Abstract
Clear cell sarcoma (CCS), also known as melanoma of soft parts, is an uncommon deep soft tissue tumor presenting typically in the lower extremities of young adults. Previous cytogenetic studies have established the specificity of the recurrent t(12;22)(q13;q12), resulting in a EWS-ATF1 fusion, for CCS. The prevalence of the EWS-ATF1 fusion in CCS remains unclear, since most genetically confirmed CCS have been reported as isolated cytogenetic or molecular diagnostic case reports. We therefore studied histologically confirmed CCS from 12 patients for the presence of EWS-ATF1 by reverse-transcriptase polymerase chain reaction (RT-PCR), using RNA extracted from either frozen (four cases) or formalin-fixed paraffin-embedded (eight cases) material. All primary tumors were located in the deep soft tissues of the extremities. Histologically, 10 cases had a typical epithelioid nested appearance. Most or all cases showed immunostaining for HMB45 (12 of 12), S-100 protein (10 of 12), and MITF (12 of 12). Ultrastructural analysis showed melanosomes in six of seven cases. The presence of an EWS-ATF1 fusion transcript was identified by RT-PCR in 11 of 12 cases (91%), all of which showed the same fusion transcript structure, namely the previously described in-frame fusion of EWS exon 8 to ATF1 codon 65. RT-PCR analysis for the melanocyte-specific splice form of the MITF transcript was positive in all cases tested (4 of 4). These data confirm that EWS-ATF1 detection can be used as a highly sensitive diagnostic test for CCS and that CCS expresses the melanocyte-specific form of the MITF transcript, further supporting its genuine melanocytic differentiation.
Clear cell sarcoma (CCS) is an unusual tumor with a predilection for the deep soft tissues of the lower extremity, and close proximity to tendon, fascia or aponeuroses. CCS preferentially affects adolescents and young adults, and is associated with a high propensity for regional or distant metastases. The histogenesis of this tumor has long been controversial. Since its original description by Enzinger in 1965 1 as a novel type of soft tissue sarcoma designated CCS, several lines of evidence supporting melanocytic differentiation have been presented. In 1983, Chung and Enzinger 2 renamed it melanoma of soft parts, to connote a proposed origin from migrated neural crest cells with a capacity for melanin synthesis. The majority of CCS show immunoreactivity for melanoma markers, such as HMB45 and contain melanosomes as demonstrated ultrastructurally. 3, 4, 5 More recently, the expression of microphthalmia transcription factor (MITF) has been shown to be a sensitive marker for both cutaneous melanomas and CCS. 6, 7, 8, 9, 10 MITF is a transcription factor with several isoforms one of which (MITF-M) is critical in the differentiation of melanocytes, while other isoforms are similarly important in the biology of retinal pigment epithelium, mast cells, and osteoclasts. 11 Despite some overlap with melanoma, a significant number of clinical and biological features are consistent with the notion that CCS represents a unique entity, rather than simply a deep form or metastatic implant of malignant melanoma, 6 most notably its cytogenetic profile.
Unlike melanomas, most CCS are characterized cytogenetically by a recurrent chromosomal translocation, t(12;22)(q13;q12), resulting in fusion of the EWS gene on 22q12 with the ATF1 gene on 12q13. 12 In the resulting chimeric protein, the C-terminal of EWS, which contains an RNA binding domain, is replaced by a functional bZIP DNA-binding and dimerization domain of ATF1, a transcription factor which is normally regulated by cAMP. 13, 14 EWS-ATF1 binds to ATF sites in cAMP-responsive promoters through the bZIP domain derived from ATF1 and its function as a constitutive transcriptional activator is dependent on an activation domain within EWS containing repetitive elements. 15 EWS-ATF1 functions as a potent constitutive activator of several cAMP-inducible promoters when assayed by transfection in cells lacking the EWS-ATF1 fusion. 13, 14 However, the in vivo targets of EWS-ATF1 in CCS cells still remain unclear. 16 Nonetheless, it appears that the viability of CCS cells is dependent on the activity of EWS-ATF1, based on a study using intracellular anti-ATF1 antibody. 17 Recent data suggest that EWS-ATF1 may operate at least partly by interfering with P53 function, through competition with the latter for the transcriptional coactivator CBP. 18
Since most of the genetically confirmed CCS have been studied by conventional karyotyping and published as isolated case reports in the cytogenetic or molecular diagnostic literature, 19, 20, 21, 22, 23, 24, 25 the exact prevalence of t(12;22)-positive cases as confirmed by molecular techniques among cases histologically diagnosed as CCS remains uncertain. In this study, we assessed the prevalence of EWS-ATF1 fusion as detected by reverse transcriptase polymerase chain reaction (RT-PCR) in a group of 12 pathologically confirmed cases of CCS. In addition, we present for the first time an RT-PCR assay for the detection of EWS-ATF1 transcripts applicable to tumor RNA extracted from formalin-fixed paraffin-embedded material and we show that CCS expresses the melanocyte-specific form of the MITF transcription factor, further supporting its melanocytic differentiation.
Materials and Methods
Histological, Immunohistochemical, and Ultrastructural Analysis
A prospectively gathered adult soft tissue sarcoma database at Memorial Sloan-Kettering Cancer Center was searched for the diagnosis of CCS, made during the 18-year period, 1982–2000. Out of 4496 cases in this database, 16 (0.3%) were diagnosed as CCS and of these, 12 cases of CCS were identified in which both pathological material for reconfirming the histological diagnosis and also adequate tumor tissue for molecular analysis were available. Before inclusion in the sarcoma database, medical charts and clinical histories were reviewed in all cases to exclude an antecedent or concurrent diagnosis of primary cutaneus melanoma.
Criteria for the microscopic diagnosis of CCS included the presence of polygonal cells with abundant clear to pale eosinophilic cytoplasm, vesicular nuclei with prominent nucleoli, and arrangement in a nested pattern. The cases were assessed for the presence of melanin pigment, multinucleated giant cells, and also for the presence of an association with tendon or aponeurotic structures.
Immunohistochemical studies for S-100 protein [Biogenex, San Ramon, CA; 1:50,000; citrate buffer] and HMB45 [DAKO, Carpinteria, CA; 1:500; citrate buffer] were done in all cases. Immunostaining with D5 (Labvision, Fremont, CA), a monoclonal antibody generated against human MITF, was also performed in all 12 cases. The D5 antibody binds a portion of MITF encoded by both melanocyte-specific (MITF-M) and non-melanocyte-specific isoforms of MITF transcripts. 11, 26
Ultrastructural examination was performed on seven tumors. Representative fresh tumor tissue was fixed in 2% glutaraldehyde, postfixed in 1% osmium tetroxide, and embedded in epoxy resin using standard procedures.
Molecular Analysis
Frozen tissue, collected under an Institutional Review Board-approved protocol, was available in 4 of the 12 cases (CCS 1, 8–10), and represented primary (CCS 8 and 10) or metastatic (CCS 1 and 9) tumor samples. Tumors were snap-frozen and stored at −70°C. Total RNA was extracted using 1 ml of Trizol reagent (Gibco BRL Inc., Gaithersburg, MD). The extraction was carried out according to instructions supplied by the manufacturer.
In the remaining eight cases, RNA was extracted from a representative paraffin block containing formalin-fixed tumor tissue. In four cases (CCS 3, 4, 6, and 11) the block available for RNA extraction was from the primary tumor, in one case (CCS 5) from a local recurrence, and in three cases (CCS 2, 7, and 12) from a metastasis. The extraction was performed as directed by the manufacturer (Paraffin Block RNA Isolation Kit; Ambion, Inc., Austin, TX) with several alterations. In an effort to yield more RNA from the extraction, four 15-um sections were cut and proteinase K was used in twice the suggested volume (200 μl) for 25 minutes. To this, 2 μl of linear acrylamide were added followed by precipitation overnight at −20°C. Every effort was taken to prevent cross-contamination. For each case, a new microtome blade was used and the microtome was wiped with 10% bleach and 70% ethanol.
In all samples, RT-PCR was first performed with the outer pair of primers consisting of a sense primer located in exon 8 of EWS and an antisense primer in ATF1 (codon 130). All primer sequences, annealing temperatures used, and expected product sizes are shown in Table 1 . The reaction was carried out using the One-Step RT-PCR kit (Qiagen, Inc., Valencia, CA) at 50°C for 30 minutes, 95°C for 15 minutes, 30 cycles of 95°C for 30 seconds, 58°C for 30 seconds, 72°C 30 seconds, and a final extension at 72°C for 7 minutes. In parallel, each sample was amplified without reverse transcriptase in a reaction set up with the HotStartTaq DNA polymerase kit (Qiagen), according to package directions, as a “no RT” negative control. All reactions also included a no RNA negative control. The adequacy of the extracted RNA was assessed by RT-PCR, using primers for PGK (phosphoglycerate kinase) transcripts, as described previously. 27
Table 1.
PCR Primers
| Primer pair | Primer name | Primer sequence | TA (°C)* | Product size |
|---|---|---|---|---|
| EWS-ATF1 Type 1 or 2 outer pair | EWSex8-F1 ATF1-R1 | GAGGCATGAGCAGAGGTGG GAAGTCCCTGTACTCCATCTGTG | 58° | Type 1: 246 bp Type 2: 183 bp |
| EWS-ATF1 Type 1 or 2 inner pair | EWSex8-F2 ATF1-R2 | GAGGAGGACGCGGTGGAATG CTGTAAGGCTCCATTTGGGGC | 64° | Type 1: 185 bp Type 2: 122 bp |
| EWS-ATF1 Type 3 outer pair | EWSex7-F1† ATF1-R1 | TCCTACAGCCAAGCTCCAAGTC See above | 58° | Type 3: 124 bp |
| EWS-ATF1 Type 3 inner pair | EWSex7-F2 ATF1-R2 | TATAGCCAACAGAGCAGCAG See above | 55° | Type 3: 63 bp |
| MITF consensus | MITF-C-F MITF-C-R | ACAACCTGATTGAACGAAG AATCTGGAGAGCAGAGACC | 48° | 303 bp |
| MITF melanocyte-specific | MITF-M-F MITF-M-R | TTATAGTACCTTCTCTTTGCC GCTTGCTGTATGTGGTACTTG | 48° | 188 bp |
The eight tumors in which the RNA was obtained from paraffin-embedded material were subjected to a second PCR reaction (nested RT-PCR). One μL of each of the RT- PCR products (including “no RT” control products) was used in a second PCR with an inner pair of primers (Table 1) . This reaction was set up using 2.5 units of AmpliTaq DNA polymerase (Perkin Elmer, Inc., Norwalk, CT), 1X PCR buffer, 1.5 mmol/L MgCl2 (Perkin Elmer), 200 μmol/L PCR nucleotide mix (Boehringer Mannheim, Corp., Indianapolis, IN) and 0.6 μmol/L each of the forward and reverse primers. Initial denaturation at 95°C for 2 minutes was followed by 95°C for 30 seconds; 64°C for 30 seconds; 72°C for 30 seconds for 25 cycles with a 7 minutes final extension at 72°C. PCR products were electrophoresed on a 2% agarose gel and bands were cut and purified with the Concert rapid gel extraction system (Gibco BRL). Products were routinely sequenced on automated sequencer.
The nested and non-nested EWS-ATF1 RT-PCR assays were designed to detect the most common type of EWS-ATF-1 fusion transcript, namely the EWS exon 8 to ATF1 codon 65 fusion, 12 which we designate type 1 and one of the molecular variants, namely the EWS exon 10 to ATF1 codon 110 fusion, 28 which we designate type 2. The only negative case for these primers was also subjected to a nested RT-PCR using nested EWS exon 7 forward primers 29 in combination with the nested ATF1 primers (Table 1) to exclude the EWS exon 7 to ATF1 codon 110 fusion described by Pellin et al, 30 which we designate type 3. The nested RT-PCR protocol used was exactly as above, except for the different annealing temperatures (Table 1) .
RT-PCR analysis for MITF mRNA expression was performed using two sets of primers (Table 1) . The MITF-C consensus primers 10 amplify a portion of the MITF transcript shared by all MITF splice forms. The MITF-M primers are designed to amplify only the melanocyte-specific isoform of MITF, which differs from other forms by the usage of an alternative exon 1 (exon 1M). 31 RT-PCR analysis for MITF transcripts was performed without nesting and therefore was only considered appropriate in cases with good quality RNA. This included three of four cases with RNA obtained from frozen tissue (CCS 9 had insufficient RNA) and one case (CCS 3) in which good quality RNA was extracted from the archival paraffin-embedded sample. The target amplicons of both pairs of MITF primers included exon boundaries, to avoid amplification of trace amounts of genomic DNA in the RNA preparations. RNA from the SK-MEL-19 melanoma cell line was used a positive control for both MITF RT-PCR assays.
Results
Clinical Data
There were seven males and five females, with a mean age of 32 (range, 20 to 69 years). All cases were located in the deep soft tissues of the extremities (see Table 2for demographics and exact locations). All tumors were deep and were excised by a wide-local resection. In seven cases the surgical margins were microscopically free. The tumors in 10 cases were localized at the time of diagnosis, while in two cases there were lung metastases at presentation as detected by staging chest CT-scans. In 11 of 12 cases follow-up information was available and the follow-up ranged from 5 to 204 months (mean of 44 months for survivors). Seven (63%) patients developed local recurrences. All five tumors with positive surgical margins eventually recurred locally. Seven of 11 (63%) patients developed regional lymph node metastases, diagnosed at intervals of 0 to 47 months (mean of 21 months) from the primary diagnosis. Distant metastases were identified in nine cases (82%), at time intervals after original diagnosis ranging from 0 (three cases) to 132 months, with a mean of 42 months. In all cases, the site of distant spread was the lung, while additional sites such as brain, bone, adrenal gland, and spleen were noted in one case each. Adjuvant chemotherapy information is listed in Table 2 . At last follow-up, there were three patients with no evidence of disease (NED), one patient alive with disease (AWD), and seven patients dead of their disease (DOD). Only one of the three NED patients did not develop any regional or distant recurrences; this after a 22-month follow-up period. The other two patients remained NED after resection of their regional (CCS 2) or distant (CCS 1) metastasis, 9 and 136 months later. All patients alive with or dead of disease had had distant metastases.
Table 2.
Summary of Clinicopathological Data, Management, and Follow-up
| CCS | Age | Sex | Site | Size (cm) | Stage at diagnosis | Surg Mg | Radiotherapy | Chemotherapy | Local recurrence (mo) | Reg LN metastasis site | Reg LN (mo) | Distant metastasis sites | Distant metastasis (mo) | Follow-up (mo) | Status |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | 25 | M | Arm | 4 | 1° | Neg | Preop: 4897cGy Postop: 1600cGy | After mets: Cis DTIC, IFN | 30 | Distal arm | 36 | Lung | 120 | 136 | NED |
| 2 | 31 | M | Knee | 3.5 | 1° | Neg | Postop ERT | — | — | Popliteal | 8 | — | — | 9 | NED |
| 3 | 25 | F | Foot | <5 | 1° | Neg | N/A | N/A | N/A | N/A | N/A | N/A | N/A | N/A | N/A |
| 4 | 23 | M | Buttock | 13.5 | 1° | Pos | — | — | 3 | Inguinal Iliac | 1 | Lung, Brain Bone, Adrn | 5 | 5 | DOD |
| 5 | 36 | M | Wrist | <5 | 1° | Pos | Postop ERT | Cis limb-perfusion | 8 | Mid-forearm | 54 | Lung | 85 | 85 | DOD |
| 6 | 36 | F | Thigh/pelvis | >10 | 1° | Pos | Preop: 5200cGy Postop: 200cGy | 5 | Inguinal | 0 | Lung | 12 | 12 | DOD | |
| 7 | 26 | M | Hand | 4.6 | 1° | Neg | — | — | — | — | — | Lung, Med LN | 32 | 39 | DOD |
| 8 | 21 | M | Thigh | 2.5 | 1° | Neg | — | — | — | — | — | — | — | 22 | NED |
| 9 | 25 | M | Arm | 7 | 1° | Neg | Preop: 5000cGy Postop: Brachy | Preop/Postop: Cis DTIC,Adr,Ifos | 47 | Cubital | 47 | Lung | 38 | 42 | DOD |
| 10 | 46 | F | Hip/buttock | 15.7 | Mets | Pos | Postop: ERT for residual disease | Preop:Cis, Vinbl, IFN, Dacarbazine Postop: Docetaxel, Gem | — | Sacral-tuberous ligament | 3 | Lung, LN, spleen | 0 | 10 | AWD |
| 11 | 69 | F | Thigh | 6.2 | Mets | Neg | — | — | — | — | — | Lung | 0 | 17 | DOD |
| 12 | 20 | F | Foot | <5 | 1° | Pos | — | — | 12 | — | — | Lung | 132 | 204 | DOD |
Cis, cisplatin; IFN, interferon; Adr, adriamycin; Ifos, ifosfamide; Vinbl, vinblastine; Gem, gemcitabine; NED, no evidence of disease; DOD, dead of disease; AWD, alive with disease; N/A, not available; ERT, external radiation; mets, metastasis; Surg Mg, surgical margins; Adrn, adrenal gland; LN, lymph node; Med, mediastinal.
Histopathological Data
On histological review, 10 cases (all but CCS 6 and 10) had a classic microscopic appearance, with a distinct compartmentalization of the tumor cells by thin fibrous septa. The ovoid tumor cells showed clear to pale eosinophilic cytoplasm, and centrally located nuclei, with minimal pleomorphism but prominent nucleoli. In 7 of these 10 cases, areas of spindling were at least focally noted. Only one case (CCS 3) showed a predominantly spindle cell morphology, arranged in nests or short fascicles. Multinucleated giant cells were also identified in 5 cases (Figure 1) . In 8 cases definite evidence of tumor invasion of tendon or aponeurotic structures was identified microscopically. Cases CCS 6 and CCS 10 showed a distinct microscopic appearance, with epithelioid cells, a higher degree of nuclear pleomorphism, anaplasia, and an alveolar pattern of growth, due to a striking loss of cell cohesion (Figure 2) . In some areas the tumor cells had a rhabdoid appearance, with bright eosinophilic cytoplasm and an eccentrically located nucleus. Case CCS 10 showed in addition scattered multinucleated giant cells and evidence of infiltration into a tendon structure.
Figure 1.
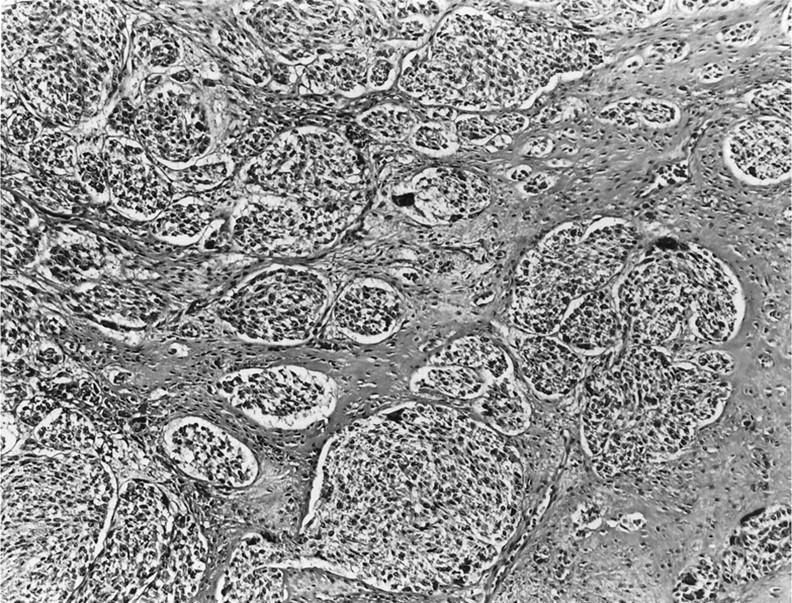
Microscopic appearance of nested epithelioid cells with clear cytoplasm and prominent nucleoli, separated by fibrous bands and showing scattered multinucleated giant cells (hematoxylin & eosin; ×10).
Figure 2.
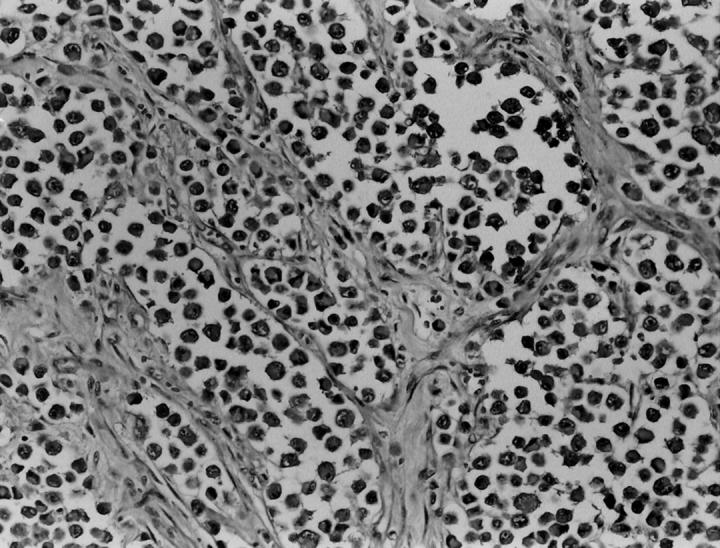
Case CCS 6 showed a distinct microscopic appearance, with epithelioid cells, showing a higher degree of nuclear pleomorphism and anaplasia, arranged in an alveolar pattern of growth, due to a striking loss of cohesion (hematoxylin & eosin; ×20).
Immunohistochemical Data
These results are summarized in Table 3 . All cases tested showed strong and diffuse immunoreactivity for HMB45 (cytoplasmic) and MITF (nuclear) (Figure 3) . Two cases were negative for S-100 protein (CCS 3 and 11), but the remaining cases were strongly and diffusely positive.
Table 3.
Summary of Immunohistochemical, Ultrastructural, and Molecular Data
| CCS | IHC findings | EM findings | RT-PCR | |||||||
|---|---|---|---|---|---|---|---|---|---|---|
| S-100 | HMB45 | MITF | Melanosomes | Glycogen pools | PCJ | BM | EWS-ATF1 | MITF | ||
| C | M | |||||||||
| 1 | + | + | + | Stage I and II | + | − | − | + | + | + |
| 2 | + | + | + | Stage II and III | − | − | − | + | ND | ND |
| 3 | − | + | + | ND | ND | ND | ND | + | + | + |
| 4 | + | + | + | Stage II possible Stage III | + | + | − | + | ND | ND |
| 5 | + | + | + | Stage II possible Stage III | − | + | + | + | ND | ND |
| 6 | + | + | + | ND | ND | ND | ND | + | ND | ND |
| 7 | + | + | + | ND | ND | ND | ND | + | ND | ND |
| 8 | + | + | + | ND | ND | ND | ND | + | + | + |
| 9 | + | + | + | Stage II | − | − | + | + | ND | ND |
| 10 | + | + | + | −* | + | − | + | + | + | + |
| 11 | − | + | + | Stage II and III | + | + | + | − | ND | ND |
| 12 | + | + | + | ND | ND | ND | ND | + | ND | ND |
Rare cytoplasmic granules having the shape and size of melanosomes, but lacking an internal structure and therefore not-diagnostic for melanosomes.
PCJ, primitive cell junctions; BM, basement membrane; ND, not done; C, MITF-consensus RT-PCR; M, RT-PCR for melanocyte-specific MITF transcript.
Figure 3.
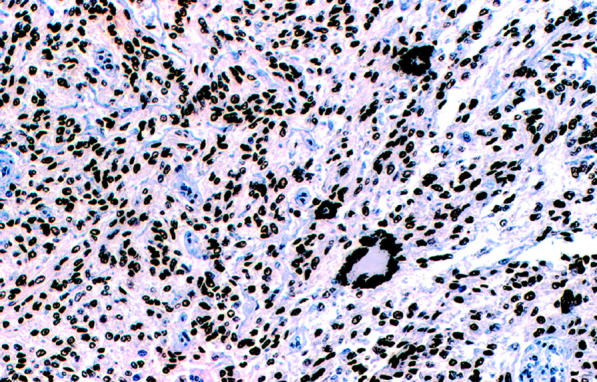
Immunohistochemical study with D5 monoclonal antibody showing strong and diffuse nuclear labeling in the tumor cells and also in the multinucleated giant cells (×20).
Ultrastructural Data
Electron microscopy revealed scattered stage II unpigmented melanosomes and rare pigmented stage III melanosomes in six of the seven cases studied (Table 3) (Figure 4) . In case CCS 10, rare cytoplasmic granules of the size and shape of melanosomes but lacking an internal structure were noted. In addition, electron-dense myelin figure-like structures indistinguishable from lysosomes were focally present in some cells. There were external lamina and rudimentary cell junctions in four and three cases, respectively. Numerous mitochondriae were identified in most cases. The presence of glycogen pools was a consistent finding only in four cases.
Figure 4.
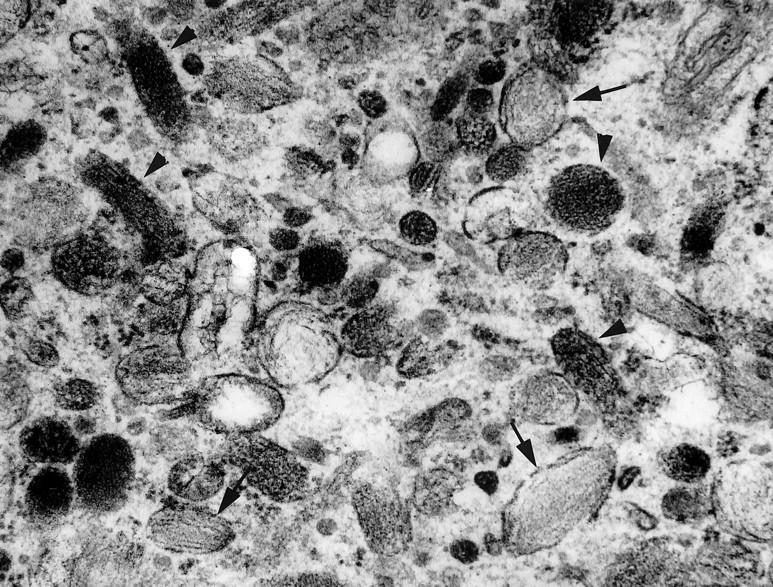
Ultrastructural appearance of case CCS 4 with few stage II melanosomes (arrows) and stage III pigmented melanosomes (arrowheads) (×29,400).
Molecular Results
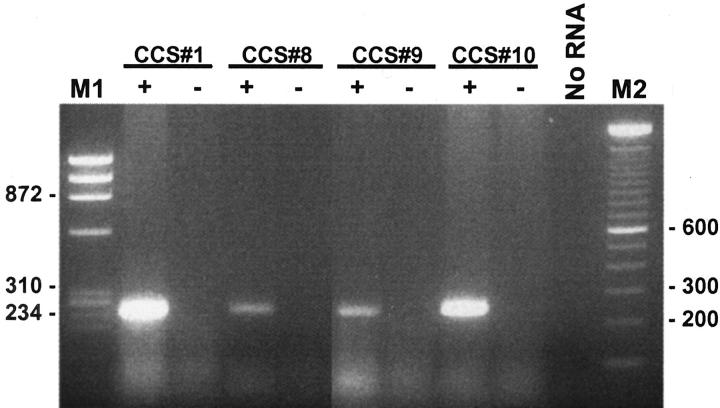
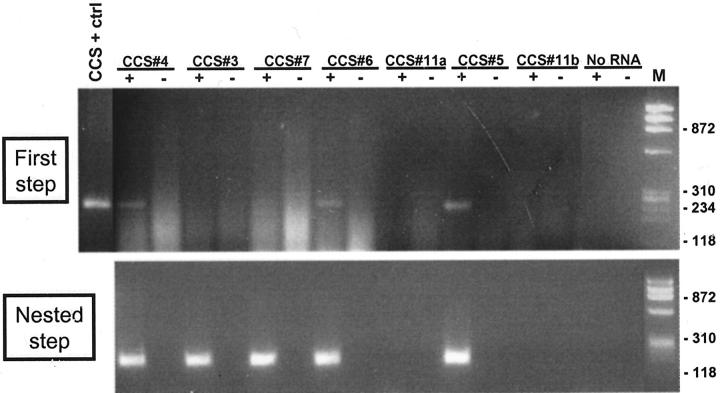
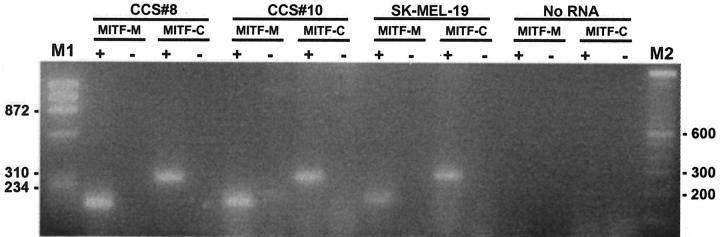
In all four cases in which RNA was extracted from frozen tissue (cases CCS 1, 8–10) a 246-bp product was identified by RT-PCR (Figure 5) using the outer pair of primers for the EWS-ATF1 type 1 or 2 fusion products (Table 1) . Of the eight cases analyzed using paraffin-extracted RNA, four showed a faint 246-bp amplified product using the outer primers (Figure 6) . After the nested PCR step using the inner primers (Table 1) , seven of eight cases showed an amplification product of 185 bp (Figure 6) . Thus, in all, 11 of 12 cases (91%) contained EWS-ATF1 fusion transcripts by RT-PCR (Table 3) . Controls lacking RT were appropriately negative in all 11 EWS-ATF1-positive cases. Paraffin block case CCS 11 was negative despite repeated assays on two RNA samples extracted from separate paraffin blocks, and despite satisfactory results with the control RT-PCR assay for PGK transcripts. Direct sequencing was performed in 11 of the 11 EWS-ATF1 positive cases and showed the same in-frame junction of exon 8 of EWS to ATF1 codon 65 in all cases. No variant fusion structure was identified. RT-PCR assays using the two pairs of primers for the MITF transcripts showed that all 4 CCS analyzed were positive with the consensus MITF primers, as well as for the melanocyte-specific MITF-M transcript (Figure 7) .
Figure 5.
Detection of EWS-ATF1 transcripts in CCS by RT-PCR using RNA extracted from frozen tissues. M1: Size marker (HaeIII digest of ΦX174); M2: 100-bp DNA ladder; ± , reverse transcriptase added/not added (the latter representing the “no RT” negative control).
Figure 6.
Detection of EWS-ATF1 transcripts in CCS by nested RT-PCR using RNA extracted from paraffin-embedded tissues. Top panel shows products of RT-PCR (first step); bottom panel shows products of nested PCR performed on products of first step. M: HaeIII digest of ΦX174. CCS + ctrl: RNA extracted from frozen tissue in CCS 1, used as positive control. CCS 11, a and b: different paraffin blocks used for RNA extraction. ±, reverse transcriptase added/not added (the latter representing the “no RT” negative control).
Figure 7.
Detection of melanocyte-specific (M) and consensus (C) MITF transcripts in CCS by RT-PCR. RNA from the SK-MEL-19 melanoma cell line provided a positive control. M1: Size marker (HaeIII digest of ΦX174); M2: 100-bp DNA ladder; ±, reverse transcriptase added/not added (the latter representing the “no RT” negative control).
Discussion
Despite an often prolonged clinical course, CCS is associated with a poor prognosis. A high incidence of metastases was identified in our EWS-ATF1 positive group; 8 of 10 (80%) patients with available clinical follow-up developed either regional lymph node or distant metastases, and seven (70%) were either alive with or dead of disease. This outcome is similar to that reported in larger clinicopathologic studies without confirmatory molecular data. 32, 33, 34
The differential diagnosis of CCS includes a variety of epithelial and mesenchymal malignancies, but CCS can be particularly difficult to distinguish from the epithelioid variant of malignant peripheral nerve sheath tumor (MPNST) or from a metastatic implant from an occult cutaneous melanoma. 35 A diagnosis of epithelioid MPNST is favored in the presence of a significant myxoid stroma and tumor cells in a cord-like arrangement. Epithelioid MPNST typically lacks HMB45 immunoreactivity or the presence of melanosomes on electron microscopic examination. Clinical presentation of a malignant melanoma as a soft tissue mass generally occurs in patients with a known history of cutaneous melanoma. Soft tissue is not a common site for metastatic melanoma, yet neither is it exceedingly rare. Histologically, malignant melanoma usually displays a much greater degree of cellular anaplasia than CCS and often shows intranuclear inclusions. However, unlike the distinction between epithelioid MPNST and CCS, the distinction between CCS and metastatic melanoma cannot be made in all problematic cases based on pathological grounds alone, and therefore may often require genetic confirmation. At the genetic level, malignant melanomas demonstrate a broad range of genetic alterations, most commonly involving chromosomes 1 and 5, as well as deletions of 6q. 36, 37 By DNA flow cytometry, most melanomas are markedly aneuploid, while most CCS are diploid or only mildly aneuploid. 38 The cytogenetic hallmark of CCS is the presence of a recurrent t(12;22)(q13;q12), resulting in the EWS-ATF1 fusion, often found as the only chromosomal abnormality. Confirming existing data in the cytogenetic literature, molecular testing shows that EWS-ATF1 is detected in almost all cases of CCS, as shown in the present study, and in no cases of cutaneous melanoma, as shown by van Roggen et al. 36 In aggregate, previous cytogenetic studies and the data of van Roggen et al 36 have established the specificity of the t(12;22)(q13;q12), and its resulting EWS-ATF1 fusion transcript, for CCS.
Despite this fundamental genetic difference, immunohistochemical and ultrastructural findings indicate that, like melanoma, CCS is a neuroectodermal tumor with melanocytic differentiation. 3, 4, 5 Similar to previous reports, we find that the majority of CCS tumors containing the EWS-ATF1 fusion also are immunoreactive for melanoma markers, such as HMB45, and contain melanosomes by ultrastructural examination. More recently, immunoreactivity for MITF has been used as a novel marker of melanocytic lineage. 7, 8, 9, 10 The MITF transcription factor is known to regulate the differentiation of melanocytes and other pigmented cells. 11 MITF immunoreactivity has been previously reported in a total of 19 out of 26 CCS (73%), 6, 7, 10 but none of these CCS were studied for EWS-ATF1. In the present study, all CCS cases tested showed diffuse and strong nuclear labeling for MITF, suggesting that the prevalence of MITF positivity may be higher in series consisting of genetically confirmed CCS.
Most immunohistochemical studies of MITF, including the present, have used the D5 and/or C5 monoclonal antibodies (Labvision, Fremont, CA). The precise epitopes bound by these antibodies are unknown but appear to lie in a portion of MITF encoded by both melanocyte-specific (MITF-M) and non-melanocyte-specific isoforms of MITF transcripts. 11, 26 Indeed, some recent immunohistochemical studies have noted reactivity of D5 antibody with a variety of non-melanocytic normal and neoplastic cells. 7, 10, 39 Therefore, we also performed RT-PCR for two portions of the MITF transcripts, a portion specific to the MITF-M isoform expressed only in melanocytes, 11 and a portion shared by all isoforms of MITF. The latter primer pair had been previously used to demonstrate by RT-PCR the ubiquitous expression of MITF in a variety of normal tissues. 10 We found that all four CCS analyzed were positive by RT-PCR with the consensus MITF primers, as expected from their immunoreactivity for MITF. Furthermore, we were able to confirm using the RT-PCR assay for the MITF-M isoform, that they expressed the melanocyte-specific form of MITF. MITF induces the expression of key melanocytic genes such as tyrosinase, TRP-1 and TRP-2, and appears sufficient to convert fibroblasts to melanocyte-like cells. 40 Hypothetically, the aberrant expression of MITF, possibly induced by EWS-ATF1-mediated transcriptional deregulation in the mesenchymal precursor cell of CCS, may be sufficient to trigger an ectopic melanocytic differentiation cascade. Indeed, there is recent evidence for direct transactivation of the MITF-M promoter by EWS-ATF1 (David E. Fisher, unpublished data).
Detection of specific fusion transcripts in sarcomas can be particularly useful not only to distinguish CCS from other morphological mimics but also to confirm the diagnosis in the setting of an unusual histology or uncommon primary site. 41 Two cases in the present study had an unusual alveolar growth pattern and rhabdoid cells with significant nuclear pleomorphism. In these cases, the detection of the EWS-ATF1 fusion transcript was useful in confirming the diagnosis.
In CCS, three hybrid transcript variants have been described so far in the literature 12, 13, 28, 30, 42, 43, 44, 45, 46 (Table 4) . The most common fusion product consists of EWS amino acid residues 1 to 325, followed by ATF1 residues 65 to 271, seen in 87% of positive cases (20 of 23) with fusion structure data. In the present study, all EWS-ATF1 positive cases had this same fusion structure, as originally described by Zucman et al, 12 which we have designated the type 1 fusion. The other two EWS-ATF1 variant fusions are rare and were previously described as single case reports. 28, 30, 44 A fusion mRNA with a junction between EWS exon 10 and ATF-1 codon 110 was identified by Speleman et al 28 and we now designate this as the type 2 fusion. The EWS exon 7–ATF1 codon 110 fusion, designated here as the type 3 fusion, was recently described in two separate case reports. 30, 44 There have been too few cases of CCS with non-type 1 EWS-ATF1 fusions to begin to comment on possible clinical or pathological correlates. However, some of the differences in the structure of the two variant fusion proteins could be functionally significant. The absence of codons 65 to 109 of ATF1 in the type 2 and type 3 fusion products would exclude the remainder of a putative activation domain in ATF1, 14 which could hypothetically affect overall transactivation by EWS-ATF1. The absence of the portion encoded by EWS exon 8 in the type 3 fusion would remove the EWS IQ domain whose phosphorylation appears to enhance EWS-ATF1 function. 47
Table 4.
Distribution of EWS-ATF1 Transcript Types Reported to Date
| Reference | Year | EWS-ATF1 fusion type* | ||||
|---|---|---|---|---|---|---|
| Type 1 | Type 2 | Type 3 | Fusion structure not specified | No fusion detected | ||
| 12 | 1993 | 3 | ||||
| 28 | 1997 | 1 | ||||
| 30 | 1998 | 1 | ||||
| 44 | 1999 | 1 | ||||
| 45 | 1999 | 1 | ||||
| 46 | 2000 | 1 | ||||
| 43 | 2000 | 1 | ||||
| 42 | 2001 | 2 | 2† | |||
| 12, 13, 45 | CCS cell lines | SU-CCS-1, DTC1 | HS-MM | |||
| Present study | 2002 | 11 | 1 | |||
| Totals | N = 28 | 20 | 1 | 2 | 2 | 3 |
Type 1, EWS ex.8 (codon 325)–ATF1 codon 65; Type 2, EWS ex.10 (codon 349)–ATF1 codon 110; Type 3, EWS ex.7 (codon 265)–ATF1 codon 110.
Assay was not designed to detect type 3 fusion.
RT-PCR assays in one CCS sample in the present series (CCS 10) were negative for EWS-ATF1. This tumor showed a typical histological appearance, was positive for MITF1 and HMB45, but negative for S-100, and showed melanosomes by electron microscopy. Although we cannot entirely rule out a soft tissue metastasis of a cutaneous melanoma in this case, the lack of a clinical history of cutaneous melanoma and the histological appearance favored the diagnosis of CCS. The interpretation of this lone negative EWS-ATF1 RT-PCR result is complicated by the possibility that the limited quality of the RNA extracted from the archival paraffin-embedded material in this case may have been insufficient to detect a novel molecular variant of EWS-ATF1 producing a larger than expected fusion product. Another possibility is that this case contains a variant translocation, replacing one of the two usual translocation partners by a related gene, as seen in several other sarcomas. 41 Finally, it is also possible that rare CCS have a different molecular pathogenesis.
In conclusion, the present study shows that the EWS-ATF1 gene fusion is found in the majority of CCS cases (91%) and thus provides a sensitive molecular diagnostic marker for CCS. Conventional RT-PCR methodology can be applied with good results for EWS-ATF1 detection using RNA extracted from frozen tissues. When using archival material nested RT-PCR can yield similar results, in the presence of adequate RNA. The expression of the melanocyte-specific MITF-M transcript is further evidence of the bona fide melanocytic differentiation of CCS.
Acknowledgments
We thank Elizabeth Weiss for technical assistance with the ultrastructural analysis.
Address reprint requests to Marc Ladanyi, M.D., Department of Pathology, Memorial Sloan-Kettering Cancer Center, New York, NY 10021. E-mail: ladanyim@mskcc.org.
Footnotes
Supported by PO1 CA47179 to M.F.B.
References
- 1.Enzinger FM: Clear cell sarcoma of tendons and aponeuroses: an analysis of 21 cases. Cancer 1965, 18:1163-1174 [DOI] [PubMed] [Google Scholar]
- 2.Chung EB, Enzinger FM: Malignant melanoma of soft parts: a reassessment of clear cell sarcoma. Am J Surg Pathol 1983, 7:403-413 [DOI] [PubMed] [Google Scholar]
- 3.Kindblom L-G, Lodding O, Angervall L: Clear cell sarcoma of tendons and aponeuroses: an immunohistochemical and electron microscopic analysis indicating neural crest origin. Virchows Arch 1983, 401:109-128 [DOI] [PubMed] [Google Scholar]
- 4.Benson JD, Kraemer B, Mackay B: Malignant melanoma of soft parts: an ultrastructural study of four cases. Ultrastruct Pathol 1985, 8:57-70 [DOI] [PubMed] [Google Scholar]
- 5.Hasegawa T, Hirose T, Kudo E, Hizawa K: Clear cell sarcoma: an immunohistochemical and ultrastructural study. Acta Pathol Jpn 1989, 39:321-327 [DOI] [PubMed] [Google Scholar]
- 6.Granter SR, Weilbaecher KN, Quigley C, Fletcher CDM, Fisher DE: Clear cell sarcoma shows immunoreactivity for microphthalmia transcription factor: further evidence for melanocytic differentiation. Mod Pathol 2001, 14:6-9 [DOI] [PubMed] [Google Scholar]
- 7.Koch MB, Shih IM, Weiss SW, Folpe AL: Microphthalmia transcription factor and melanoma cell adhesion molecule expression distinguish desmoplastic/spindle cell melanoma from morphologic mimics. Am J Surg Pathol 2001, 25:58-64 [DOI] [PubMed] [Google Scholar]
- 8.King R, Googe PB, Weilbaecher KN, Mihm Jr MC, Fisher DE: Microphthalmia transcription factor expression in cutaneous benign, malignant melanocytic, and nonmelanocytic tumors. Am J Surg Pathol 2001, 25:51–57 [DOI] [PubMed]
- 9.Miettinen M, Fernandez M, Franssila K, Gatalica Z, Lasota J, Sarlomo-Rikala M: Microphthalmia transcription factor in the immunohistochemical diagnosis of metastatic melanoma: comparison with four other melanoma markers. Am J Surg Pathol 2001, 25:205-211 [DOI] [PubMed] [Google Scholar]
- 10.Busam KJ, Iversen K, Coplan KC, Jungbluth AA: Analysis of microphthalmia transcription factor expression in normal tissues and tumors, and comparison of its expression with S-100 protein, gp100, and tyrosinase in desmoplastic malignant melanoma. Am J Surg Pathol 2001, 25:197-204 [DOI] [PubMed] [Google Scholar]
- 11.Tachibana M: MITF: a stream flowing for pigment cells. Pigment Cell Res 2000, 13:230-240 [DOI] [PubMed] [Google Scholar]
- 12.Zucman J, Delattre O, Desmase C, Epstein AL, Stenman G, Speleman F, Fletcher CDM, Aurias A, Thomas G: EWS and ATF-1 gene fusion induced by t(12;22) in malignant melanoma of soft parts. Nat Genet 1993, 4:341-345 [DOI] [PubMed] [Google Scholar]
- 13.Brown AD, Lopez-Terrada D, Denny CT, Lee KAW: Promoters containing ATF-binding sites are deregulated in cells that express the EWS/ATF1 oncogene. Oncogene 1995, 10:1749-1756 [PubMed] [Google Scholar]
- 14.Fujimura Y, Ohno T, Siddique H, Lee L, Rao VN, Reddy ESP: The EWS-ATF-1 gene involved in malignant melanoma of soft parts with t(12;22) chromosome translocation, encodes a constitutive transcriptional activator. Oncogene 1996, 12:159-167 [PubMed] [Google Scholar]
- 15.Feng L, Lee KA: A repetitive element containing a critical tyrosine residue is required for transcriptional activation by the EWS/ATF1 oncogene. Oncogene 2001, 20:4161-4168 [DOI] [PubMed] [Google Scholar]
- 16.Li KKC, Lee KAW: MMSP tumor cells expressing the EWS-ATF1 oncogene do not support cAMP-inducible transcription. Oncogene 1996, 16:1325-1331 [DOI] [PubMed] [Google Scholar]
- 17.Bosilevac JM, Olsen RJ, Bridge JA, Hinrichs SH: Tumor cell viability in clear cell sarcoma requires DNA binding activity of the EWS/ATF1 fusion protein. J Biol Chem 1999, 274:34811-34818 [DOI] [PubMed] [Google Scholar]
- 18.Fujimura Y, Siddique H, Lee L, Rao VN, Reddy ESP: EWS-ATF-1 chimeric protein in soft tissue clear cell sarcoma associates with CREB-binding protein and interferes with p53-mediated trans-activation function. Oncogene 2001, 20:6653-6659 [DOI] [PubMed] [Google Scholar]
- 19.Bridge JA, Borek DA, Neff JR, Huntrakoon M: Chromosomal abnormalities in clear cell sarcoma: implications for histogenesis. Am J Clin Pathol 1990, 93:26-31 [DOI] [PubMed] [Google Scholar]
- 20.Bridge JA, Sreekantaiah C, Neff JR, Sandberg AA: Cytogenetic findings in clear cell sarcoma of tendons and aponeuroses: malignant melanoma of soft parts. Cancer Genet Cytogenet 1991, 52:101-106 [DOI] [PubMed] [Google Scholar]
- 21.Speleman F, Colpaert C, Goovaerts G, Leroy JG, Van Mark E: Malignant melanoma of soft parts: further cytogenetic characterization. Cancer Genet Cytogenet 1992, 60:176-179 [DOI] [PubMed] [Google Scholar]
- 22.Stenman G, Kindblom L-G, Angervall L: Reciprocal translocation t(12;22)(q13;q13) in clear cell sarcoma of tendons and aponeuroses. Genes Chromosomes Cancer 1992, 4:122-127 [DOI] [PubMed] [Google Scholar]
- 23.Fletcher JA: Translocation t(12;22)(q13–14;q12) is a nonrandom aberration in soft tissue clear cell sarcoma. Genes Chromosomes Cancer 1992, 184 [DOI] [PubMed]
- 24.Peulve P, Michot C, Vannier JP, Tron P, Hemet J: Clear cell sarcoma with t(12;22)(q13–14;q12). Genes Chromosomes Cancer 1991, 3:400-402 [DOI] [PubMed] [Google Scholar]
- 25.Travis JA, Bridge J: Significance of both numerical and structural chromosomal abnormalities in clear cell sarcoma. Cancer Genet Cytogenet 1992, 64:104-106 [DOI] [PubMed] [Google Scholar]
- 26.Hemesath TJ, Price ER, Takemoto C, Badalian T, Fisher DE: MAP kinase links the transcription factor microphthalmia to c-kit signalling in melanocytes. Nature 1998, 391:298-301 [DOI] [PubMed] [Google Scholar]
- 27.Argani P, Perez-Ordonez B, Xiao H, Caruana SM, Huvos AG, Ladanyi M: Olfactory neuroblastoma is not related to the Ewing family of tumors: absence of EWS/FLI1 gene fusion and MIC2 expression. Am J Surg Pathol 1998, 22:391-398 [DOI] [PubMed] [Google Scholar]
- 28.Speleman F, Delattre O, Peter M, Hauben E, Van Roy N, Van Marck E: Malignant melanoma of soft parts (clear cell sarcoma): confirmation of EWS and ATF1 gene fusion caused by a t(12;22) translocation. Mod Pathol 1997, 10:496-499 [PubMed] [Google Scholar]
- 29.Delattre O, Zucman J, Plougastel B, Desmaze C, Melot T, Peter M, Kovar H, Joubert I, de Jong P, Rouleau G: Gene fusion with an ETS DNA-binding domain caused by chromosome translocation in human tumors. Nature 1992, 359:162-165 [DOI] [PubMed] [Google Scholar]
- 30.Pellin A, Monteagudo C, Lopez-Gines C, Carda C, Boix J, Llombart-Bosch A: New type of chimeric fusion product between the EWS and ATF1 genes in clear cell sarcoma (malignant melanoma of soft parts). Genes Chromosomes Cancer 1998, 23:358-360 [PubMed] [Google Scholar]
- 31.Udono T, Yasumoto K, Takeda K, Amae S, Watanabe K, Saito H, Fuse N, Tachibana M, Takahashi K, Tamai M, Shibahara S: Structural organization of the human microphthalmia-associated transcription factor gene containing four alternative promoters. Biochim Biophys Acta 2000, 1491:205-219 [DOI] [PubMed] [Google Scholar]
- 32.Montgomery EA, Meis JM, Ramos AG, Frisman DM, Martz KL: Clear cell sarcoma of tendons and aponeuroses: a clinicopathologic study of 58 cases with analysis of prognostic markers. Int J Surg Pathol 1993, 1:89-100 [Google Scholar]
- 33.Lucas DR, Nascimento AG, Sim FH: Clear cell sarcoma of soft tissues: Mayo Clinic experience with 35 cases. Am J Surg Pathol 1992, 16:1197-1204 [DOI] [PubMed] [Google Scholar]
- 34.Sara AS, Evans HL, Benjamin RS: Malignant melanoma (clear cell sarcoma): a study of 17 cases, with emphasis on prognostic factors. Cancer 1990, 65:367-374 [DOI] [PubMed] [Google Scholar]
- 35.D’Amore ESG, Ninfo V: Clear cell tumors of the somatic soft tissues. Semin Diagn Pathol 1997, 14:270-280 [PubMed] [Google Scholar]
- 36.van Roggen JFG, Mooi WJ, Hogendoorn PCW: Clear cell sarcoma of tendons and aponeuroses (malignant melanoma of soft parts) and cutaneous melanoma: exploring the histogenetic relationship between these two clinicopathologic entities. J Pathol 1998, 186:3-7 [DOI] [PubMed] [Google Scholar]
- 37.Muir PD, Gunz FW: A cytogenetic study of 8 human melanoma cell lines. Pathology 1979, 11:597-606 [DOI] [PubMed] [Google Scholar]
- 38.El-Naggar AK, Ordonez NG, Sara A, McLemore D, Batsakis JG: Clear cell sarcomas and metastatic soft tissue melanomas: a flow cytometric comparison and prognostic implications. Cancer 1991, 67:2173-2179 [DOI] [PubMed] [Google Scholar]
- 39.Granter SR, Weilbaecher KN, Quigley C, Fletcher CD, Fisher DE: Microphthalmia transcription factor: not a sensitive or specific marker for the diagnosis of desmoplastic melanoma and spindle cell (non-desmoplastic) melanoma. Am J Dermatopathol 2001, 23:185-189 [DOI] [PubMed] [Google Scholar]
- 40.Tachibana M, Takeda K, Nobukuni Y, Urabe K, Long JE, Meyers KA, Aaronson SA, Miki T: Ectopic expression of MITF, a gene for Waardenburg syndrome type 2, converts fibroblasts to cells with melanocyte characteristics. Nat Genet 1996, 1914:50-54 [DOI] [PubMed] [Google Scholar]
- 41.Ladanyi M, Bridge JA: Contribution of molecular genetic data to the classification of sarcomas. Hum Pathol 2000, 31:532-538 [DOI] [PubMed] [Google Scholar]
- 42.Langezaal SM, van Roggen JFG, Cleton-Jansen AM, Baelde JJ, Hogendoorn PC: Malignant melanoma is genetically distinct from clear cell sarcoma of tendons and aponeurosis (malignant melanoma of soft parts). Br J Cancer 2001, 84:535-538 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Fukuda T, Kakihara T, Baba K, Yamaki T, Yamaguchi T, Suzuki T: Clear cell sarcoma arising in the transverse colon. Pathol Int 2000, 50:412-416 [DOI] [PubMed] [Google Scholar]
- 44.Ohba Y, Suzuki H, Hiraga H, Ito T, Sawa H, Nagai M, Satoh SI, Iwaki H, Nagashima K: Melanotic peritoneal sarcomatosis originating from clear cell sarcoma. Pathol Int 1999, 49:653-657 [DOI] [PubMed] [Google Scholar]
- 45.Sonobe H, Takeuchi T, Taguchi T, Shimizu K, Iwata J, Furihata M, Ohtsuki Y: Further characterization of the human clear cell sarcoma cell line HS-MM demonstrating a specific t(12;22)(q13;q12) translocation and hybrid EWS-ATF1 transcript. J Pathol 1999, 187:594-597 [DOI] [PubMed] [Google Scholar]
- 46.Naito N, Kawai A, Ouchida M, Dan’ura T, Morimoto Y, Ozaki T, Shimizu K, Inoue H: A reverse transcriptase-polymerase chain reaction assay in the diagnosis of soft tissue sarcomas. Cancer 2000, 89:1992-1998 [DOI] [PubMed] [Google Scholar]
- 47.Olsen RJ, Hinrichs SH: Phosphorylation of the EWS IQ domain regulates transcriptional activity of the EWS-ATF1 and EWS-FLI1 fusion proteins. Oncogene 2001, 20:1756-1764 [DOI] [PubMed] [Google Scholar]