Abstract
We describe the use of fluorescent-labeled primers to analyze T-cell receptor γ gene rearrangements (TCRγGR) using capillary electrophoresis in the ABI Prism 310 Genetic Analyzer. We also compare the performance with denaturing gradient gel electrophoresis (DGGE). In a single multiplex polymerase chain reaction (PCR) we amplified TCRγGR with primers for all known groups of variable region genes, and joining region genes described in lymphoid neoplasms. Ten reactive samples, followed by five cell lines and 25 tumor samples with 41 individual TCRγGR (due to many biallelic rearrangements) previously identified by DGGE, were analyzed to validate the technique. The capillary electrophoresis protocol has 92% concordance for both TCR clonal status (23 of 25) and 95% concordance in the number of individual TCRγGR (38 of 41) identified by DGGE. The reproducible sensitivity for detecting TCRγGR diluted in reactive lymphoid DNA is 2% in clinical applications. Discrimination of predominant rearrangements requires a minimum ratio of two times the height of the normal distribution of polyclonal peaks. Capillary electrophoresis can provide results within 60 minutes for each specimen after PCR is complete. Capillary electrophoresis provides a faster result than sequence-based separation methods and gives an archival electronic record. Fluorescent labeling allows the identification of both the variable and joining gene segments used in a TCRγGR. The effectiveness of capillary electrophoresis is similar to DGGE.
TCRγ Detection Methods
There have been various polymerase chain reaction (PCR)-based methods described during the past decade for the analysis of T-cell receptor γ gene rearrangements (TCRγGR). These have included agarose gel electrophoresis, 1 polyacrylamide gel electrophoresis, 2 denaturing gradient gel electrophoresis (DGGE), 3, 4, 5, 6 single-stranded conformation polymorphism electrophoresis (SSCP), 7, 8 heteroduplex analysis in mutation detection enhancing gels, 9, 10 temperature gradient gel electrophoresis, 11, 12 fluorescent analysis in polyacrylamide gels, 13, 14, 15, 16, 17 and most recently via capillary electrophoresis (CE). 18, 19, 20, 21, 22, 23, 24, 25 These methods can be divided into two broad groups: those which separate rearrangements based on length only and those which maximize separation of TCRγGR based on the effect of specific sequence conformations of the DNA during electrophoresis. We describe a capillary electrophoresis protocol which can identify both variable and joining gene segments in TCRγGR amplified by PCR, and compare the results with DGGE.
T-Cell Receptor γ Gene
There are 11 variable region genes, divided among four groups, and five joining region genes organized in three groups that rearrange in T cells, (eg, V3-J2) and have been described in lymphoid tumors. 26, 27, 28, 29 (Figure 1) The functional segments include V2–5,8,9,11, and the details of the genomic structure have been well described. There is no diversity region between the variable and joining gene segments. This simplicity in TCRγ has resulted in the development of multiple PCR methods to amplify all TCRγGRs.
Figure 1.
Schematic of gene segments in the T-cell receptor γ gene. V, Variable region; J, Joining region; GP, Group; C1, Constant region.
Experience with DGGE
We have described and used a high resolution GC-clamped technique of DGGE in our clinical and research laboratories with excellent results over the past six years. 5 In addition, we have explored the utilization of fluorescent-labeled primers in the variable region 13 for detecting TCRγGR using Gene Scan software on the gel-based method of the ABI 373 and similarly reported by Simon. 14 However, use of the ABI 373 gel-based instrument provided no time savings compared to DGGE.
Goal of the Capillary Electrophoresis Assay
Currently in the Nebraska Health System’s Molecular Diagnostics Laboratory we use the ABI Prism 310 Genetic Analyzer for short tandem repeat analysis in forensic, paternity testing, and bone marrow engraftment studies. We hypothesized that capillary electrophoresis could be used effectively to detect TCRγGR compared to DGGE. Our goal was to develop a fluorescent-labeled assay on the capillary electrophoresis system with faster throughput, and that eliminated the need to pour the complex DGGE gels, which take hours to assemble and a full day (6.5 hours) for electrophoresis and analysis.
Materials and Methods
TCRγGR Samples Studied
Ten polyclonal samples (nine reactive frozen tissue samples and one specimen of peripheral blood lymphocytes) with no monoclonal TCRγ GR were used to establish the polyclonal pattern in normal samples. Five cell lines (with biallelic TCRγ previously identified by Southern blot analysis), and 25 selected patient samples with monoclonal TCRγ gene rearrangements identified by DGGE were used to validate the capillary electrophoresis protocol (Table 1) . The 25 samples include 11 cases of peripheral T-cell lymphoma, three cases of mycosis fungoides, three cases of atypical lymphoid hyperplasia, one case of anaplastic large cell lymphoma, two cases of acute lymphoblastic leukemia, one case of acute undifferentiated leukemia, one case of large granular lymphocytosis, one case of mixed cellularity Hodgkin’s disease, and two cases of B-cell lymphoma. Fifteen cases were amplified from frozen tissue, three cases from blood/bone marrow, and seven cases from paraffin-embedded tissue.
Table 1.
Comparison of TCRγGR Results in Cell Lines and Tumor Specimens by Two Methods
| A: Cell lines | DGGE | CE | Variable gene segment | Joining gene segment | ||
|---|---|---|---|---|---|---|
| Status | No. RR | Status | No. RR | |||
| CEM | pos | 2 | pos | 2 | Vγ3, Vγ4 | 2-Jγ1/Jγ2 |
| HSB2 | pos | 2 | pos | 2 | Vγ9, Vγ10 | 2-Jγ1/Jγ2 |
| 8402 | pos | 2 | pos | 2 | Vγ4, Vγ10 | 2-Jγ1/Jγ2 |
| MOLT4 | pos | 2 | pos | 2 | Vγ2, Vγ10 | 2-JγP1/JγP2 |
| Jurkat | pos | 2 | pos | 2 | Vγ8, Vγ11 | 2-Jγ1/Jγ2 |
| Cell line totals B: Clinical specimens | 5/5 | 10/10 | 5/5 | 10/10 | ||
| Selected TCRγGR positive cases for validation set | 25/25 | 31/31 | 23/25 | 28/31 | ||
| Total: All TCRγGR | 30/30 | 41/41 | 28/30 | 38/41 | ||
CE, capillary electrophoresis; DGGE, denaturing gradient gel electrophoresis; No., number; RR, individual rearrangements per case may be 1 or more; pos, positive, Status, clonal for TCRγGR.
TCRγGR PCR
The designed average target size of PCR products of all TCRγGR is 190 nucleotides (nt) with a normal distribution of TCRγGR occurring between 165 and 215 nt. Each variable primer is located about 140 nt from the junction and each joining primer is located about 50 nt from the junction, which produces a tight normal distribution of product sizes. Two primers were used for the group 1 (Vγ2–8) variable genes (Table 2) ; one for Vγ2,4–8 and in one for Vγ3, which varies in sequence from Vγ2 at the target site of the primers for Group 1. The addition of a Vγ3 specific primer to the multiplex reaction resulting in successful amplification of sequence proven Vγ3 TCRγGR in CEM and case 1.38 (Table 3) . A single primer was used for each of group 2 (Vγ9), group 3 (Vγ10), and group 4 (Vγ11) variable regions (Table 2) . For the joining region genes, a fluorescent-labeled primer was used for each of the three groups Jγ1/Jγ2, JγP, JγP1/JγP2 using sequences previously described by our group. 5, 13 All eight primers were combined in one tube in a multiplex PCR reaction containing labeled joining primers (Table 2) . An optional tube can be used with labeled variable primers and unlabeled joining primers. The PCR mixture included 10 mmol/L Tris HC1, 1.5 mmol/L MgC12, gelatin, 1.25 units Taq polymerase (Promega Corp., Madison, WI), 100 μmol/L dNTPs, and 0.6 μmol/L of each primer. Samples were heated initially to 94°C for 10 minutes, followed by 30 cycles at 94°C for 75 seconds, 60°C for 75 seconds, 72°C for 10 seconds, plus a 1-second extension each cycle. A terminal extension at 60°C for 45 minutes was performed. Cell lines with known TCRγ GR are used as controls (Table 1) . For JγP, Beaubier et al 20 have reported that the cell line SUP-B15 contains the JγP segment in the TCRγGR, however, this cell line was not available in our institution, therefore a T-cell lymphoma was used to confirm the amplification of the Jγ PGR.
Table 2.
Primers for TCRγGR
| Primer | Jγ-labeled PCR | Vγ-labeled PCR | Sequence 5′-3′ | Reference |
|---|---|---|---|---|
| Vγ2* | NED | ACTCCAGGGTTGTGTTGGGAATCA | 27, 28 | |
| Vγ3 | NED | CCGCAAGGGATGTGTTGGAATCA | 27, 28 | |
| Vγ9 | JOE | ACGGCACTGTCAGAAAGGAATC | 27, 28 | |
| Vγ10 | ROX | AATCCGCAGCTCGACGCAGCA | 26, 29 | |
| Vγ11 | FAM | GGCTCAAGATTGCTCAGGTGG | 29 | |
| Jγ1/Jγ2 | NED | TAC CTG TGA CAA CAA GTG TTG TTC | 5 | |
| JγP | FAM | AAG CTT TGT TCC GGG ACC AAA TAC | 5 | |
| JγP1/JγP2 | JOE | GAA GTT ACT ATG AGC T/CTA GTC CCT T | 5 |
FAM, JOE and NED are fluorochromes attached to the 5′ end of the Jγ primers which were established in a previous DGGE protocol. 5 The Vγ primers were newly designed for this assay from published TCRγ variable gene sequences.
, Vγ2 also has sequence homology with gene segments Vγ4-8.
Table 3.
Sequenced TCRγGR Analyzed by Capillary Electrophoresis.
| DNA | Size | V-gene | Vγ* | N† | Jγ‡ | J-gene |
|---|---|---|---|---|---|---|
| 4.93 | 190 bp | Vγ4 | TGGGATG | CTTCC | TTATTATAAGAA | Jγ1/Jγ2 |
| CEM | 185 bp | Vγ3 | TGGGACAG | TCCC | ATAAGAAACT | Jγ1/Jγ2 |
| 1.38 | 185 bp | Vγ3 | TGGGACAGG | CGGATGAG | TTTATTTATAAGAA | Jγ1/Jγ2 |
, Vγ sequence at 3′ prime end of variable segment near the junction;
, nucleotides inserted in the junction;
, Jγ sequence at 5′ end of joining segment.
The use of a Vγ3 specific primer successfully amplified the TCRγGR in CEM and Case 1.38.
DNA Extraction
DNA was extracted from fresh/frozen tissue with proteinase K digestion, followed by phenol/chloroform purification as previously described. 30 Paraffin-embedded tissue was cleared of paraffin by xylene followed by 100% ethanol rinse followed by overnight digestion in proteinase K (0.5 μg/ml) to form a tissue lysate.
Analysis on the ABI Prism System
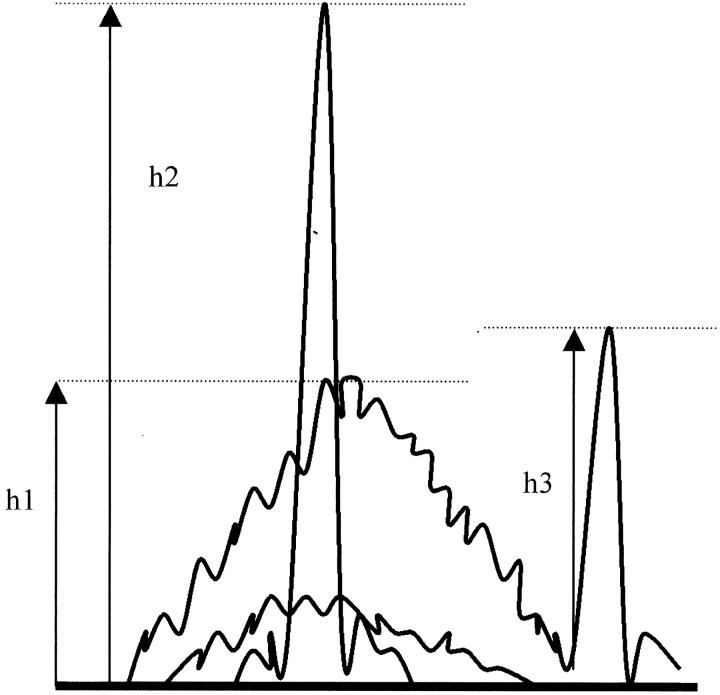
The quality and quantity of amplified PCR product obtained was estimated from a 2% 3:1 agarose gel (Sigma, St. Louis, MO) to determine the dilution factor for the product before analysis by capillary electrophoresis on a ABI Prism 310 Genetic analyzer (Perkin Elmer Applied Biosystems, Foster City, CA). Performance optimized polymer-4 (POP-4) was used as a separation matrix in the capillary. One to 2 μl from a 100 μl PCR reaction was electrokinetically injected at 15,000 volts for 3 to 5 seconds. Electrophoresis run conditions were 15,000 volts at 60°C for 24 minutes. The internal size standard GS350 labeled with Rox gives reference peaks at 75, 100, 139, 150, 160, 200, 300, and 350 nt. The three dyes used for joining region primer labeling were Fam (JγP-blue), Joe (JγP1/Jγ P2-green), and Ned (Jγ1/Jγ2-black). The first injection run of a fluorescent-labeled Rox size standard was used only to optimize the equipment before collection of data in subsequent runs. Each specimen takes approximately 30 minutes for all steps on the CE. The data were stored electronically and the products were analyzed automatically using Gene Scan software (Perkin Elmer Applied Biosystems, Foster City, CA). The intra-run precision SD for our ABI Prism 310 Genetic analyzer is <0.15 bases from size ranges of 103 to 316 bases. A positive TCRγGR was identified when a rearranged clone produced a peak height (h2) that was greater than two times the height of the highest polyclonal background (h1) using the formula: ratio of peak to background (RPB) = h2/h1 (Figure 2) . Polyclonal populations give a normal distribution spanning 190 nucleotides with a range from 165 to 215 (Figure 3A) . The presence of three colored lines in the electropherogram help ensure that all Jγ primers are amplifying TCRγGR. The dyes used for optional variable region labeling were Ned (Vγ2–8 and Vγ3-black), Joe (Vγ9-green), Rox (Vγ10-red), and Fam (Vγ11-blue). To analyze the Vγ-labeled products, two injections of the PCR product were performed as described by Vega and colleagues. 23 One injection had the Rox size standard added and one injection was performed without the Rox size standard. While the injection with Rox allows size determination of the peaks, the injection without Rox allows accurate evaluation of peaks without interference by the 200 nt standard.
Figure 2.
The height (h2) of the peak must exceed the height (h1) of the polyclonal background by a ratio of more than 2.0 (RPB = h2/h1) to define a clonal result. Evaluation of bialellic TCRγGR in cell lines where one peak (h3) is outside the normal distribution of polyclonal T cells suggests that the ratio of such peaks (h3) are better assessed by adding h3 to h1 to calculate h2. Otherwise one peak will be called positive and one peak will be called negative.
Figure 3.
A: Polyclonal TCRγ gene rearrangements are demonstrated by the normal distribution pattern obtained from peripheral blood lymphocytes. Labeling of the TCRγGRs is as follows: the common joining region genes Jγ1/Jγ2-black, JγP-blue, and JγP1/JγP2-green. B: Molt4 cell line with biallelic Vγ2-JγP1/JγP2, Vγ10-JγP1/JγP2 TCRγGR. Molt4 DNA was diluted 50:50 in peripheral blood lymphocyte DNA before PCR. The green peaks identify that the JγP1/JγP2 joining segment is used in both TCRγGR. GR = gene rearrangement. Peak height of a clonal peak should be a minimum of two times the height of the polyclonal background. C: Fluorescent analysis of dual TCRγGR (Vγ2-JγP and Vγ9-JγP1/JγP2) in a peripheral T-cell lymphoma, demonstrating the capability of identifying different joining region genes used in each rearrangement. JγP-blue; JγP1/JγP2-green. D: Sensitivity study by serial dilution of TCRγGR (Vγ3-Jγ1/Jγ2; Vγ4-Jγ1/Jγ2) CEM in peripheral blood lymphocytes. A detection sensitivity of at least 0.5% was obtained with the CEM cell line DNA serially diluted in peripheral blood lymphocyte DNA. Below 0.5%, the ratio (RPB) of the peak height of the TCRγGRs will fall below two times of the polyclonal background. E: Discriminating a clonal peak (right arrow) from the background polyclonal T-cells in this case is difficult. Neither of the two peak heights (left arrows) is greater than two times the maximum height (at 1200) of the main polyclonal background. This illustrates the potential problem that can occur with identifying the significance of oligoclonal peaks. F: Polyclonal TCRγ gene rearrangements are demonstrated in peripheral blood lymphocytes using variable region primers labeled as follows: the common variable region genes of Vγ2–8 and Vγ3-black. The lesser common genes are Vγ9-green; Vγ10-red; Vγ11-blue. G: Fluorescent analysis identifies the variable gene used in dual TCRγGR (Vγ2 and Vγ10). in a peripheral T-cell lymphoma., Vγ2-black; Vγ10-red. x axis, nucleotide length; y axis, intensity of signal; Rox-labeled size standard at 200 nt.
Results
All 10 reactive samples were demonstrated to be polyclonal by both DGGE and CE. All known TCGγGR in the cell lines were identified by both methods. There was 100% correlation between CE and DGGE for the number and identification of known joining segments used in the TCRγGR of the five cell lines (Table 1) . The analysis of cell lines established the capability of the CE protocol to identify rearrangements with each group of the variable region gene segments (Groups 1–4), and the joining region segments, except for JγP, which is not present in the cell lines tested. For JγP, tumor samples with JγP rearrangements were included in the validation set. Rearrangements with JγP1/JγP2 region genes are illustrated in Figures 3B and 3C . Figure 3C also shows a TCRγGR with JγP. Figure 3D illustrates the presence of biallelic Jγ1/Jγ2 TCRγGR. Figure 3E shows the polyclonal results with the labeled variable region primers and Figure 3F demonstrates the capability of Vγ gene segment identification.
Sensitivity
Twenty-three of 25 (92%) of the cases in the validation set were found to have clonal TCRγGR by capillary electrophoresis compared to the gold standard of DGGE. In two cases, the rearrangement seen in DGGE was not identified with CE due to the greater quantitative sensitivity (0.1%) of DGGE in our lab. A cell line or tumor may have two (biallelic) TCRγGR in up to 75% of cases. 31 Our DGGE analysis has previously shown that 64% (16/25) of cases had biallelic TCRγGR. Thirty-eight of 41 individual TCRγGR were identified by CE for a sensitivity of 95% in TCRγGR identification. Two gene rearrangements did not produce a peak high enough to meet our criteria for CE. In another case, two gene rearrangements were identified as one in CE since these were the same length.
The quantitative detection sensitivity for capillary electrophoresis may reach 0.5% under carefully controlled conditions in our research laboratory (Figure 3D) , however 2% is obtained in repeated clinical assays during daily use. This reproducible level of sensitivity is sufficient for routine clinical screening of rearrangements as neoplastic cell populations below 2% are somewhat difficult to identify by morphology. The use of the patient’s tumor-specific-labeled variable segment primer and the labeled joining primer allows identification of follow-up tumor involvement to 0.5%.
The quantitative detection sensitivity of 0.1% for DGGE is the result of the greater ability of DGGE to widely separate sequences which vary by single nucleotides. Patient-specific primers targeted at the junctional sequence of the tumor would be needed for minimal residual disease detection at the 1 × 105 to 106 level.
Discussion
The primers previously described in the DGGE protocol will effectively work in a multiplex reaction in a single tube. 5 However, the DGGE primers targeted to the variable region genes resulted in amplifications of different length products, ranging from 140 nt (Vγ11) to 260 nt (Vγ1–8). We choose not to adopt the variable region primers from the DGGE protocol for this length-based CE protocol because the distribution of PCR products would be scattered at four different regions in the electropherogram resulting in difficulties in interpretation. Secondly, the 260 nt products for Group 1 is near the threshold of effective amplification from paraffin-embedded tissues. A higher degree of success is obtained when amplifying targets that are less than 200 nt in most assays on paraffin-embedded tissues. Therefore, we designed the primers to produce approximately 190 nt products.
Basis of Method
In capillary electrophoresis of fluorescent-labeled products, high resolution is achieved in separating TCRγGR that differ in length by single nucleotides. Oda et al 18 first evaluated the use of fluorescent detection of TCRγGR with a limited number of Tγ primers using a Beckman capillary electrophoresis instrument. They described similar findings for identifying polyclonal and clonal populations with a YO-PRO-1 dye intercalated in double-stranded DNA. 18
For our comparative study with DGGE, we chose to label the 5′ end of the joining region primers with individual fluorescent dyes similar to previous descriptions. 15, 20, 21, 24, 32 Fluorescent labels have also been described on the 5′ end of the variable region primer 13, 19, 22, 23, 25 . By doing so, the CE protocol allows the identification of both gene segments used in a TCRγGR. In the polyacrylamide gel-based protocols, fluorescent-labeled products were detected with a laser scanning system. 15, 16 Gene Scan software was used to analyze the peak size and intensity in relative fluorescent units.
Incomplete primer sets were reported in many TCRγ protocols using fluorescent gels or CE. 15, 17, 18, 19, 20, 22, 32 Beaubier 20 acknowledged that a comprehensive set of primers is needed to detect all TCRγGR identified by Southern blot analysis, and is supported by the recent multi-institutional study covering multiple TCRγGR methods by Arber et al. 33 Our CE protocol includes a comprehensive set of primers and was validated with cell lines with known TCRγGR in a manner similar to our DGGE protocol.
A second PCR tube can also be set up with the variable region primers labeled with fluorochromes and non-labeled joining region primers as described in our optional set of primers (Table 2) . Since our assay is performed in duplicate in the clinical lab, as with most gene rearrangement assays, there is no added cost to have one tube labeled with joining primers and one with variable primers. The identification of both the variable region and joining region genes in a two-tube reaction is useful for research applications or for better identification of the clone in follow-up biopsies. However, for ease of interpretation, laboratories may find that a single color of fluorochrome (eg, Fam) on each of the joining region primers, or on each of the variable region primers, would perform satisfactorily.
Defining Criteria for a Clonal Population by Ratio
We recommend that a minimum 2.0 ratio (RPB) between the peak height of the single or biallelic peak and the highest normal distribution of the background polyclonal T-cells is defined as a clonal result (Figure 2) as previously suggested. 32 The second criteria is the peak(s) must be reproducible in duplicate analyses at the same nucleotide size to avoid false positive results. 17 When only a normal distribution is identified or the peak ratio of a single peak is less than 2.0, a polyclonal result is reported. The pattern of the polyclonal background, number of peaks, and location in the polyclonal background are also considered in the interpretation of the results. A bell-shape curve should be present for the polyclonal background, otherwise the problem of amplifying few T cells makes interpretation difficult. A bell-shaped curve also suggests an adequate amount of T-cell DNA was in the reaction.
Whereas single or biallelic peaks represent a monoclonal population, multiple peaks represent either an oligoclonal expansion or few T cells in the specimen (Figure 3E) . In our clinical experience since establishing the assay, difficulties in interpretation can occur when multiple small peaks are present in the 2.0 to 3.0 range. When multiple peaks fail to meet the criteria for monoclonality, they may be referred to as oligoclonal peaks, and the result for the case is reported as indeterminate. Other criteria for oligoclonal results include cases where the peak(s) exceeds 2.0, but the nucleotide size of the peak varies between duplicate tubes, or four or more reproducible peaks are seen with ratios above 2.0. Oligoclonal expansions or small clonal peaks can be seen in peripheral blood lymphocytes of the elderly, and in biopsies of eczema and psoriasis. 17, 34 Correlation of CE data with the number of T cells in 3-mm biopsies of paraffin-embedded skin or intestine is paramount to correct interpretation for ratios around the RPB criteria of 2.0. When samples have few T cells, the electropherogram will frequently shows multiple peaks of an oligoclonal pattern.
Optimal Primer Design
We believe that it is imperative to use primers to all of the variable and joining region genes that result in rearrangements in T-cell lymphomas. There are two main groups of published protocols: one type amplifies the most common TCRγGR using a primer for the Group 1 variable region gene segments and a primer for the common rearranged joining region genes Jγ1/Jγ2. This can be expected to detect about 75 to 80% of TCRγGR, which is a low degree of qualitative sensitivity. However, there are other rearrangements that occur with the uncommon variable and joining region genes. 31 Protocols with comprehensive sets of TCRγ primers have demonstrated successful amplification of the highest percentage of TCRγGR. 1, 5, 6, 21, 23, 24, 25, 35, 36 Qualitative sensitivity in detecting all TCRγGR is necessary to assist in accurate morphological diagnosis. Quantitative sensitivity is appropriate for detecting recurrent disease.
Effectiveness of Capillary Electrophoresis:
The advantages of this capillary electrophoresis protocol are several. The PCR set-up time is essentially the same as other TCRγGR protocols, affected only by the number of tubes used, however, the analysis time is much shorter. Previously, we used an agarose screening gel to detect the size of the amplicons before running DGGE since the length of amplified products cannot be determined in DGGE. The polyacrylamide gel for DGGE took one to two hours to make and 6.5 hours to assemble, electrophorese, and stain. After the CE system is set up (30 minutes), each specimen can be analyzed in 30 minutes in an automated format overnight. This allows for a shorter turnaround time for specimens by eliminating the lengthy DGGE procedure. The CE protocol, with shorter PCR products (190 nt), will theoretically amplify more samples from paraffin-embedded tissues than protocols with 260 nt or longer products.
We sequenced several TCRγGRs to establish that the new primer set for the variable genes would amplify previously detected Vγ3 GR. In Table 3 , the gene segments of variable and joining regions are identified, as well as the nucleotide inserts or deletions to verify Vγ3 amplification in the CE protocol. We have shown that all known variable and joining segments of TCRγGR can be amplified in the CE protocol.
We have shown that capillary electrophoresis has a high concordance with DGGE, as has been previously shown. 23, 24 The two cases which were not diagnosed as positive by capillary electrophoresis had low peak heights (RPB less than 2.0), thus they did not reach the criteria for a positive result (data not shown). These cases were atypical hyperplasias that were suspicious for peripheral T-cell lymphoma, however, no further lymphadenopathy developed. Thus, the small clones identified by DGGE may have had no clinical significance.
There are two limitations to a CE protocol for TCRγGR because it separates DNA by length and not by sequence. Fluorescent analysis may not separate the rare biallelic TCRγGR which are the same length and use the same joining or variable gene. This has no clinical significance because detection of one peak diagnoses the case as clonal. Monitoring for low level disease is more difficult because one cannot ensure clonal identity of a small peak when products are only separated by length. In DGGE or single stranded conformation polymorphism, co-migrating bands most often reflect identical sequence composition. To help overcome this latter limitation of CE, we have described the use of both labeled variable and joining gene primers in separate PCR reactions.
Conclusions
Fluorescent detection by capillary electrophoresis can be performed much faster than DGGE. A lab using capillary electrophoresis for other assays could quickly adopt a TCRγGR protocol. We have described a fluorescent-labeled protocol where primers amplify all TCRγGR in one tube. Secondly, we have shown multiple fluorescent labels allow identification of both groups of joining and the variable gene segments in the rearrangements. This is useful in comparing follow-up specimens for recurrent disease. Third, the protocol amplifies 190 nt products, which is usable in DNA extracted from paraffin-embedded tissues. The sensitivity of detecting TCRγGR by fluorescent analysis is nearly equivalent to DGGE. As long as a laboratory is aware of the minor limitations of capillary electrophoresis, the method can be an effective tool in the diagnosis of TCRγ GR.
Acknowledgments
We thank Deb Lytle for technical assistance and Katrina Matthews for manuscript preparation.
Address reprint requests to Timothy C. Greiner, M.D., University of Nebraska Medical Center, Department of Pathology and Microbiology, 983135 Nebraska Medical Center, Omaha, NE 68198-3135. E-mail: tgreiner@unmc.edu.
Footnotes
Supported in part by American Cancer Society CDA-95–42 (T. C. G.)
References
- 1.Trainor KJ, Brisco MJ, Wan JH, Neoh S, Grist S, Morley AA: Gene rearrangement in B- and T-lymphoproliferative disease detected by the polymerase chain reaction. Blood 1991, 78:192-196 [PubMed] [Google Scholar]
- 2.McCarthy KP, Sloane JP, Kabarowski JH, Matutes E, Wiedemann LM: The rapid detection of clonal T-cell proliferations in patients with lymphoid disorders. Am J Pathol 1991, 138:821-828 [PMC free article] [PubMed] [Google Scholar]
- 3.Bourguin A, Tung R, Galili N, Sklar J: Rapid, nonradioactive detection of clonal T-cell receptor gene rearrangements in lymphoid neoplasms. Proc Natl Acad Sci USA 1990, 87:8536-8540 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Wood GS, Tung RM, Haeffner AC, Crooks CF, Liao S, Orozco R, Veelken H, Kadin ME, Koh H, Heald P, Uluer AZ: Detection of clonal T-cell receptor γ gene rearrangements in early mycosis fungoides/Sezary syndrome by polymerase chain reaction and denaturing gradient gel electrophoresis (PCR/DGGE). J Invest Dermatol 1994, 103:34-41 [DOI] [PubMed] [Google Scholar]
- 5.Greiner TC, Raffeld M, Lutz C, Dick F, Jaffe ES: Analysis of T cell receptor-γ gene rearrangements by denaturing gradient gel electrophoresis of GC-clamped polymerase chain reaction products: correlation with tumor-specific sequences. Am J Pathol 1995, 146:46-55 [PMC free article] [PubMed] [Google Scholar]
- 6.Theodorou I, Bigorgne C, Delfau MH, Lahet C, Cochet G, Vidaud M, Raphael M, Gaulard P, Farcet JP: VJ rearrangements of the TCR γ locus in peripheral T-cell lymphomas: analysis by polymerase chain reaction and denaturing gradient gel electrophoresis. J Pathol 1996, 178:303-310 [DOI] [PubMed] [Google Scholar]
- 7.Volkenandt M, Wienecke R, Koch OM, Buer J, Probst M, Atzpodien J, Horikoshi T, Danenberg K, Danenberg P, Bertino JR: Conformational polymorphisms of cRNA of T-cell receptor genes as a clone-specific molecular marker for cutaneous lymphoma. J Invest Dermatol 1993, 101:514-516 [DOI] [PubMed] [Google Scholar]
- 8.Signoretti S, Murphy M, Cangi MG, Puddu P, Kadin ME, Loda M: Detection of clonal T-cell receptor γ gene rearrangements in paraffin-embedded tissue by polymerase chain reaction and nonradioactive single-strand conformational polymorphism analysis. Am J Pathol 1999, 154:67-75 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bottaro M, Berti E, Biondi A, Migone N, Crosti L: Heteroduplex analysis of T-cell receptor γ gene rearrangements for diagnosis and monitoring of cutaneous T-cell lymphomas. Blood 1994, 83:3271-3278 [PubMed] [Google Scholar]
- 10.Chhanabhai M, Adomat SA, Gascoyne RD, Horsman DE: Clinical utility of heteroduplex analysis of TCR γ gene rearrangements in the diagnosis of T-cell lymphoproliferative disorders. Am J Clin Pathol 1997, 108:295-301 [DOI] [PubMed] [Google Scholar]
- 11.Scheller U, Muche JM, Sterry W, Lukowsky A: Detection of clonal T cells in cutaneous T-cell lymphoma by polymerase chain reaction: comparison of mutation detection enhancement—polyacrylamide gel electrophoresis, temperature gradient gel electrophoresis and fragment analysis of sequencing gels. Electrophoresis 1998, 19:653-658 [DOI] [PubMed] [Google Scholar]
- 12.Alkan S, Cosar E, Ergin M, Hsi E: Detection of T-cell receptor-γ gene rearrangement in lymphoproliferative disorders by temperature gradient gel electrophoresis. Arch Pathol Lab Med 2001, 125:202-207 [DOI] [PubMed] [Google Scholar]
- 13.Greiner TC: Analysis of TCR gene rearrangements by two methods: denaturing gradient gel electrophoresis and laser-scanning of fluorescent-labeled products. Oksenberg JR Austin TX Landes R. G. eds. The Human Antigen T Cell Receptor: Selected Protocols and Applications. 1997:407-431
- 14.Simon M, Kind P, Kaudewitz P, Krokowski M, Graf A, Prinz J, Puchta U, Medeiros LJ, Sander CA: Automated high-resolution polymerase chain reaction fragment analysis: a method for detecting T-cell receptor γ-chain gene rearrangements in lymphoproliferative diseases. Am J Pathol 1998, 152:29-33 [PMC free article] [PubMed] [Google Scholar]
- 15.Kerlan-Candon S, Soua Z, Lefranc MP, Clot J, Eliaou JF: Detection of antigen receptor gene rearrangements in lymphoproliferative malignancies by fluorescent polymerase chain reaction. Tissue Antigens 1998, 51:20-29 [DOI] [PubMed] [Google Scholar]
- 16.Ayling JF, Iland HJ: High-resolution analysis of gene rearrangements in lymphoid malignancies. Pathology 1999, 31:252-256 [DOI] [PubMed] [Google Scholar]
- 17.Dippel E, Assaf C, Hummel M, Schrag HJ, Stein H, Goerdt S, Orfanos CE: Clonal T-cell receptor γ-chain gene rearrangement by PCR-based GeneScan analysis in advanced cutaneous T-cell lymphoma: a critical evaluation. J Pathol 1999, 188:146-154 [DOI] [PubMed] [Google Scholar]
- 18.Oda RP, Wick MJ, Rueckert LM, Lust JA, Landers JP: Evaluation of capillary electrophoresis in polymer solutions with laser-induced fluorescence detection for the automated detection of T-cell gene rearrangements in lymphoproliferative disorders. Electrophoresis 1996, 17:1491-1498 [DOI] [PubMed] [Google Scholar]
- 19.Miller JE, Wilson SS, Jaye DL, Kronenberg M: An automated semi-quantitative B and T cell clonality assay. Mol Diagn 1999, 4:101-117 [DOI] [PubMed] [Google Scholar]
- 20.Beaubier NT, Hart AP, Bartolo C, Willman CL, Viswanatha DS: Comparison of capillary electrophoresis and polyacrylamide gel electrophoresis for the evaluation of T and B cell clonality by polymerase chain reaction. Diagn Mol Pathol 2000, 9:121-131 [DOI] [PubMed] [Google Scholar]
- 21.Delabesse E, Burtin ML, Millien C, Madonik A, Arnulf B, Beldjord K, Valensi F, Macintyre EA: Rapid, multifluorescent TCRG Vγ and Jγ typing: application to T-cell acute lymphoblastic leukemia and to the detection of minor clonal populations. Leukemia 2000, 14:1143-1152 [DOI] [PubMed] [Google Scholar]
- 22.Lee SC, Berg KD, Racke FK, Griffin CA, Eshleman JR: Pseudo-spikes are common in histologically benign lymphoid tissues. J Mol Diagn 2000, 2:145-152 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Vega F, Medeiros LJ, Jones D, Abruzzo LV, Lai R, Manning J, Dunmire V, Luthra R: A novel four-color PCR assay to assess T-cell receptor γ gene rearrangements in lymphoproliferative lesions. Am J Clin Pathol 2001, 116:17-24 [DOI] [PubMed] [Google Scholar]
- 24.Luo V, Lessin SR, Wilson RB, Rennert H, Tozer C, Benoit B, Leonard DG: Detection of clonal T-cell receptor γ gene rearrangements using fluorescent-based PCR and automated high-resolution capillary electrophoresis. Mol Diagn 2001, 6:169-179 [DOI] [PubMed] [Google Scholar]
- 25.Meier VS, Rufle A, Gudat F: Simultaneous evaluation of T- and B-cell clonality, t(11;14) and t(14;18), in a single reaction by a four-color multiplex polymerase chain reaction assay and automated high-resolution fragment analysis: a method for the rapid molecular diagnosis of lymphoproliferative disorders applicable to fresh frozen and formalin-fixed, paraffin-embedded tissues, blood, and bone marrow aspirates. Am J Pathol 2001, 159:2031-2043 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Forster A, Huck S, Ghanem N, Lefranc MP, Rabbitts TH: New subgroups in the human T cell rearranging V γ gene locus. EMBO J 1987, 6:1945-1950 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lefranc MP, Forster A, Rabbitts TH: Rearrangement of two distinct T-cell γ-chain variable-region genes in human DNA. Nature 1986, 319:420-422 [DOI] [PubMed] [Google Scholar]
- 28.LeFranc MP, Forster A, Baer R, Stinson MA, Rabbitts TH: Diversity and rearrangement of the human T cell rearranging γ genes: nine germ-line variable genes belonging to two subgroups. Cell 1986, 45:237-246 [DOI] [PubMed] [Google Scholar]
- 29.Huck S, Lefranc MP: Rearrangements to the JP1, JP and JP2 segments in the human T-cell rearranging γ gene (TRG γ) locus. FEBS Lett 1987, 224:291-296 [DOI] [PubMed] [Google Scholar]
- 30.Greiner TC, Moynihan MJ, Chan WC, Lytle DM, Pedersen A, Anderson JR, Weisenburger DD: p53 mutations in mantle cell lymphoma are associated with variant cytology and predict a poor prognosis. Blood 1996, 87:4302-4310 [PubMed] [Google Scholar]
- 31.Theodorou I, Raphael M, Bigorgne C, Fourcade C, Lahet C, Cochet G, Lefranc MP, Gaulard P, Farcet JP: Recombination pattern of the TCR γ locus in human peripheral T-cell lymphomas. J Pathol 1994, 174:233-242 [DOI] [PubMed] [Google Scholar]
- 32.Sprouse JT, Werling R, Hanke D, Lakey C, McDonnel L, Wood BL, Sabath DE: T-cell clonality determination using polymerase chain reaction (PCR) amplification of the T-cell receptor γ-chain gene and capillary electrophoresis of fluorescently labeled PCR products. Am J Clin Pathol 2000, 113:838-850 [DOI] [PubMed] [Google Scholar]
- 33.Arber DA, Braziel RM, Bagg A, Bijwaard KE: Evaluation of T-cell receptor testing in lymphoid neoplasms: results of a multicenter study of 29 extracted DNA and paraffin-embedded samples. J Mol Diagn 2001, 3:133-140 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Posnett DN, Sinha R, Kabak S, Russo C: Clonal populations of T cells in normal elderly humans: the T cell equivalent to “benign monoclonal gammapathy.”. J Exp Med 1994, 179:609-618 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Goudie RB, Karim SN, Mills K, Alcorn M, Lee FD: A sensitive method of screening for dominant T cell clones by amplification of T cell γ gene rearrangements with the polymerase chain reaction. J Pathol 1990, 162:191-196 [DOI] [PubMed] [Google Scholar]
- 36.Lorenzen J, Jux G, Zhao-Hohn M, Klockner A, Fischer R, Hansmann ML: Detection of T-cell clonality in paraffin-embedded tissues. Diagn Mol Pathol 1994, 3:93-99 [DOI] [PubMed] [Google Scholar]