] [Google Scholar]
- 49.Pierce KE, Rice JE, Sanchez JA, Brenner C, Wangh LJ: Real-time PCR using molecular beacons for accurate detection of the Y chromosome in single human blastomeres. Mol Hum Reprod 2000, 6:1155-1164 [DOI] [PubMed] [Google Scholar]
- 50.Kuo HC, Mackie Ogilvie C, Handyside AH: Chromosomal mosaicism in cleavage-stage human embryos and the accuracy of single-cell genetic analysis. J Assist Reprod Genet 1996, 15:276-280 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Harper JC, Coonen E, Handyside AH, Winston RML, Hopman AHN, Delhanty JDA: Mosaicism of autosomes and sex chromosomes in morphologically normal, monospermic preimplantation human embryos. Prenat Diagn 1995, 15:41-49 [DOI] [PubMed] [Google Scholar]
- 52.Wells D, Sherlock JK: Strategies for preimplantation genetic diagnosis of single gene disorders by DNA amplification. Prenat Diagn 1998, 18:1389-1401 [PubMed] [Google Scholar]
- 53.Daniels R, Holding C, Kontogianni E, Monk M: Single-cell analysis of unstable genes. J Assist Reprod Genet 1996, 13:163-169 [DOI] [PubMed] [Google Scholar]
- 54.Drury KC, Liu MC, Kipersztok S, Williams RS: Reduced incidence of allele dropout using SYBR Green I and PCR identification of X and Y amelogenin gene in preimplantation genetic diagnosis (PGD). Fertil Steril 1997, 68:S151–S152 (Abstract)
- 55.Sermon K, De Vos A, Van de Velde H, Seneca S, Lissens W, Joris H, Vandervorst M, Van Steirteghem A, Liebaers I: Fluorescent PCR and automated fragment analysis for the clinical application of preimplantation genetic diagnosis of myotonic dystrophy (Steinert’s disease). Mol Hum Reprod 1998, 4:791-796 [DOI] [PubMed] [Google Scholar]
- 56.Eldadah ZA, Grifo JA, Dietz HC: Marfan syndrome as a paradigm for transcript-targeted preimplantation diagnosis of heterozygous mutations. Nat Med 1995, 1:798-803 [DOI] [PubMed] [Google Scholar]
- 57.Daniels R, Adjaye J, Bolton V, Monk M: Detection of a novel splice variant of the hypoxanthine-guanine phosphoribosyl transferase gene in human oocytes and preimplantation embryos: implications for a RT-PCR-based preimplantation diagnosis of Lesch-Nyhan syndrome. Mol Hum Reprod 1998, 4:785-789 [DOI] [PubMed] [Google Scholar]
- 58.Tsai YH: Cost-effective screening methods for various single gene defects in single cells using high magnesium and total ionic strength and restriction enzymes. Prenat Diagn 2000, 20:979-985 [DOI] [PubMed] [Google Scholar]
- 59.Schulze E, Bettendorf M, Maser-Gluth C, Decker M, Schwabe U: Allele dropout using PCR-based diagnosis for the splicing mutation in intron 2 of the CYP21B gene: successful amplification with a Taq/Pwo-polymerase mixture. Endocr Res 1998, 24:637-641 [DOI] [PubMed] [Google Scholar]
- 60.Ao A, Wells D, Handyside AH, Winston RM, Delhanty JD: Preimplantation genetic diagnosis of inherited cancer: familial adenomatous polyposis coli. J Assist Reprod Genet 1998, 15:140-144 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Rechitsky S, Strom C, Verlinsky O, Amet T, Ivakhnenko V, Kukharenko V, Kuliev A, Verlinsky Y: Accuracy of preimplantation diagnosis of single-gene disorders by polar body analysis of oocytes. J Assist Reprod Genet 1999, 16:192-198 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Findlay I, Urquhart A, Quirke P, Sulivan K, Rutherford A, Lilford R: Simultaneous DNA “fingerprinting,” diagnosis of sex and single-gene defect status from single cells. Mol Hum Reprod 1995, 10:1005-1013 [PubMed] [Google Scholar]
- 63.Lewis CM, Pinel T, Whittaker JC, Handyside AH: Controlling misdiagnosis errors in preimplantation genetic diagnosis: a comprehensive model encompassing extrinsic and intrinsic sources of error. Hum Reprod 2001, 16:43-50 [DOI] [PubMed] [Google Scholar]
- 64.Ray PF, Kaeda JS, Bingham J, Roberts I, Handyside AH: Preimplantation genetic diagnosis of beta-thalassemia major. Lancet 1996, 347:1696. [DOI] [PubMed] [Google Scholar]
- 65.El-Hashemite N, Wells D, Delhanty JD: Preimplantation genetic diagnosis of beta-thalassaemia. Lancet 1996, 348:620-621 [DOI] [PubMed] [Google Scholar]
- 66.Thornhill AR, Pickering SJ, Whittock N, Caller J, Andritsos V, Handyside AH, Braude PR, Eady RA, McGrath J: Preimplantation genetic diagnosis of compound heterozygous mutations leading to ablation of Plakophilin-1 (PKP1) and resulting in skin fragility ectodermal dysplasia syndrome. Prenat Diagn 2000, 20:1055-1062 [DOI] [PubMed] [Google Scholar]
- 67.Van de Velde H, De Vos A, Sermon K, Staessen C, De Rycke M, Van Assche E, Lissens W, Vandervorst M, Van Ranst H, Liebaers I, Van Steirteghem A: Embryo implantation after biopsy of one or two cells from cleavage-stage embryos with a view to preimplantation genetic diagnosis. Prenat Diagn 2000, 20:1030-1037 [DOI] [PubMed] [Google Scholar]
- 68.Sago H, Kim HS, Goldberg JD, Chung JH, Pedersen RA, Lebo RV: Dual blastomere analysis improves reliability of preimplantation trembler mouse diagnosis. Hum Genet 1997, 101:223-228 [DOI] [PubMed] [Google Scholar]
- 69.Kim HS, Klimanskaya IV, Damsky CK, Pedersen RA, Lebo RV: Preimplantation diagnosis of the beta-1 integrin knockout mutation as a model for aneuploid gene testing. Hum Genet 1999, 105:480-488 [DOI] [PubMed] [Google Scholar]
- 70.Garvin A, Holzgreve W, Hahn S: Highly accurate analysis of heterozygous loci by single cell PCR. Nucleic Acids Res 1998, 26:3468-3472 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Li HH, Gyllensten UB, Cui XF, Saiki RK, Erlich HA, Arnheim N: Amplification and analysis of DNA sequences in single human sperm and diploid cells. Nature 1988, 335:414-417 [DOI] [PubMed] [Google Scholar]
- 72.Holding C, Monk M: Diagnosis of beta-thalassemia by DNA amplification in single blastomeres from mouse preimplantation embryos. Lancet 1989, 2:532-535 [DOI] [PubMed] [Google Scholar]
- 73.Moutou C, Viville S: Improvement of preimplantation genetic diagnosis (PGD) for the cystic fibrosis mutation delta F508 by fluorescent polymerase chain reaction. Prenat Diagn 1999, 19:1248-1250 [PubMed] [Google Scholar]
- 74.Hattori M, Yoshioka K, Sakaki Y: High-sensitive fluorescent DNA sequencing and its application for detection and mass-screening of point mutations. Electrophoresis 1992, 13:560-565 [DOI] [PubMed] [Google Scholar]
- 75.Handyside AH, Lesko JG, Tarin JJ, Winston RM, Hughes MR: Birth of a normal girl after in vitro fertilization and preimplantation diagnostic testing for cystic fibrosis. N Engl J Med 1992, 327:905-909 [DOI] [PubMed] [Google Scholar]
- 76.Ioulianos A, Wells D, Harper JC, Delhanty JD: A successful strategy for preimplantation diagnosis of medium-chain acyl-CoA dehydrogenase (MCAD) deficiency. Prenat Diagn 2000, 20:593-598 [DOI] [PubMed] [Google Scholar]
- 77.Moutou C, Gardes N, Rongieres C, Ohl J, Bettahar-Lebugle K, Wittemer C, Gerlinger P, Viville S: Allele-specific amplification for preimplantation genetic diagnosis (PGD) of spinal muscular atrophy. Prenat Diagn 2001, 21:498-503 [DOI] [PubMed] [Google Scholar]
- 78.Van de Velde H, Sermon K, De Vos A, Lissens W, Joris H, Vandervorst M, Van Steirteghem A, Liebaers I: Fluorescent PCR and automated fragment analysis in preimplantation genetic diagnosis for 21-hydroxylase deficiency in congenital adrenal hyperplasia. Mol Hum Reprod 1999, 5:691-696 [DOI] [PubMed] [Google Scholar]
- 79.Pertl B, Weitgasser U, Kopp S, Kroisel PM, Sherlock J, Adinolfi M: Rapid detection of trisomies 21 and 18 and sexing by quantitative fluorescent multiplex PCR. Hum Genet 1996, 98:55-59 [DOI] [PubMed] [Google Scholar]
- 80.Sherlock J, Cirigliano V, Petrou M, Tutschek B, Adinolfi M: Assessment of quantitative fluorescent multiplex PCR performed on single cells. Ann Hum Genet 1998, 62:9-23 [DOI] [PubMed] [Google Scholar]
- 81.Blake D, Tan SL, Ao A: Assessment of multiplex fluorescent PCR for screening single cells for trisomy 21 and single gene defects. Mol Hum Reprod 1999, 5:1166-1175 [DOI] [PubMed] [Google Scholar]
- 82.Eggerding FA, Iovannisci DM, Brinson E, Grossman P, Winn-Deen ES: Fluorescence-based oligonucleotide ligation assay for analysis of cystic fibrosis transmembrane conductance regulator gene mutations. Hum Mutat 1995, 5:153-165 [DOI] [PubMed] [Google Scholar]
- 83.Kuliev A, Rechitsky S, Verlinsky O, Ivakhnenko V, Evsikov S, Wolf G, Angastiniotis M, Georghiou D, Kukharenko V, Strom C, Verlinsky Y: Preimplantation diagnosis of thalassemias. J Assist Reprod Genet 1998, 15:219-225 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Paunio T, Reima I, Syvanen AC: Preimplantation diagnosis by whole-genome amplification, PCR amplification, and solid-phase minisequencing of blastomere DNA. Clin Chem 1996, 42:1382-1390 [PubMed] [Google Scholar]
- 85.Zhang L, Cui X, Schmitt K, Hubert R, Navidi W, Arnheim N: Whole genome amplification form a single cell: implications for genetic-analysis. Proc Natl Acad Sci USA 1992, 89:5847-5851 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Wells D, Sherlock JK, Handyside AH, Delhanty JD: Detailed chromosomal and molecular genetic analysis of single cells by whole genome amplification and comparative genomic hybridisation. Nucleic Acids Res 1999, 27:1214-1218 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Gibbons WE, Gitlin SA, Lanzendorf SE: Strategies to respond to polymerase chain reaction deoxyribonucleic acid amplification failure in a preimplantation genetic diagnosis program. Am J Obstet Gynecol 1995, 172:1088-1095 [DOI] [PubMed] [Google Scholar]
- 88.Schaaff F, Wedemann H, Schwinger E: Analysis of sex and delta F508 in single amniocytes using primer extension preamplification. Hum Genet 1996, 98:158-161 [DOI] [PubMed] [Google Scholar]
- 89.Xu K, Tang Y, Grifo JA, Rosenwaks Z, Cohen J: Primer extension preamplification for detection of multiple genetic loci from single human blastomeres. Hum Reprod 1993, 8:2206-2210 [DOI] [PubMed] [Google Scholar]
- 90.Snabes MC, Chong SS, Subramanian SB, Kristjansson K, DiSepio D, Hughes MR: Preimplantation single-cell analysis of multiple genetic loci by whole-genome amplification. Proc Natl Acad Sci USA 1994, 91:6181-6185 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Kristjansson K, Chong SS, Van den Veyver IB, Subramanian S, Snabes MC, Hughes MR: Preimplantation single cell analyses of dystrophin gene deletions using whole genome amplification. Nat Genet 1994, 6:19-23 [DOI] [PubMed] [Google Scholar]
- 92.Sermon K, Lissens W, Joris H, Van Steirteghem A, Liebaers I: Adaptation of the primer extension preamplification (PEP) reaction for preimplantation diagnosis: single blastomere analysis using short PEP protocols. Mol Hum Reprod 1996, 2:209-212 [DOI] [PubMed] [Google Scholar]
- 93.Telenius H, Carter NP, Bebb CE, Nordenskjold M, Ponder BA, Tunnacliffe A: Degenerate oligonucleotide-primed PCR: general amplification of target DNA by a single degenerate primer. Genomics 1992, 13:718-725 [DOI] [PubMed] [Google Scholar]
- 94.Wells D, Delhanty JD: Comprehensive chromosomal analysis of human preimplantation embryos using whole genome amplification and single cell comparative genomic hybridization. Mol Hum Reprod 2000, 6:1055-1062 [DOI] [PubMed] [Google Scholar]
- 95.Voullaire L, Slater H, Williamson R, Wilton L: Chromosome analysis of blastomeres from human embryos by using comparative genomic hybridization. Hum Genet 2000, 106:210-217 [DOI] [PubMed] [Google Scholar]
- 96.Wilton L, Williamson R, McBain J, Edgar D, Voullaire L: Birth of a healthy infant after preimplantation confirmation of euploidy by comparative genomic hybridization. N Engl J Med 2001, 21:1537-1541 [DOI] [PubMed] [Google Scholar]
- 97.Focault F, Praz F, Jaulin C, Amor-Gueret M: Experimental limits of PCR analysis of (CA)n repeat alterations. Trends Genet 1996, 12:450-452 [DOI] [PubMed] [Google Scholar]
- 98.Scobie G, Woodroffe B, Fishel S, Kalsheker N: Identification of the five most common cystic fibrosis mutations in single cells using a rapid and specific differential amplification system. Mol Hum Reprod 1996, 2:203-207 [DOI] [PubMed] [Google Scholar]
- 99.Strom CM, Rechitsky S, Wolf G, Cieslak J, Kuliev A, Verlinsky Y: Preimplantation diagnosis of autosomal dominant retinitis pigmentosum using two simultaneous single cell assays for a point mutation in the rhodopsin gene. Mol Hum Reprod 1998, 4:351-355 [DOI] [PubMed] [Google Scholar]
- 100.Cserhalmi-Friedman PB, Tang Y, Adler A, Krey L, Grifo JA, Christiano AM: Preimplantation genetic diagnosis in two families at risk for recurrence of Herlitz junctional epidermolysis bullosa. Exp Dermatol 2000, 9:290-297 [DOI] [PubMed] [Google Scholar]
- 101.Ray PF, Harper JC, Ao A, Taylor DM, Winston RM, Hughes M, Handyside AH: Successful preimplantation genetic diagnosis for sex -linked Lesch-Nyhan syndrome using specific diagnosis. Prenat Diagn 1999, 19:1237-1241 [DOI] [PubMed] [Google Scholar]
- 102.Xu K, Shi ZM, Veeck LL, Hughes MR, Rosenwaks Z: First unaffected pregnancy using preimplantation genetic diagnosis for sickle cell anemia. JAMA 281:1701–1706 [DOI] [PubMed]
- 103.Verlinsky Y, Rechitsky S, Schoolcraft W, Strom C, Kuliev A: Preimplantation diagnosis for Fanconi anemia combined with HLA matching. JAMA 2001, 285:3130-3133 [DOI] [PubMed] [Google Scholar]
- 104.Ray PF, Gigarel N, Bonnefont JP, Attie T, Hamamah S, Frydman N, Vekemans M, Frydman R, Munnich A: First specific preimplantation genetic diagnosis for ornithine transcarbamylase deficiency. Prenat Diagn 2000, 20:1048-1054 [DOI] [PubMed] [Google Scholar]
- 105.De Vos A, Sermon K, Van de Velde H, Joris H, Vandervorst M, Lissens W, De Paepe A, Liebaers I, Van Steirteghem A: Two pregnancies after preimplantation genetic diagnosis for osteogenesis imperfecta type I and type IV. Hum Genet 2000, 106:605-613 [DOI] [PubMed] [Google Scholar]
- 106.Goossens V, Sermon K, Lissens W, Vandervorst M, Vanderfaeillie A, De Rijcke M, De Vos A, Henderix P, Van De Velde H, Van Steirteghem A, Liebaers I: Clinical application of preimplantation genetic diagnosis for cystic fibrosis. Prenat Diagn 2000, 20:571-581 [DOI] [PubMed] [Google Scholar]
- 107.Blaszczyk A, Tang YX, Dietz HC, Adler A, Berkeley AS, Krey LC, Grifo JA: Preimplantation genetic diagnosis of Marfan’s syndrome. J Assist Reprod Genet 1998, 15:281-284 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Dreesen JC, Bras M, de Die-Smulders C, Dumoulin JC, Cobben JM, Evers JL, Smeets HJ, Geraedts JP: Preimplantation genetic diagnosis of spinal muscular atrophy. Mol Hum Reprod 1998, 4:881-885 [DOI] [PubMed] [Google Scholar]
- 109.Fallon L, Harton GL, Sisson ME, Rodriguez E, Field LK, Fugger EF, Geltinger M, Sun Y, Dorfmann A, Schoener C, Bick D, Schulman J, Levinson G, Black SH: Preimplantation genetic diagnosis for spinal muscular atrophy type I. Neurology 1999, 53:1087-1090 [DOI] [PubMed] [Google Scholar]
- 110.Daniels G, Pettigrew R, Thornhill A, Abbs S, Lashwood A, O’Mahoney F, Mathew C, Handyside A, Braude P: Six unaffected livebirths following preimplantation diagnosis for spinal muscular atrophy. Mol Hum Reprod 2001, 7:995-1000 [DOI] [PubMed] [Google Scholar]
- 111.Gibbons WE, Gitlin SA, Lanzendorf SE, Kaufmann RA, Slotnick RN, Hodgen GD: Preimplantation genetic diagnosis for Tay-Sachs disease: successful pregnancy after pre-embryo biopsy and gene amplification by polymerase chain reaction. Fertil Steril 1995, 63:723-728 [DOI] [PubMed] [Google Scholar]
- 112.Orita M, Suzuki Y, Sekiya T, Hayashi K: Rapid and sensitive detection of point mutations and DNA polymorphisms using the polymerase chain reaction. Genomics 1989, 5:874-879 [DOI] [PubMed] [Google Scholar]
- 113.El-Hashemite N, Wells D, Delhanty JD: Single cell detection of beta-thalassaemia mutations using silver stained SSCP analysis: an application for preimplantation diagnosis. Mol Hum Reprod 1997, 3:693-698 [DOI] [PubMed] [Google Scholar]
- 114.Sheffield VC, Cox DR, Lerman LS, Myers RM: Attachment of a 40-base-pair G + C-rich sequence (GC-clamp) to genomic DNA fragments by the polymerase chain reaction results in improved detection of single-base changes. Proc Natl Acad Sci USA 1989, 86:232-236 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Kanavakis E, Vrettou C, Palmer G, Tzetis M, Mastrominas M, Traeger-Synodinos J: Preimplantation genetic diagnosis in 10 couples at risk for transmitting beta-thalassaemia major: clinical experience including the initiation of six singleton pregnancies. Prenat Diagn 1999, 19:1217-1222 [DOI] [PubMed] [Google Scholar]
- 116.Holding C, Bentley D, Roberts R, Bobrow M, Mathew C: Development and validation of laboratory procedures for preimplantation diagnosis of Duchenne muscular dystrophy. J Med Genet 1993, 30:903-909 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Pickering SJ, McConnell JM, Johnson MH, Braude PR: Use of a polymorphic dinucleotide repeat sequence to detect non-blastomeric contamination of the polymerase chain reaction in biopsy samples for preimplantation diagnosis. Hum Reprod 1994, 9:1539-1545 [DOI] [PubMed] [Google Scholar]
- 118.Walsh PS, Fildes NJ, Reynolds R: Sequence analysis and characterization of stutter products at the tetranucleotide repeat locus vWA. Nucleic Acids Res 1996, 24:2807-2812 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Harton GL, Tsipouras P, Sisson ME, Starr KM, Mahoney BS, Fugger EF, Schulman JD, Kilpatrick MW, Levinson G, Black SH: Preimplantation genetic testing for Marfan syndrome. Mol Hum Reprod 1996, 2:713-715 [DOI] [PubMed] [Google Scholar]
- 120.Apessos A, Abou-Sleiman PM, Harper JC, Delhanty JD: Preimplantation genetic diagnosis of the fragile X syndrome by use of linked polymorphic markers. Prenat Diagn 2001, 21:504-511 [DOI] [PubMed] [Google Scholar]
- 121.Sermon K, Lissens W, Messiaen L, Bonduelle M, Vandervorst M, Van Steirteghem A, Liebaers I: Preimplantation genetic diagnosis of Marfan syndrome with the use of fluorescent polymerase chain reaction and the automated laser fluorescence DNA sequencer. Fertil Steril 1999, 71:163-166 [DOI] [PubMed] [Google Scholar]
- 122.Dreesen JC, Jacobs LJ, Bras M, Herbergs J, Dumoulin JC, Geraedts JP, Evers JL, Smeets HJ: Multiplex PCR of polymorphic markers flanking the CFTR gene: a general approach for preimplantation genetic diagnosis of cystic fibrosis. Mol Hum Reprod 2000, 6:391-396 [DOI] [PubMed] [Google Scholar]
- 123.Eftedal I, Schwartz M, Bendtsen H, Andersen AN, Ziebe S: Single intragenic microsatellite preimplantation genetic diagnosis for cystic fibrosis provides positive allele identification of all CFTR genotypes for informative couples. Mol Hum Reprod 2001, 7:307-312 [DOI] [PubMed] [Google Scholar]
- 124.Braude PR, De Wert GM, Evers-Kiebooms G, Pettigrew RA, Geraedts JP: Non-disclosure preimplantation genetic diagnosis for Huntington’s disease: practical and ethical dilemmas. Prenat Diagn 1998, 18:1422-1426 [DOI] [PubMed] [Google Scholar]
- 125.Lee SH, Kwak IP, Cha KE, Park SE, Kim NK, Cha KY: Preimplantation diagnosis of non-deletion Duchenne muscular dystrophy (DMD) by linkage polymerase chain reaction analysis. Mol Hum Reprod 1988, 4:345-349 [DOI] [PubMed] [Google Scholar]
- 126.Sutterlin M, Sleiman PA, Onadim Z, Delhanty J: Single cell detection of inherited retinoblastoma predisposition. Prenat Diagn 1999, 19:1231-1236 [PubMed] [Google Scholar]
- 127.Sermon K, Lissens W, Joris H, Seneca S, Desmyttere S, Devroey P, Van Steirteghem A, Liebaers I: Clinical application of preimplantation diagnosis for myotonic dystrophy. Prenat Diagn 1997, 17:925-932 [PubMed] [Google Scholar]
- 128.Sermon K, Seneca S, Vanderfaeillie A, Lissens W, Joris H, Vandervorst M, Van Steirteghem A, Liebaers I: Preimplantation diagnosis for fragile X syndrome based on the detection of the non-expanded paternal and maternal CGG. Prenat Diagn 1999, 19:1223-1230 [PubMed] [Google Scholar]
- 129.Dreesen JC, Geraedts JP, Dumoulin JC, Evers JL, Pieters MH: RS46(DXS548) genotyping of reproductive cells: approaching preimplantation testing of the fragile-X syndrome. Hum Genet 1995, 96:323-329 [DOI] [PubMed] [Google Scholar]
- 130.Piyamongkol W, Harper JC, Sherlock JK, Doshi A, Serhal PF, Delhanty JD, Wells D: A successful strategy for preimplantation genetic diagnosis of myotonic dystrophy using multiplex fluorescent PCR. Prenat Diagn 2001, 21:223-232 [DOI] [PubMed] [Google Scholar]
- 131.Thornhill AR, Monk M: Cell recycling of a single human cell for preimplantation diagnosis of X-linked disease and dual sex determination. Mol Hum Reprod 1996, 2:285-289 [DOI] [PubMed] [Google Scholar]
- 132.Pettigrew R, Kuo HC, Scriven P, Rowell P, Pal K, Handyside A, Braude P, Ogilvie CM: A pregnancy following PGD for X-linked autosomal dominant incontinentia pigmenti (Bloch-Sulzberger syndrome): case report. Hum Reprod 2000, 15:2650-2652 [DOI] [PubMed] [Google Scholar]
- 133.Rechitsky S, Freidine M, Verlinsky Y, Strom CM: Allele dropout in sequential PCR and FISH analysis of single cells (cell recycling). J Assist Reprod Genet 1996, 13:115-124 [DOI] [PubMed] [Google Scholar]
- 134.Pals G, Young C, Mao HS, Worsham MJ: Detection of a single base substitution in a single cell using the LightCycler. J Biochem Biophys Methods 2001, 47:121-129 [DOI] [PubMed] [Google Scholar]
- 135.Underhill PA, Jin L, Lin AA, Mehdi SQ, Jenkins T, Vollrath D, Davis RW, Cavalli-Sforza LL, Oefner PJ: Detection of numerous Y chromosome biallelic polymorphisms by denaturing high-performance liquid chromatography. Genome Res 1997, 7:996-1005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Drury KC, Liu MC, Lilleberg S, Kipersztok S, Williams RS: Results on single cell PCR for Huntington’s gene and WAVE product analysis for preimplantation genetic diagnosis. Mol Cell Endocrinol 2001, 183 (Suppl 1):S1-S4 [DOI] [PubMed] [Google Scholar]
- 137.Hughes TR, Roberts CJ, Dai H, Jones AR, Meyer MR, Slade D, Burchard J, Dow S, Ward TR, Kidd MJ, Friend SH, Marton MJ: Widespread aneuploidy revealed by DNA microarray expression profiling. Nat Genet 2000, 25:333-337 [DOI] [PubMed] [Google Scholar]
- 138.Harper JC, Wells D: Recent advances and future developments in PGD. Prenat Diagn 1999, 19:1193-1199 [DOI] [PubMed] [Google Scholar]
- 139.Burkardt HJ: Standardization and quality control of PCR analyses. Clin Chem Lab Med 2000, 38:87-91 [DOI] [PubMed] [Google Scholar]
- 140.Simoni M: Molecular diagnosis of Y chromosome microdeletions in Europe: state-of-the-art and quality control. Hum Reprod 2001, 16:402-409 [DOI] [PubMed] [Google Scholar]
- 141.Verlinsky Y, Munne S, Simpson JL, Kuliev A, Ao A, Ray P, Sermon K, Martin R, Strom C, Van Steirteghem A, Veiga A, Drury K, Williams S, Ginsberg N, Wilton L: Current status of preimplantation diagnosis. J Assist Reprod Genet 1997, 14:72-75 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Viville S, Pergament D: Results of a survey of the legal status and attitudes towards preimplantation genetic diagnosis conducted in 13 different countries. Prenat Diagn 1998, 18:1374-1380 [DOI] [PubMed] [Google Scholar]
- 143.Fasouliotis SJ, Schenker JG: Preimplantation genetic diagnosis principles and ethics. Hum Reprod 1998, 13:2238-2245 [DOI] [PubMed] [Google Scholar]
- 144.Beyleveld D: Is embryo research and preimplantation genetic diagnosis ethical? Forensic Sci Int 2000, 113:461-475 [DOI] [PubMed] [Google Scholar]
- 145.Draper H, Chadwick R: Beware! Preimplantation genetic diagnosis may solve some old problems but it also raises new ones. J Med Ethics 1999, 25:114-120 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.King DS: Preimplantation genetic diagnosis and the “new” eugenics. J Med Ethics 1999, 25:176-182 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.American Society for Reproductive Medicine and Society for Assisted Reproductive Technology Practice Committee Report: Preimplantation Genetic Diagnosis. June, 2001