Abstract
Synovial sarcomas comprise approximately 5% of soft tissue sarcomas and occur primarily in young adults. The t(X;18) (p11.2;q11.2) has been demonstrated to be highly characteristic of synovial sarcomas, and the resulting SYT-SSX fusion transcripts have been shown to be useful diagnostic markers. We have developed a real-time, reverse transcriptase-polymerase chain reaction (RT-PCR) multiplex assay for the identification of the primary fusion transcript types (SYT-SSX1 and SYT-SSX2) from formalin-fixed, paraffin-embedded (FFPE) tissues. Twenty-nine of 30 (96.7%) histologically diagnosed FFPE synovial sarcomas were positive for the presence of either the SYT-SSX1 or SYT-SSX2 fusion transcripts. Ten of 16 (62.5%) and five of 16 (31.25%) monophasic fibrous synovial sarcomas were positive for SYT-SSX1 and SYT-SSX2, respectively. One of 16 (6.25%) monophasic fibrous synovial sarcomas was negative for either SYT-SSX fusion transcript. Twelve of 14 (85.7%) and 2 of 14 (14.3%) biphasic synovial sarcomas were positive for SYT-SSX1 and SYT-SSX2, respectively. All 13 non-synovial sarcomas tested were negative for SYT-SSX1 and SYT-SSX2 fusion transcripts. This method is a relatively simple and rapid procedure for the detection of the t(X;18)(p11.2;q11.2).
Synovial sarcomas comprise approximately 5 to 10% of soft tissue sarcomas. These tumors occur in a broad age range and have a wide anatomical distribution but preferentially affect the para-articular regions in young adults. There are four recognized subtypes of synovial sarcoma: biphasic tumors consist of spindle-shaped cells admixed with epithelial cells and variable numbers of epithelioid (“transitional”) cells; monophasic fibrous tumors contain spindled cells and variable numbers of epithelioid cells but lack a recognizable epithelial element; monophasic epithelial tumors are defined as consisting entirely or almost entirely of epithelial tumor cells (this subtype is extremely rare); and poorly differentiated tumors consist of highly atypical epithelioid or spindled cells with increased nuclear-to-cytoplasmic ratios and prominent mitotic activity (typically greater than or equal to 10 mitoses/10 high power fields). 1, 2 Thelast subtype is often admixed with one of the first two tumor types and is important to recognize because it is associated with a poor prognosis. 3, 4 Most synovial sarcomas are readily recognized because of their distinctive clinical and histopathological features. In instances where classification is difficult, immunohistochemistry can be helpful, because synovial sarcomas commonly express keratins and epithelial membrane antigen. However, a small percentage of synovial sarcomas (primarily poorly differentiated and some monophasic fibrous examples) have minimal or no reactivity for “epithelial” markers. 5, 6, 7, 8, 9 In these cases it can be difficult to confidently rule out a diagnosis of fibrosarcoma, malignant peripheral nerve sheath tumor (MPNST) or, in selected instances, a peripheral primitive neuroectodermal tumor (pPNET); therefore, more reliable methods are necessary for the diagnosis of synovial sarcoma. 1, 8, 10, 11, 12, 13, 14, 15, 16
A characteristic t(X;18) (p11;q11) reciprocal translocation is detectable in > 90% of synovial sarcomas. 17 This translocation results from the fusion of the proximal portion of the SYT gene at 18q11 to the distal portion of primarily one of two genes, SSX1 and SSX2, that comprise part of a highly homologous five gene family at Xp11. Also, there have been a few reports describing a variant fusion between the SYT gene and the SSX4 gene. 18, 19
The t(X;18) translocation is amenable to detection by both fluorescence in situ hybridization (FISH) and reverse-transcriptase polymerase chain reaction (RT-PCR) on formalin-fixed, paraffin-embedded tissues (FFPE). Identification of the t(X;18) translocation by FISH requires the use of both chromosome X and 18 sequence specific and centromeric probes. These probes do not allow for the determination of the fusion type without additional hybridizations using probes for the specific SSX gene. 6, 20, 21, 22, 23 RT-PCR can also identify the fusion type with probes located on the SSX region of the fusion, through restriction digestion of the PCR products, use of specific reverse primers for each fusion type, or by direct sequencing. 7, 11, 24, 25, 26, 27 Peter et al 28 have recently reported the use of real-time RT-PCR for the detection of gene fusions in solid tumors, but the method does not distinguish the fusion transcript types.
Recent studies have demonstrated a correlation between the type of fusion (SYT-SSX1 vs SYT-SSX2) and proliferative activity and/or metastasis-free survival. 24, 26, 29 This suggests that the fusion type may prove to be a valuable prognostic factor that could influence treatment and overall patient care. Reported methods for the identification of the fusion type consist of RT-PCR using reverse primers specific for SSX1 or SSX2 followed by gel visualization, RT-PCR followed by restriction digestion and gel visualization, RT-PCR followed by Southern blot using specific probes, and RT-PCR followed by sequencing. All of these methods generally require several days to complete.
An alternative method for the identification of SYT-SSX fusion transcripts is the utilization of real-time RT-PCR. We describe an assay that is both highly sensitive and specific. Real-time PCR utilizes probes labeled with two dyes, a reporter and a quencher, which are in close proximity on the intact probe, resulting in quenching of the reporter fluorescence by fluorescent resonance energy transfer (FRET). 30 When the probe binds to the specific PCR product, it is cleaved by the 5′–exonuclease activity of Taq polymerase separating the reporter from the quencher, resulting in increased fluorescence from the reporter dye. The ability of the instrument to measure fluorescence from several dyes simultaneously allows for multiplex amplifications, with simultaneous detection of different targets in the same reaction. The instrument analyzes the fluorescence data generated during the reaction and calculates the cycle number at which fluorescence crosses a threshold value determined by analysis of data from early cycles in the amplification process. This cycle number, the CT value, is related to the quantity of specific target in the reaction, with larger quantities of starting material leading to lower CT values. By carrying out the amplification and detection in the presence of two sequence specific probes, labeled with two distinct reporter dyes, differentiation between the two primary fusion types is quickly and easily attained, resulting in decreased turn-around time and labor.
Materials and Methods
Case Selection
Forty-three formalin-fixed, paraffin-embedded (FFPE) tumors (30 synovial sarcomas and 13 non-synovial sarcomas) were obtained from the archives of the Armed Forces Institute of Pathology. The non-synovial sarcomas consisted of five Ewing’s sarcoma/pPNET and two rhabdomyosarcomas, and six small cell sarcomas, not otherwise specified. The Bax-1 cell line, containing the SYT-SSX2 fusion, was a gift from R. Wickert, University of Nebraska Medical Center, and seven non-synovial sarcoma cell lines (Ewing’s sarcomas, alveolar rhabdomyosarcomas, liposarcoma, and neuroblastoma) were obtained from American Type Culture Collection (Manassas, VA). The FFPE cases were from between 1988 and 2000. The diagnoses on the synovial sarcoma cases were reevaluated according to current histological criteria. Tumors that lacked a biphasic component were required to have acceptable histology and keratin-positive cells for inclusion in this study. Fourteen cases were diagnosed as biphasic synovial sarcomas, five of which contained a poorly differentiated element, and 16 were diagnosed as monophasic fibrous synovial sarcomas, two of which had a poorly differentiated component.
RNA Extraction
RNA extracted from six 6-μm sections was placed in a 1.5-ml microcentrifuge tube and the samples were deparaffinized by the addition of 800 μl of Hemo-DE (Scientific Safety Solvents, Keller, TX) and 400 of μl absolute ethanol. The tissue fragments were pelleted by centrifugation, the supernatant was decanted, and the pellet washed with 1 ml of absolute ethanol. The supernatant was discarded after centrifugation and the samples were air dried. The tissue pellets were digested overnight at 55°C in an extraction buffer containing 20 mmol/L Tris-hydrochloride (Sigma-Aldrich, St. Louis, MO), pH 7.6/20 mmol/L ethylenediamine tetraacetic acid (EDTA) (Sigma-Aldrich)/10% sodium dodecyl sulfate (SDS), and 0.5 mg/ml Proteinase K. 31 RNA was purified using TRIzol LS (Life Technologies, Grand Island, NY) according to the manufacturer’s instructions. RNA was purified from cell lines using the TRIzol reagent. After isopropanol precipitation the RNA pellet was hydrated in 30 to 50 μl of diethyl-pyrocarbonate-treated H2O (Research Genetics, Huntsville, AL), incubated at 55°C for 10 minutes, and stored at −70°C until use.
Reverse Transcriptase-Polymerase Chain Reaction
Assays were performed in MicroAmp optical reaction tubes and caps (Applied Biosystems, Foster City, CA). Two and 10 μl of RNA were reverse transcribed in a 20-μl reaction consisting of 1X PCR Buffer II (Applied Biosystems), 1.5 mmol/L MgCl2, 10 mmol/L dithiothreitol, 6 U of RNase Inhibitor (Life Technologies), 0.5 mmol/L each of 2-deoxynucleoside 5′-triphosphate (Promega Corp., Madison, WI), 100 U of Moloney murine leukemia virus (Life Technologies), and 0.5 μg of random primers (Life Technologies). The reactions were incubated for 60 minutes at 37°C, heated for 5 minutes to 95°C, and the resulting cDNA was stored at 4°C until use.
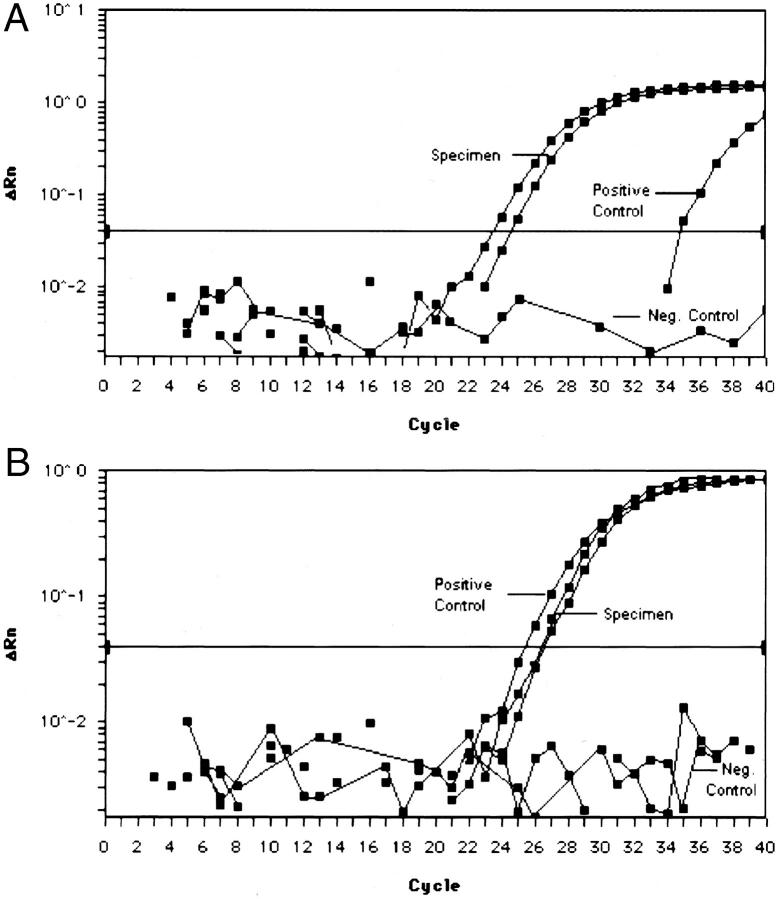
PCR was performed in a 50-μl reaction containing 10 μl of the reverse transcription reaction, 1X Universal PCR Master Mix (Applied Biosystems), 15 pmol of each primer and 2.5 pmol of each probe. The samples were placed in the ABI Prism 7700 Sequence Analyzer, which was set to detect both 6-FAM and VIC reporter dyes simultaneously (Figure 1) . To increase resolution between the two dyes, the spectral compensation feature was used. A control RT-PCR reaction for β-2-microglobulin (β2M) was used to evaluate the samples for the presence of amplifiable RNA. The β2M reactions were performed separately using the remaining 10 μl of cDNA as described previously. 32 After initial incubations at 50°C for 2 minutes and 95°C for 10 minutes, the samples were amplified by running 40 cycles of 95°C for 15 seconds followed by 60°C for 1 minute. We have established criteria in our laboratory of a CT ≤ 38 for the sample to be determined as positive for both the translocation and the amplification control.
Figure 1.
Amplification plot for exonuclease-based RT-PCR assay for SYT-SSX fusion in FFPE specimens. Graphs demonstrate fluorescence emmision data (ΔRn) during each cycle. A: SYT-SSX1 FFPE specimen (two levels of sample) and FFPE-positive control. B: SYT-SSX2 FFPE specimen (two levels of sample) and Bax-1-positive control cell line.
Sensitivity
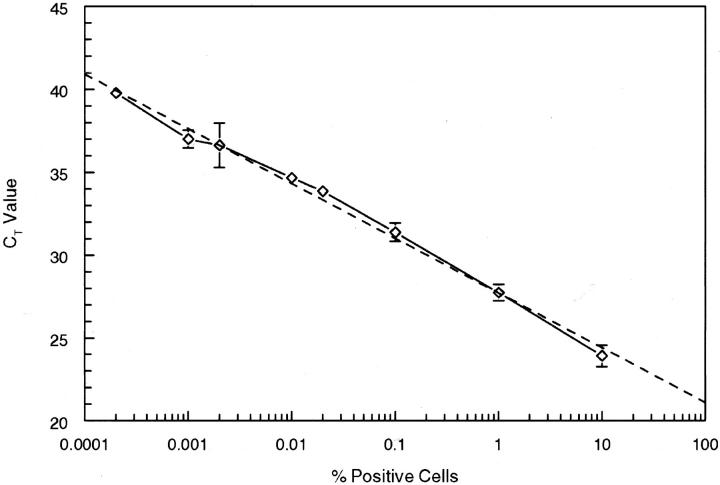
Bax-1 and RD-ES cells were cultured at 37°C in a 5% CO2 atmosphere in RPMI 1640 media (Gibco/Life Technologies, Ltd., Grand Island, NY) supplemented with 20% fetal calf serum (FCS), 2 mmol/L of l-glutamine, 0.1 mmol/L sodium pyruvate, 1X minimal essential medium (MEM) non-essential amino acids, 1X MEM vitamins, and penicillin/streptomycin (100 U/ml, 100 mg/ml, respectively) (Gibco). Cells were pelleted and washed with 1X Dulbecco’s phosphate buffered saline (PBS), counted, and adjusted to 1 × 105 cells/ml. For the experiment shown in Figure 2 , serial dilutions of Bax-1 synovial sarcoma cells were prepared in RD-ES (Ewing’s sarcoma) cells before RNA isolation. Cells (1 × 105) were pelleted in sterile 1.5-ml microcentrifuge tubes and RNA was extracted using TRIzol as described above, resuspended in 50 μl of DEPC-treated dH2O, and stored at −70°C.
Figure 2.
Percent of cells containing the SYT-SSX2 fusion diluted into SYT-SSX-negative cells. Each point represents the mean of two levels (1 and 5 μl) RNA from five separate RT-PCR amplifications from two separate lysate extractions.
Primer and Probe Design
Primer and probe sequences for the SYT-SSX1, SYT-SSX2 fusions, and β-2-microglobulin (β2M) are presented in Table 1 . Primers and probes were designed using the Primer Express software (Applied Biosystems) and yield an expected product size of 98 base pairs (bp). The SYT-SSX1 probe was labeled with the reporter dye 6-FAM (6-carboxyfluorescein), and the SYT-SSX2 and β2M probes were labeled with the reporter dye VIC. Probes were purchased from Integrated DNA Technologies (Coralville, IN) or Applied Biosystems.
Table 1.
Primer and Probe Sequences
| t(X;18)/SYT-SSX | |
| SYT | 5′ AGA GGC CTT ATG GAT ATG ACC AGA T 3′ |
| SSX | 5′ C(A/G)T TTT GTG GGC CAG ATG C C 3′ |
| SSX1 probe | 5′ [6-FAM] TCC CTT CGA ATC ATT TTC GTC CTC TGC T [TAMRA] 3′ |
| SSX2 probe | 5′ [VIC] TCT GGC ACT TCC TCC GAA TCA TTT CCT T [TAMRA] 3′ |
| β2-Microglobulin | |
| β2M-246F | 5′ TGA CTT TGT CAC AGC CCA AGA TA 3′ |
| β2M-330R | 5′ AAT CCA AAT GCG GCA TCT TC 3′ |
| β2M-275R | 5′ [VIC] TGA TGC TGC TTA CAT GTC TCG ATC CCA [TAMRA] 3′ |
Sequencing
The PCR products from the two controls were excised and purified from a 2% agarose gel (SeaKem, FMC Corporation, Rockland, ME) containing ethidium bromide. PCR products were then cut from the agarose gel and extracted using silica beads (GeneClean, Bio101, La Jolla, CA). Sequencing of the PCR products was performed using the Perkin-Elmer Big-Dye Terminator cycle-sequencing kit on an ABI Prism 377 automated sequencer (Applied Biosystems).
Results
The synovial sarcoma cases consisted of a nearly equal distribution of monophasic and biphasic tumors, 53.3% and 46.7%, respectively. Twenty-nine of 30 (96.7%) FFPE synovial sarcomas demonstrated an SYT-SSX transcript. Both biphasic (12 of 14; 85.7%) and monophasic fibrous (10 of 16; 62.5%) tumors were positive for the presence of the SYT-SSX1 transcript. All 5 biphasic tumors with a poorly differentiated component and 1 of the monophasic fibrous tumors with a poorly differentiated component were positive for the SYT-SSX1 transcript. The SYT-SSX2 transcript was detected in 2 of 14 (14.3%) biphasic and 5 of 16 (31.25%) monophasic tumors. One of the SYT-SSX2-positive monophasic fibrous tumors had a poorly differentiated element. One tumor (a keratin-positive monophasic fibrous synovial sarcoma) was negative for both the SYT-SSX1 and SYT-SSX2 transcripts. The PCR product of this case was visualized on a 2.5% agarose gel to determine whether a SYT-SSX4 transcript might be present, a rare finding in synovial sarcomas. 18, 19 This fusion transcript can be amplified with the primers used in the assay, but cannot be detected by either of our probes. No discrete band was seen. None of the 13 non-synovial sarcomas demonstrated the presence of a SYT-SSX1 or a SYT-SSX2 transcript. Examples of the amplification profiles for the different specimens and respective positive controls are shown in Figure 1 , A and B.
The PCR products of the SYT-SSX2-positive Bax-1 cell line and the SYT-SSX1-positive FFPE tumor specimen, used as positive controls, were sequenced to verify the transcript type (data not shown). Both were shown to be variant transcripts, possessing 87-bp and 28-bp inserts, respectively, which appear to be derived from the X chromosome and situated between the SYT and SSX sequences. The 87-bp insert found in the Bax-1 cell line corresponds with that published previously. 33
To determine the sensitivity of the assay, freshly cultured Bax-1 cells were serially diluted into freshly cultured t(X;18) negative RD-ES cells (Figure 2) . RNA was isolated and tested for the presence of the t(X;18) translocation with the real-time assay. A dilution corresponding to one synovial sarcoma cell in 100,000 RD-ES cells was consistently positive in a series of five separate assays. The R value of the resulting graph is 0.997 and the trend line (dashed) indicates an amplification efficiency of near 100%.
Discussion
Molecular approaches in the area of clinical diagnostics have been shown to be of considerable utility for the identification of tumor-specific sarcoma translocations, such as those occurring in Ewing’s sarcoma, alveolar rhabdomyosarcoma, desmoplastic small round cell tumor, clear cell sarcoma, and synovial sarcoma. Atypical or poorly differentiated variants of these sarcomas have been shown to mimic other tumor types or be difficult to diagnose based on histological appearance and immunohistochemistry. 15, 16, 34, 35, 36, 37 Methods that permit the use of routinely processed histological or archival material have distinct advantages, including the ability to make direct correlation with the hematoxylin and eosin and immunohistochemical stains and the ability to perform retrospective studies, based on archival FFPE tissue.
In this study, we demonstrate a simple and rapid method for the identification of the t(X;18) translocation in synovial sarcomas. The use of real-time RT-PCR in clinical diagnostics is rapidly becoming more popular since the method is overall technically less demanding because it does not require the PCR products to be analyzed by gel electrophoresis, with or without a confirmatory Southern blot to distinguish between the fusion types. RT-PCR has the distinct advantage over FISH and conventional cytogenetics as a method of analysis in that RT-PCR is faster. FISH and conventional cytogenetic analysis is time consuming, laborious, and expensive. RT-PCR is a rapid, less laborious, and less costly method. As long as adequate precautions are undertaken, the risk of contamination can be minimized. However, RT-PCR using restriction digestion, Southern blot hybridization, or sequencing of PCR products to identify the fusion type can still take several days to complete. Real-time RT-PCR has the advantage because the amplification and detection occur simultaneously. Results can be provided within 3.5 hours of extraction of the RNA, without further manipulation of the PCR products.
A study by Peter et al 28 reports the utilization of real-time RT-PCR for the detection of the SYT-SSX fusion in synovial sarcomas. Their method, however, still requires conventional RT-PCR and a confirmatory Southern blot with specific probes to determine the fusion type. Also, the minimum product size amplified with their primers is approximately 252 bp, which is often beyond the upper limit in size for what can be amplified by PCR from archival tissue. 31
In our assay, SSX1 and SSX2 specific probes, differing by five bases and which are labeled with different reporter dyes (6-FAM and VIC), are used to discriminate between the SYT-SSX1 and the SYT-SSX2 fusions. Due to the relative infrequency of the SYT-SSX4 fusion, which has only been described in two cases to date, we chose not to include a probe for this fusion. 18, 19 The primers do recognize the fusion however, so if it is desired, the PCR product from negative cases can be run on an agarose or polyacrylamide gel to determine whether a band of the appropriate size (approximately 98 bp) is present. Transcript variants described to date are characterized as containing inserted sequences of varying lengths between the SYT and SSX junction or as having lost portions of the SYT gene. 6, 33, 38, 39, 40 As such, the primers and probes will recognize potential variant transcripts (such as the positive controls used in this study) with little difficulty. We have also shown this method to be highly sensitive and reproducible. We were able to reproduce in five separate runs the detection of one positive cell from the Bax-1 cell line in 1 × 105 SYT-SSX-negative RD-ES cells (0.001%) (see Figure 2 ). The high sensitivity of this method makes it suitable for use in monitoring of patients during treatment and for the detection of minimal residual disease. Detection of the SYT-SSX fusion in the peripheral blood of at least one patient indicates the feasibility of this approach. 41 The level of sensitivity observed may not necessarily be achieved with archival tissue, however, the small product size increases the likelihood of obtaining amplifiable RNA from FFPE, poorly fixed, or problematic specimens.
The t(X;18)(p11.2;q11.2) translocation has been shown to be present in >90% of synovial sarcomas and appears to be highly specific for this tumor. 1, 17, 34, 42, 43, 44, 45, 46 Of the synovial sarcoma cases that we tested in this study, 96.7% (29 of 30) were positive for the presence of either fusion transcript. None of the non-synovial sarcoma cases or cell lines tested showed the presence of either SYT-SSX transcript. In this study, 23% of the cases (7 of 30) were found to have a poorly differentiated element. Five were biphasic and two were monophasic fibrous tumors, and all biphasic tumors containing a poorly differentiated element exhibited the SYT-SSX1 fusion. Two of the biphasic tumors tested were found to be SYT-SSX2 positive. Most biphasic tumors have been reported to be SYT-SSX1 positive, while monophasic tumors may demonstrate either SYT-SSX1 or SYT-SSX2 fusion transcripts. Several studies however, have also reported SYT-SSX2 transcripts in biphasic tumors. 1, 6, 8, 16, 19, 21, 24, 25, 26, 27, 33, 47, 48 This may be due to the differences in diagnostic criteria used as to whether the presence of either epithelial or epithelioid areas and/or open glandular spaces is used in the subtyping the tumor.
O’Sullivan recently reported the presence of the t(X;18) translocation in 75% of MPNSTs. These findings are at variance with other studies that fail to identify the t(X;18) translocation in MPNSTs. 7, 42, 49, 50, 51 This disagreement may be due to antigen expression criteria used to diagnose the tumors that did not meet histological criteria for “classic” synovial sarcoma or MPNST or the lack of confirmation as to the presence of the translocation by another methodology. 5, 6, 51, 52, 53, 54 This issue might be effectively addressed by applying the methodology described here to archival material and is being investigated in an ongoing study.
In conclusion, the purpose of this study is to demonstrate a new method for ascertaining the presence of the t(X;18) translocation in archival FFPE synovial sarcomas. Current methodologies used to identify the presence of the t(X;18) translocation typically require several days and additional manipulation of the samples and/or PCR products to identify the fusion type. The use of real-time RT-PCR enables rapid detection and identification of the presence of the t(X;18) translocation and fusion type. The principle advantages of this method are: the decreased turn around time; the decreased risk of cross-contamination between specimens due to the lack of further manipulation of the PCR products required; high sensitivity; and high specificity.
Address reprint requests to Karen E. Bijwaard, Department of Cellular Pathology and Genetics, Armed Forces Institute of Pathology, 1413 Research Blvd., Bldg. 101, Room 1057D, Rockville, MD 20850. E-mail: bijwaard@afip.osd.mil.
Footnotes
Supported by intramural funds of the Armed Forces Institute of Pathology.
References
- 1.Tsuji S, Hisaoka M, Morimitsu Y, Hashimoto H, Shimajiri S, Komiya S, Ushijima M, Nakamura T: Detection of SYT-SSX fusion transcripts in synovial sarcoma by reverse transcription-polymerase chain reaction using archival paraffin-embedded tissues. Am J Pathol 1998, 153:1807-1812 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ladanyi M: The emerging molecular genetics of sarcoma translocations. Diagn Mol Pathol 1995, 4:162-173 [DOI] [PubMed] [Google Scholar]
- 3.Bergh P, Meis-Kindblom JM, Gherlinzoni F, Berlin O, Bacchini P, Bertoni F, Gunterberg B, Kindblom LG: Synovial sarcoma: identification of low and high risk groups. Cancer 1999, 85:2596-2607 [DOI] [PubMed] [Google Scholar]
- 4.Meis-Kindblom JM, Stenman G, Kindblom LG: Differential diagnosis of small round cell tumors. Semin Diagn Pathol 1996, 13:213-241 [PubMed] [Google Scholar]
- 5.Folpe AL, Schmidt RA, Chapman D, Gown AM: Poorly differentiated synovial sarcoma: immunohistochemical distinction from primitive neuroectodermal tumors and high-grade malignant peripheral nerve sheath tumors. Am J Surg Pathol 1998, 22:673-682 [DOI] [PubMed] [Google Scholar]
- 6.dos Santos NR, de Bruijn DR, van Kessel AG: Molecular mechanisms underlying human synovial sarcoma development. Genes Chromosomes Cancer 2001, 30:1-14 [DOI] [PubMed] [Google Scholar]
- 7.Hiraga H, Nojima T, Abe S, Sawa H, Yamashiro K, Yamawaki S, Kaneda K, Nagashima K: Diagnosis of synovial sarcoma with the reverse transcriptase-polymerase chain reaction: analyses of 84 soft tissue and bone tumors. Diagn Mol Pathol 1998, 7:102-110 [DOI] [PubMed] [Google Scholar]
- 8.van de Rijn M, Barr FG, Xiong QB, Hedges M, Shipley J, Fisher C: Poorly differentiated synovial sarcoma: an analysis of clinical, pathologic, and molecular genetic features. Am J Surg Pathol 1999, 23:106-112 [DOI] [PubMed] [Google Scholar]
- 9.Jorgensen LJ, Lyon H, Myhre-Jensen O, Nordentoft A, Sneppen O: Synovial sarcoma: an immunohistochemical study of the epithelial component. APMIS 1994, 102:191-196 [PubMed] [Google Scholar]
- 10.Argani P, Askin FB, Colombani P, Perlman EJ: Occult pulmonary synovial sarcoma confirmed by molecular techniques. Pediatr Dev Pathol 2000, 3:87-90 [DOI] [PubMed] [Google Scholar]
- 11.Argani P, Faria PA, Epstein JI, Reuter VE, Perlman EJ, Beckwith JB, Ladanyi M: Primary renal synovial sarcoma: molecular and morphologic delineation of an entity previously included among embryonal sarcomas of the kidney. Am J Surg Pathol 2000, 24:1087-1096 [DOI] [PubMed] [Google Scholar]
- 12.Hisaoka M, Hashimoto H, Iwamasa T, Ishikawa K, Aoki T: Primary synovial sarcoma of the lung: report of two cases confirmed by molecular detection of SYT-SSX fusion gene transcripts. Histopathology 1999, 34:205-210 [DOI] [PubMed] [Google Scholar]
- 13.Kim DH, Sohn JH, Lee MC, Lee G, Yoon GS, Hashimoto H, Sonobe H, Ro JY: Primary synovial sarcoma of the kidney. Am J Surg Pathol 2000, 24:1097-1104 [DOI] [PubMed] [Google Scholar]
- 14.Oizumi S, Igarashi K, Takenaka T, Yamashiro K, Hiraga H, Fujino T, Horimoto M: Primary pericardial synovial sarcoma with detection of the chimeric transcript SYT-SSX. Jpn Circ J 1999, 63:330-332 [DOI] [PubMed] [Google Scholar]
- 15.Masui F, Matsuno Y, Yokoyama R, Nakanishi Y, Hasegawa T, Kanai Y, Beppu Y, Hirohashi S, Fujii K, Shimoda T: Synovial sarcoma, histologically mimicking primitive neuroectodermal tumor/Ewing’s sarcoma at distant sites. Jpn J Clin Oncol 1999, 29:438-441 [DOI] [PubMed] [Google Scholar]
- 16.Willeke F, Mechtersheimer G, Schwarzbach M, Weitz J, Zimmer D, Lehnert T, Herfarth C, von Knebel Doeberitz M, Ridder R: Detection of SYT-SSX1/2 fusion transcripts by reverse transcriptase-polymerase chain reaction (RT-PCR) is a valuable diagnostic tool in synovial sarcoma. Eur J Cancer 1998, 34:2087-2093 [DOI] [PubMed] [Google Scholar]
- 17.Sreekantaiah C, Ladanyi M, Rodriguez E, Chaganti RS: Chromosomal aberrations in soft tissue tumors: relevance to diagnosis, classification, and molecular mechanisms. Am J Pathol 1994, 144:1121-1134 [PMC free article] [PubMed] [Google Scholar]
- 18.Skytting B, Nilsson G, Brodin B, Xie Y, Lundeberg J, Uhlen M, Larsson O: A novel fusion gene, SYT-SSX4, in synovial sarcoma. J Natl Cancer Inst 1999, 91:974-975 [DOI] [PubMed] [Google Scholar]
- 19.Mancuso T, Mezzelani A, Riva C, Fabbri A, Dal Bo L, Sampietro G, Perego P, Casali P, Zunino F, Sozzi G, Pierotti MA, Pilotti S: Analysis of SYT-SSX fusion transcripts and bcl-2 expression and phosphorylation status in synovial sarcoma. Lab Invest 2000, 80:805-813 [DOI] [PubMed] [Google Scholar]
- 20.de Leeuw B, Suijkerbuijk RF, Olde Weghuis D, Meloni AM, Stenman G, Kindblom LG, Balemans M, van den Berg E, Molenaar WM, Sandberg AA, Geurts van Kessel A: Distinct Xp11: 2 breakpoint regions in synovial sarcoma revealed by metaphase and interphase FISH: relationship to histologic subtypes. Cancer Genet Cytogenet 1994, 73:89-94 [DOI] [PubMed] [Google Scholar]
- 21.Shipley J, Crew J, Birdsall S, Gill S, Clark J, Fisher C, Kelsey A, Nojima T, Sonobe H, Cooper C, Gusterson B: Interphase fluorescence in situ hybridization and reverse transcription polymerase chain reaction as a diagnostic aid for synovial sarcoma. Am J Pathol 1996, 148:559-567 [PMC free article] [PubMed] [Google Scholar]
- 22.Geurts van Kessel A, dos Santos NR, Simons A, de Bruijn D, Forus A, Fodstad O, Myklebost O, Balemans M, Baats E, Olde Weghuis D, Suijkerbuijk RF, van den Berg E, Molenaar WM, de Leeuw B: Molecular cytogenetics of bone and soft tissue tumors. Cancer Genet Cytogenet 1997, 95:67-73 [DOI] [PubMed] [Google Scholar]
- 23.Zilmer M, Harris CP, Steiner DS, Meisner LF: Use of nonbreakpoint DNA probes to detect the t(X;18) in interphase cells from synovial sarcoma: implications for detection of diagnostic tumor translocations. Am J Pathol 1998, 152:1171-1177 [PMC free article] [PubMed] [Google Scholar]
- 24.Inagaki H, Nagasaka T, Otsuka T, Sugiura E, Nakashima N, Eimoto T: Association of SYT-SSX fusion types with proliferative activity and prognosis in synovial sarcoma. Mod Pathol 2000, 13:482-488 [DOI] [PubMed] [Google Scholar]
- 25.Lasota J, Jasinski M, Debiec-Rychter M, Szadowska A, Limon J, Miettinen M: Detection of the SYT-SSX fusion transcripts in formaldehyde-fixed, paraffin-embedded tissue: a reverse transcription polymerase chain reaction amplification assay useful in the diagnosis of synovial sarcoma. Mod Pathol 1998, 11:626-633 [PubMed] [Google Scholar]
- 26.Nilsson G, Skytting B, Xie Y, Brodin B, Perfekt R, Mandahl N, Lundeberg J, Uhlen M, Larsson O: The SYT-SSX1 variant of synovial sarcoma is associated with a high rate of tumor cell proliferation and poor clinical outcome. Cancer Res 1999, 59:3180-3184 [PubMed] [Google Scholar]
- 27.Antonescu CR, Kawai A, Leung DH, Lonardo F, Woodruff JM, Healey JH, Ladanyi M: Strong association of SYT-SSX fusion type and morphologic epithelial differentiation in synovial sarcoma. Diagn Mol Pathol 2000, 9:1-8 [DOI] [PubMed] [Google Scholar]
- 28.Peter M, Gilbert E, Delattre O: A multiplex real-time PCR assay for the detection of gene fusions observed in solid tumors. Lab Invest 2001, 81:905-912 [DOI] [PubMed] [Google Scholar]
- 29.Kawai A, Woodruff J, Healey JH, Brennan MF, Antonescu CR, Ladanyi M: SYT-SSX gene fusion as a determinant of morphology and prognosis in synovial sarcoma. N Engl J Med 1998, 338:153-160 [DOI] [PubMed] [Google Scholar]
- 30.Holland PM, Abramson RD, Watson R, Gelfand DH: Detection of specific polymerase chain reaction product by utilizing the 5′—-3′ exonuclease activity of Thermus aquaticus DNA polymerase. Proc Natl Acad Sci USA 1991, 88:7276-7280 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Krafft AE, Duncan BW, Bijwaard KE, Taubenberger JK, Lichy JH: Optimization of the isolation and amplification of RNA from formalin-fixed, paraffin-embedded tissue: the Armed Forces Institute of Pathology experience and literature review. Mol Diagn 1997, 2:217-230 [DOI] [PubMed] [Google Scholar]
- 32.Bijwaard KE, Aguilera NS, Monczak Y, Trudel M, Taubenberger JK, Lichy JH: Quantitative real-time reverse transcription-PCR assay for cyclin D1 expression: utility in the diagnosis of mantle cell lymphoma. Clin Chem 2001, 47:195-201 [PubMed] [Google Scholar]
- 33.Crew AJ, Clark J, Fisher C, Gill S, Grimer R, Chand A, Shipley J, Gusterson BA, Cooper CS: Fusion of SYT to two genes, SSX1 and SSX2, encoding proteins with homology to the Kruppel-associated box in human synovial sarcoma. EMBO J 1995, 14:2333-2340 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Argani P, Zakowski MF, Klimstra DS, Rosai J, Ladanyi M: Detection of the SYT-SSX chimeric RNA of synovial sarcoma in paraffin-embedded tissue and its application in problematic cases. Mod Pathol 1998, 11:65-71 [PubMed] [Google Scholar]
- 35.Cohen IJ, Stark B, Avigad S: Synovial sarcoma mimicking desmoplastic small round-cell tumor: critical role for molecular diagnosis. Med Pediatr Oncol 2000, 34:234. [DOI] [PubMed] [Google Scholar]
- 36.Cole P, Ladanyi M, Gerald WL, Cheung NK, Kramer K, LaQuaglia MP, Kushner BH: Synovial sarcoma mimicking desmoplastic small round-cell tumor: critical role for molecular diagnosis. Med Pediatr Oncol 1999, 32:97-101 [DOI] [PubMed] [Google Scholar]
- 37.Folpe AL, Hill CE, Parham DM, O’Shea PA, Weiss SW: Immunohistochemical detection of FLI-1 protein expression: a study of 132 round cell tumors with emphasis on CD99-positive mimics of Ewing’s sarcoma/primitive neuroectodermal tumor. Am J Surg Pathol 2000, 24:1657-1662 [DOI] [PubMed] [Google Scholar]
- 38.Sanders ME, van de Rijn M, Barr FG: Detection of a variant SYT-SSX1 fusion in a case of predominantly epithelioid synovial sarcoma. Mol Diagn 1999, 4:65-70 [DOI] [PubMed] [Google Scholar]
- 39.Safar A, Wickert R, Nelson M, Neff JR, Bridge JA: Characterization of a variant SYT-SSX1 synovial sarcoma fusion transcript. Diagn Mol Pathol 1998, 7:283-287 [DOI] [PubMed] [Google Scholar]
- 40.Fligman I, Lonardo F, Jhanwar SC, Gerald WL, Woodruff J, Ladanyi M: Molecular diagnosis of synovial sarcoma and characterization of a variant SYT-SSX2 fusion transcript. Am J Pathol 1995, 147:1592-1599 [PMC free article] [PubMed] [Google Scholar]
- 41.Hashimoto N, Myoui A, Araki N, Asai T, Sonobe H, Hirota S, Yoshikawa H: Detection of SYT-SSX fusion gene in peripheral blood from a patient with synovial sarcoma. Am J Surg Pathol 2001, 25:406-410 [DOI] [PubMed] [Google Scholar]
- 42.Fletcher CD, Dal Cin P, de Wever I, Mandahl N, Mertens F, Mitelman F, Rosai J, Rydholm A, Sciot R, Tallini G, van den Berghe H, Vanni R, Willen H: Correlation between clinicopathological features and karyotype in spindle cell sarcomas: a report of 130 cases from the CHAMP study group. Am J Pathol 1999, 154:1841-1847 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Clark J, Rocques PJ, Crew AJ, Gill S, Shipley J, Chan AM, Gusterson BA, Cooper CS: Identification of novel genes, SYT and SSX, involved in the t(X;18)(p11.2;q11.2) translocation found in human synovial sarcoma. Nat Genet 1994, 7:502-508 [DOI] [PubMed] [Google Scholar]
- 44.Dei Tos AP, Dal Cin P: The role of cytogenetics in the classification of soft tissue tumours. Virchows Arch 1997, 431:83-94 [DOI] [PubMed] [Google Scholar]
- 45.Bridge JA, Sandberg AA: Cytogenetic and molecular genetic techniques as adjunctive approaches in the diagnosis of bone and soft tissue tumors. Skeletal Radiol 2000, 29:249-258 [DOI] [PubMed] [Google Scholar]
- 46.Fletcher JA: Cytogenetics and molecular biology of soft tissue tumors. Monogr Pathol 1996, 38:37-64 [PubMed] [Google Scholar]
- 47.Kasai T, Shimajiri S, Hashimoto H: Detection of SYT-SSX fusion transcripts in both epithelial and spindle cell areas of biphasic synovial sarcoma using laser capture microdissection. Mol Pathol 2000, 53:107-110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Inagaki H, Murase T, Otsuka T, Eimoto T: Detection of SYT-SSX fusion transcript in synovial sarcoma using archival cytologic specimens. Am J Clin Pathol 1999, 111:528-533 [DOI] [PubMed] [Google Scholar]
- 49.O’Sullivan MJ, Kyriakos M, Zhu X, Wick MR, Swanson PE, Dehner LP, Humphrey PA, Pfeifer JD: Malignant peripheral nerve sheath tumors with t(X;18): a pathologic and molecular genetic study. Mod Pathol 2000, 13:1253-1263 [DOI] [PubMed] [Google Scholar]
- 50.van de Rijn M, Barr FG, Collins MH, Xiong QB, Fisher C: Absence of SYT-SSX fusion products in soft tissue tumors other than synovial sarcoma. Am J Clin Pathol 1999, 112:43-49 [DOI] [PubMed] [Google Scholar]
- 51.Guillou L, Coindre J, Gallagher G, Terrier P, Gebhard S, de Saint Aubain Somerhausen N, Michels J, Jundt G, Vince DR, Collin F, Trassard M, Le Doussal V, Benhattar J: Detection of the synovial sarcoma translocation t(X;18) (SYT;SSX) in paraffin-embedded tissues using reverse transcriptase-polymerase chain reaction: a reliable and powerful diagnostic tool for pathologists. a molecular analysis of 221 mesenchymal tumors fixed in different fixatives. Hum Pathol 2001, 32:105-112 [DOI] [PubMed] [Google Scholar]
- 52.Hibshoosh H, Lattes R: Immunohistochemical and molecular genetic approaches to soft tissue tumor diagnosis: a primer. Semin Oncol 1997, 24:515-525 [PubMed] [Google Scholar]
- 53.Smith TA, Machen SK, Fisher C, Goldblum JR: Usefulness of cytokeratin subsets for distinguishing monophasic synovial sarcoma from malignant peripheral nerve sheath tumor. Am J Clin Pathol 1999, 112:641-648 [DOI] [PubMed] [Google Scholar]
- 54.Ladanyi M, Woodruff JM, Scheithauer BW, Bridge JA, Barr FG, Goldblum JR, Fisher C, Perez-Atayde A, Dal Cin P, Fletcher CD, Fletcher JA: Re: O’Sullivan MJ, Kyriakos M, Zhu X, Wick MR, Swanson PE, Dehner LP, Humphrey PA, Pfeifer JD: Malignant peripheral nerve sheath tumors with T(X;18): a pathologic and molecular genetic study. Mod Pathol 2000, 13:1336–1346, Mod Pathol 2001, 14:733–737 [DOI] [PubMed]