Abstract
Fusion genes consisting of TLS/FUS and CHOP or EWS and CHOP are characteristic markers for myxoid/round cell liposarcomas (MLS/RCLS). Several different structures of the fusion genes were reported in the case of the TLS/FUS-CHOP form, whereas only one type of structure has so far been found for the EWS-CHOP form, which consisted of exons 1 to 7 of the EWS and exons 2 to 4 of the CHOP gene. Here we describe a novel type of EWS-CHOP fusion gene in two cases of MLS/RCLS, which were found in a consecutive analysis of 21 cases. This fusion gene consisted of exons 1 to 10 of the EWS and exons 2 to 4 of the CHOP gene. The two cases with this fusion gene shared several clinical features, such as a large tumor mass, rapid and invasive growth, and local recurrence within 12 months after surgical resection. Histopathological findings also showed common features characterized by the diffuse proliferation of small spindle cells with a primitive mesenchymal appearance. The association of these clinical and histopathological features suggests a distinct biological property for this rare type of fusion product.
Tumor-specific fusion genes resulting from reciprocal chromosomal translocations have been detected in many bone and soft tissue tumors, and are now regarded as a major factor in the development of these tumors. 1 One of the common features of the fusion genes found in sarcomas is structural diversity, which is created by two factors: the component genes themselves, one is usually common and the other variable; and genomic breakpoints or alternative splicing in the components. The diversity of fusion genes is most striking in Ewing’s sarcoma. So far five different partners of the EWS gene have been reported, 2, 3, 4, 5, 6 and at least 12 different structures were found in the case of EWS-Fli1 fusion gene, 7, 8 which is the most prevalent form of the fusion gene in Ewing’s sarcoma. 8 In some tumors, the diversity seems to have biological significance, and several particular types of fusion genes were demonstrated to be associated with certain phenotypes such as prognosis or histological subtype. 8, 9, 10, 11, 12
In the case of MLS/RCLS, either the TLS/FUS (hereafter simply called TLS)-CHOP or EWS-CHOP fusion gene, the result of a t(12;16)(q13;p11) or a t(12;22)(q13;q12) translocation, respectively, 13, 14, 15 was found in almost all cases 16, 17, 18 and the two are now considered a hallmark of this type of liposarcoma. Previous data indicated that the TLS-CHOP fusion gene was far more prevalent (95 to 98%) than the EWS-CHOP gene, 16, 17, 18 having at least nine different structures due to breakpoints or alternative splicing. 19 On the other hand, only five cases of MLS/RCLS harboring the EWS-CHOP fusion gene have been reported in the literature, all of which shared the same structure consisting of exons 1 to 7 of the EWS gene and exons 2 to 4 of the CHOP gene (designated as the type 1 EWS-CHOP fusion gene in this report). 15, 20, 21 No definite correlation has been reported between any particular type of TLS-CHOP or EWS-CHOP fusion gene and the clinicopathological features of MLS/RCLS. In this report, we describe the results of fusion gene analyses of 21 cases of MLS/RCLS, in which we found a novel type of EWS-CHOP fusion gene in two cases.
Materials and Methods
Tumor Samples and Nucleic Acid Extraction
Twenty-one cases of MLS/RCLS were examined in this study, 13 of which were used previously in breakpoint analyses of the TLS-CHOP fusion gene. 22, 23 Clinical and histopathological features of each case are presented in Table 1 . Tumor tissues were frozen immediately after surgical resection and stored at −80°C until nucleic acid extraction. High molecular weight genomic DNA and total RNA were extracted from tissues as previously described. 22
Table 1.
Clinical, Histopathological, and Molecular Findings of MLS/RCLSs in This Study
| Patient ID | Location | Age | Histology* | Tumor size | Local recurrence† (months) | Distant metastases† (months) | Follow-up period (months) | Final result‡ | Fusion gene | Subtype |
|---|---|---|---|---|---|---|---|---|---|---|
| 373 | buttock | 53 | classical | 20 × 15 × 10 | 11 | ND | 59 | NED | TLS-CHOP | 1 |
| 410 | knee | 46 | classical | 14 × 14 × 0 | ND | ND | 70 | CDF | TLS-CHOP | 1 |
| 427 | leg | 48 | classical with R | 9 × 7 × 4 | ND | ND | 65 | CDF | TLS-CHOP | 1 |
| 455 | thigh | 40 | classical | 15 × 12 × 12 | ND | ND | 66 | CDF | TLS-CHOP | 1 |
| 559 | leg§ | 41 | classical | 13 × 10 × 6 | 9 | 3 | 19 | DOD | TLS-CHOP | 1 |
| 227 | thigh | 36 | classical | 10 × 5 × 5 | ND | ND | 102 | CDF | TLS-CHOP | 2 |
| 282 | thigh | 27 | classical | 23 × 10 × 10 | ND | ND | 25 | CDF | TLS-CHOP | 2 |
| 287 | thigh | 58 | classical | 12 × 6 × 5 | 39 | ND | 140 | NED | TLS-CHOP | 2 |
| 374 | thigh | 32 | classical with R | 10 × 10 × 8 | 60 | 149 | 150 | DOD | TLS-CHOP | 2 |
| 376 | thigh | 43 | classical | 17 × 11 × 8 | ND | ND | 72 | CDF | TLS-CHOP | 2 |
| 518 | buttock | 58 | classical | 5 × 4 × 4 | 16 | ND | 68 | NED | TLS-CHOP | 2 |
| 534 | groin | 27 | classical | 6 × 4 × 3 | 64 | ND | 151 | NED | TLS-CHOP | 2 |
| 555 | thigh | 33 | classical | 16 × 8 × 7 | ND | ND | 44 | CDF | TLS-CHOP | 2 |
| 386 | groin | 54 | classical | 4 × 3 × 2 | ND | ND | 52 | CDF | TLS-CHOP | 3 |
| 469 | knee | 48 | classical with R | 12 × 10 × 9 | ND | ND | 50 | CDF | TLS-CHOP | 4 |
| 509 | thigh | 62 | classical | 18 × 10 × 10 | ND | ND | 52 | CDF | TLS-CHOP | 4 |
| 180 | thigh | 24 | classical | 5 × 5 × 4 | 108 | ND | 289 | NED | TLS-CHOP | 5 |
| 307 | thigh | 26 | classical with R | 13 × 11 × 8 | ND | ND | 10 | CDF | EWS-CHOP | 1 |
| 753 | thigh | 48 | classical with R | 8 × 6 × 4 | 49 | ND | 54 | NED | EWS-CHOP | 1 |
| 682 | thigh | 26 | unusual | 35 × 20 × 20 | 12 | 10 | 16 | AWD | EWS-CHOP | 2 |
| 685 | retroperitoneal | 16 | unusual | 30 × 25 × 20 | 8 | ND | 15 | AWD | EWS-CHOP | 2 |
Classical, classical MLS; classical with R, classical MLS with round cell portion; unusual, MLS with unusual findings. See details in text.
ND, not detected.
CDF, continuous disease free; NED, no evidence of disease; AWD, alive with disease; DOD, dead of disease.
Synchronous multifocal lesions in shoulder, chest wall, and lung.
Histopathological Analysis
Hematoxylin and eosin-stained glass slides obtained from several representative areas in each tumor were available in all cases, and reviewed by two of the authors (Y.N. and J.T.).
Reverse Transcriptase-Polymerase Chain Reaction (RT-PCR)
Strict precautions against contamination were taken and negative controls were always included. cDNAs were synthesized from 1–3 μg of total RNA by Superscript II reverse transcriptase (Life Technologies, Inc., Rockville, MD) using oligo d(T) primer or random primer, and 0.5–1.0 μl of the cDNA products was amplified by rTaq polymerase (Toyobo, Osaka, Japan). TLS-CHOP fusion transcripts were amplified using primers TLS294F (5′-CAGAGCTCCCAATCGTCTTACGG-3′) and CHOP176R (5′-GAGAAAGGCAATGACTCAGCTGCC-3′), while EWS-CHOP fusion transcripts were amplified using primers EWS501F (5′-CCAGCCCAGCCTAGGATATGGACA-3′) and CHOP194R (5′-CTGGACAGTGTCCCGAAGGAGAAA-3′). After purification, the PCR products were cloned into TA vector (Invitrogen, Carlsbad, CA) and sequenced with ALF express (Amersham-Pharmacia, Piscataway, NJ) as previously reported, 22 or directly sequenced with an ABI Prism 377 Genetic Analyzer using the BigDye Terminator Cycle Sequencing Ready Reaction Kit (Applied Biosystems, Foster City, CA).
Genomic Long Distance PCR
Genomic DNA (100 ng) was amplified by LATaq polymerase (Takara Shuzo Co., Shiga, Japan). TLS-CHOP genomic fusion fragments encompassing the breakpoints were amplified using primers TLS431 (5′-CAGCCAGCAGCCTAGCTATG-3′) and CHOP105 (5′-CTGCTTTCAGGTGTGGTGATGTAT-3′) or TLS847 (5′-GGGACCAA-GGATCACGTCATGACT-3′) and CHOP105. Amplified fragments were sequenced according to the same methods used for RT-PCR products.
Results
Detection of Fusion Transcripts
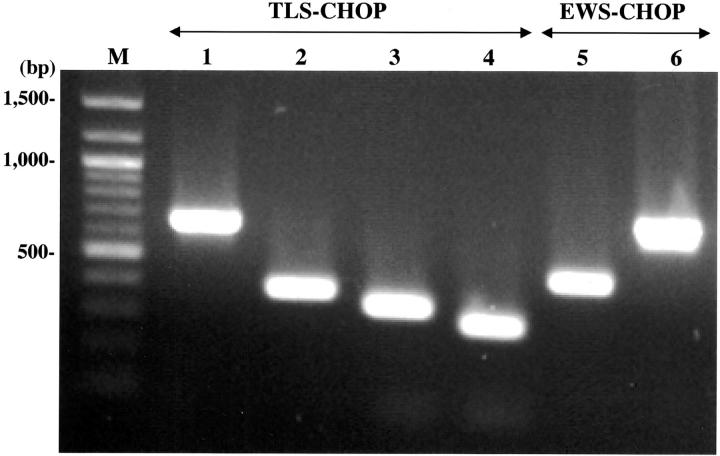
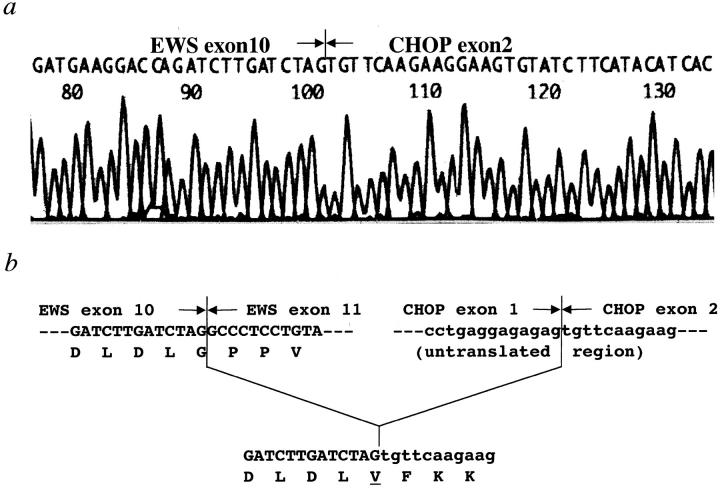
RNA of appropriate quality was available in 18 of 21 cases, and reverse-transcribed cDNAs from each case were used to amplify either the TLS-CHOP or EWS-CHOP fusion transcript by one-step PCR. Among these 18 cases, TLS-CHOP fusion transcripts were detected in 14 cases, including four types of fragments of different sizes (Figure 1 , lanes 1–4). Sequence analyses of the amplified fragments revealed five cases with the type 1, six cases with the type 2, two cases with the type 4, and one case with the type 5 TLS-CHOP transcript according to the classification used in a recent report. 19 EWS-CHOP transcripts were detected in the remaining four cases, showing two fragments of different size (Figure 1 , lanes 5–6). The shorter fragment was detected in two cases including KS307, in which we failed to detect the TLS-CHOP transcript despite an apparent rearrangement of the CHOP gene by Southern blotting in a previous report. 22 Sequence analysis of this fragment revealed the fusion of exon 7 of the EWS gene with exon 2 of the CHOP gene, which was identical with the structure of the EWS-CHOP fusion gene found in five previously reported cases. 15, 20, 21 The structure of the longer fragment detected in the other two cases showed an in-frame fusion of exon 10 of the EWS gene to exon 2 of the CHOP gene (Figure 2) , which had not previously been reported (designated as a type 2 EWS-CHOP fusion gene in this report).
Figure 1.
Detection of fusion transcripts by RT-PCR. cDNAs were amplified by TLS 294F and CHOP176R (lanes 1–4) or EWS501F and CHOP194R (lanes 5–6). M, molecular size marker; lane 1, type 1 transcript (653 bp); lane 2, type 2 transcript (377 bp); lane 3, type 4 transcript (329 bp); lane 4, type 5 transcript (284 bp); lane 5, type 1 transcript (441 bp), lane 6, type 2 transcript (693 bp).
Detection of Genomic Fusion Fragments
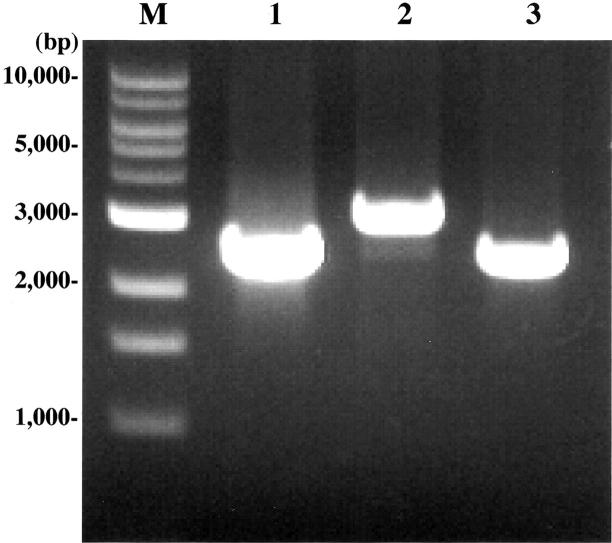
For three cases in which the quality of RNA was poor, we carried out PCR to detect genomic TLS-CHOP fragments encompassing the fusion point. All three cases showed an amplified fragment of different size on one-step PCR (Figure 3) . Sequence analyses of each fragment revealed that in two cases (KS227 and KS287), intron 5 of the TLS gene was fused to intron 1 of the CHOP gene, which presumably resulted in the fusion between exon 5 of the TLS and exon 2 of the CHOP gene, creating the type 2 TLS-CHOP fusion gene. 22 The third case (KS386) proved to have a fusion of intron 8 of the TLS gene and intron 1 of the CHOP gene, which presumably resulted in the fusion of exon 8 of the TLS gene with exon 2 of the CHOP gene. This structure was classified as a type 3 TLS-CHOP fusion gene according to the classification in a recent report. 19
Figure 3.
Detection of the TLS-CHOP genomic fusion fragments by PCR. Genomic DNAs derived from KS227 (lane 1), KS287 (lane 2), and KS386 (lane 3) were amplified by the following primers: KS227 and KS287, TLS431F, and CHOP105R; KS386, TLS847F, and CHOP105R. M, molecular size marker.
Relationship between Subtypes of Fusion Genes and Clinicopathological Findings
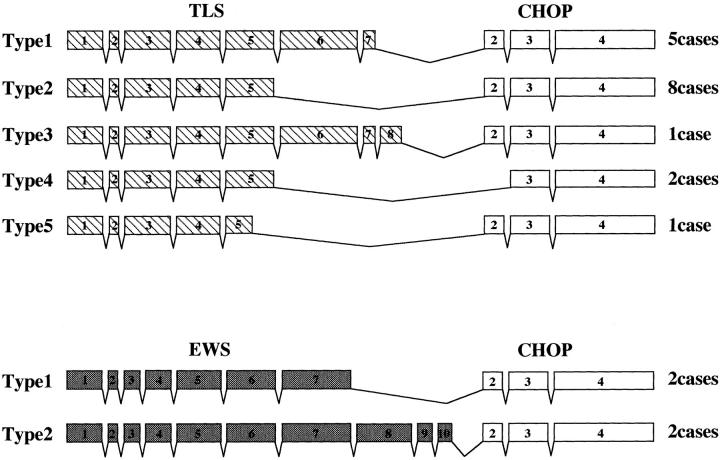
Seventeen of 21 cases (81%) were found to have TLS-CHOP fusion genes, including five cases with the type 1, eight cases with the type 2, one case with the type 3, two cases with the type 4, and one case with the type 5 gene (Figure 4) . The remaining four cases (19%) had EWS-CHOP fusion genes, two each with the type 1 and type 2 fusion genes (Figure 4) . No significant association was found between the type of fusion product and any clinical factor (Table 1) .
Figure 4.
Structure of TLS-CHOP and EWS-CHOP fusion genes. The structure of each fusion gene is schematically represented. Hatched, gray, and open boxes represent exons of the TLS, EWS, and CHOP genes, respectively.
Histopathologically, 19 of 21 tumors showed classical MLS features to some extent (Table 1) . Among these 19 tumors, 14 were solely composed of such features (designated as “classical” in Table 1 ). Features of RCLS were found in the remaining five cases, for which the fusion gene was a type 1 EWS-CHOP gene in two cases, and type 1, 2, and 4 TLS-CHOP genes in one case each (designated as “classical with R” in Table 1 ). Because the clinical and histopathological findings of the remaining two cases harboring the type 2 EWS-CHOP fusion gene were distinct from those of other cases, we describe these cases in detail.
Case KS682
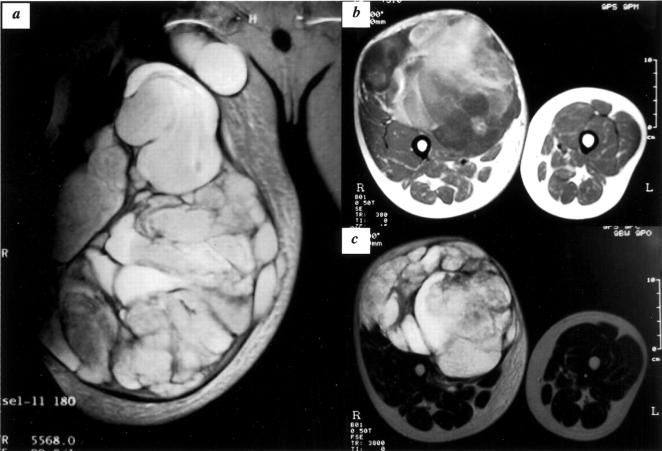
A 26-year-old male visited a hospital for a medical evaluation of a rapidly growing mass on his right thigh in November 2000. Magnetic resonance imaging (MRI) revealed a huge mass occupying the entire anterior aspect of the thigh extending in the proximal direction and crossing the inguinal ligament (Figure 5) . The patient was referred to Kyoto Prefectural University of Medicine in January 2001, and an open biopsy was performed. Histologically mononuclear small round cells proliferated diffusely in the edematous stroma together with a small amount of collagen fibers, providing a relatively cellular appearance (Figure 6A) . Microcystic change due to prominent stromal edema or myxoid change was seen in several foci (Figure 6B) . Neoplastic cells had a primitive mesenchymal appearance, hyperchromatic nucleus, and scanty eosinophilic cytoplasm of fibrillar quality. A single nucleolus was generally prominent and mitosis was relatively frequent (1 to 5 in 10 high power fields). Several cells showed lipoblastic features with cytoplasmic vacuoles (Figure 6B) , but a capillary network with a fine plexiform pattern was not typical. RT-PCR analysis revealed a novel type of EWS-CHOP fusion transcript as described in the previous session. He was treated with neoadjuvant chemotherapy for MLS/RCLS, which reduced the size of the tumor by 40%. The patient underwent the surgical resection of tumors followed by postoperative chemotherapy, and was free from disease until 10 months after surgery, when he started suffering from back pain and a gradually progressing paraplegia. MRI of the thoracic spine revealed a metastatic lesion in the second thoracic vertebral body, which was compressing the spinal cord. Two months later, local recurrence was detected proximal to the site of index operation.
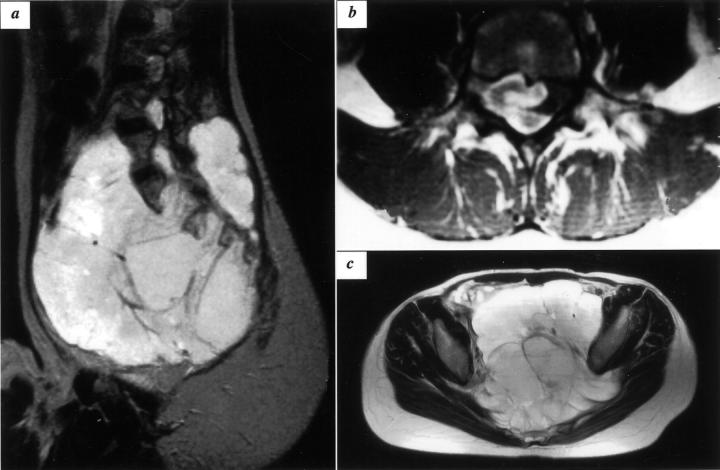
Figure 5.
MRI findings of KS682. Coronal T2-weighted image shows a large mass with high-signal intensity, extending proximally across the inguinal ligament (a). The entire anterior aspect of the thigh is occupied by tumors as shown in axial T2- and T1-weighted images (b and c, respectively).
Figure 6.
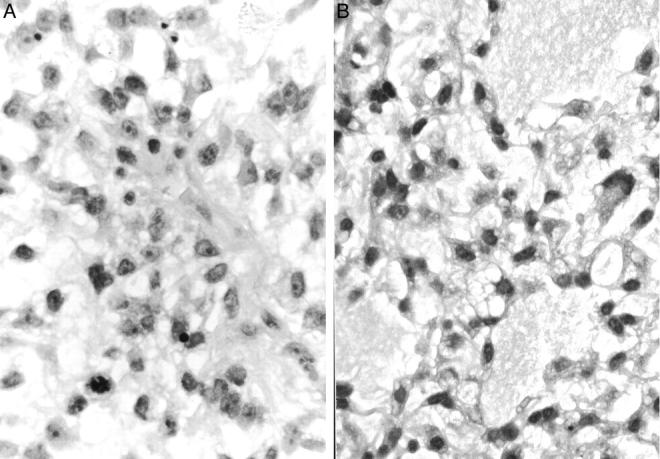
Histopathological findings of biopsy specimens of KS682. A: Round mesenchymal cells show undifferentiated features with prominent nucleoli (hematoxylin and eosin, original magnification, ×780). B: Cystic change with myxoid material is seen in several foci, and some cells show lipoblastic features with vacuoles in the cytoplasm (hematoxylin and eosin, original magnification, ×780).
Case KS685
A 16-year-old female had endured gradually increasing lower back pain for six months before seeking medical attention in September 1999. MRI of lumbar vertebrae revealed a mass in the lumbar paravertebral muscles and also in the epidural space at the fourth lumbar vertebra. An open biopsy was performed, and a tentative pathological diagnosis of intramuscular myxoma was made. The patient underwent no further examination until the pain became severe and bladder and bowel dysfunction occurred in February 2000, when the second MRI revealed a huge retroperitoneal mass occupying almost the entire pelvic cavity, connecting with the lumber lesion through the neural foramens in the sacrum (Figure 7) . She was referred to Kyoto University Hospital, and underwent further biopsy. Histologically, the tumor was composed of a diffuse and monotonous proliferation of small round mononuclear cells with an edematous background (Figure 8A and B) . Like the tumor in case KS682, individual neoplastic cells had undifferentiated or primitive mesenchymal features with a round to oval or slightly angulated hyperchromatic nucleus and a narrow eosinophilic cytoplasm frequently extending an elongated dendritic cytoplasmic process in the transparent stroma, where extracellular matrix including collagen fiber was not prominent (Figure 8A and B) . A fine plexiform capillary network was not characteristic, and typical lipoblasts were not identified (Figure 8A and B) . Mitotic figures were infrequent. RT-PCR analysis revealed that this tumor had the same EWS-CHOP fusion transcript as case KS682. Six cycles of neoadjuvant chemotherapy for MLS/RCLS was administered, which proved to be very effective, reducing the tumor size by approximately 80%. The patient underwent surgical resection of the residual tumor in August 2000. After the chemotherapy, the tumor showed prominent necrosis with degenerative changes, and there were only small foci of lipogenic quality with histological features of atypical lipoma or well-differentiated lipoma-like liposarcoma. Nuclear atypia was not prominent, and myxoid change with a plexiform vascular network pattern was not identified. Despite four cycles of postoperative chemotherapy, local recurrence was detected by MRI in January 2001, the tumor was subsequently resected, and postoperative local radiotherapy was administered.
Figure 7.
MRI findings of KS685. Sagittal (a) and axial (b) T2-weighted images show a large mass occupying the entire pelvic cavity. The axial T1-weighted image indicates tumor invasion into the epidural space at the fourth lumber vertebra (c).
Figure 8.
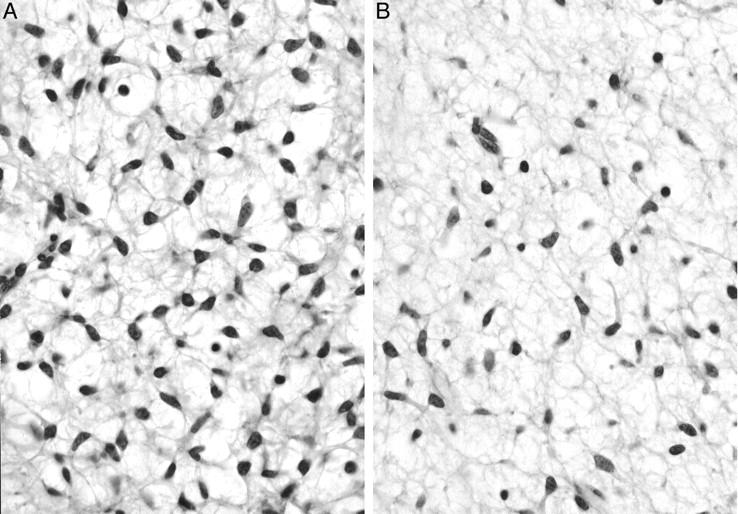
Histopathological findings of biopsy specimens of KS685. A: Small spindle cells with a primitive mesenchymal appearance proliferate diffusely in an edematous background. Dendritically elongated cytoplasms form a network-like structure making vacuolar space in the myxoid stroma. Unequivocal lipoblasts, however, are not identified (hematoxylin and eosin, original magnification, ×780). B: Myxoid change in the stroma is prominent. Plexiform fine vascular network, however, is not identified (hematoxylin and eosin, original magnification, ×780).
Discussion
Classical MLS, the most common subtype of liposarcoma, is classified as a low-grade sarcoma. Patients with MLS have a low risk of metastasis and prolonged survival. RCLS, by contrast, is a high-grade subtype of liposarcoma. 24 It is, however, now accepted that RCLS is a poorly differentiated form of classical MLS, as myxoid and round cell areas are not infrequently found within the same tumor, and the proportion of round cell components often increases in association with the progression of the disease. 25 Recent molecular findings have strengthened this assertion, showing that the two share common oncogenic fusion genes consisting of either TLS and CHOP or EWS and CHOP genes. 26 Because almost all MLS/RCLSs were found to have either of these two fusion genes, 16, 17, 18 and these events occurred exclusively in MLS/RCLSs, 16, 27 it is likely that these fusion genes play an essential role in the development of MLS/RCLS. However, it is unclear whether the structural variation in these fusion genes has any bearing on the clinical and/or histopathological variation in MLS/RCLS. Among the 21 cases examined in this study, TLS-CHOP fusion genes were found in 17 cases and had five different structures. There seemed to be no definite association of any type of TLS-CHOP fusion gene with a particular clinical feature. Among five cases with a type 1 TLS-CHOP fusion gene, four were either continuous disease free (CDF) or no evidence of disease (NED) at the last follow-up, whereas the clinical course in the last case (KS559) was extremely aggressive, showing synchronous multifocal tumors at four different anatomical sites (Table 1) . The fusion gene in a case of multifocal MLS/RCLS reported by Schneider-Stock 28 was a type 2 TLS-CHOP gene. It is thus clear that the fusion type of TLS-CHOP is not the sole determinant of clinical course. Several reports showed that the fusion gene found in RCLSs was preferentially a type 2 TLS-CHOP gene, 17, 26, 29 although the proportion of round cells in each sample was accurately described in one report. 26 In our series, only one case (KS374) may be regarded as pure RCLS, in which the round cell portion occupied more than 90% of the area examined (data not shown), and the fusion gene detected in this case was also a type 2 TLS-CHOP gene. Because the entire resected specimen was not available for the histopathological analysis in most cases, we will not refer to this issue further in this study.
Only five cases of MLS/RCLS with a type 1 EWS-CHOP fusion gene have been reported to date. 15, 20, 21 From the description in each report, two cases had round cell portions, one case was classified as high-grade MLS/RCLS, and two cases were classified as low-grade MLS/RCLS. The clinical course in these cases was not remarkable, except the case with multifocal MLS/RCLS. 20 Therefore, it seemed that there was no clear difference between tumors with the TLS-CHOP fusion gene and tumors with the type 1 EWS-CHOP gene in terms of clinical and histopathological features. Several reports have indicated that the structural difference among fusion products is related to clinical phenotype in certain kinds of sarcomas. 8, 9, 10, 11, 12 In the case of Ewing’s sarcoma, the survival of patients bearing a type 1 EWS-Fli1 fusion gene containing exons 1 to 7 of the EWS and exons 6 to 9 of the Fli1 gene is markedly better than that of patients bearing other fusion types. 8, 12 Lin et al 30 proposed that this difference was related to the difference in the transactivation activity of each fusion protein as detected by reporter assay. In their study, it was shown that when the EWS component (exons 1 to 7) of the type 1 EWS-Fli1 fusion gene was replaced with exons 1 to 10 of the EWS gene, the transactivation activity increased approximately twofold. 30 Interestingly, the difference in EWS components between these two fusion products is identical to that between the type 1 and type 2 EWS-CHOP fusion products. The CHOP protein is known to function as a transcriptional factor by forming a heterodimer with other C/EBP family members, 31 but the oncological function of TLS-CHOP or EWS-CHOP fusion protein was not yet clear. Because these cases are the first two with a type 2 EWS-CHOP fusion gene, the number is too small to suggest that it is indeed associated with a more aggressive behavior. It is of interest whether the novel EWS-CHOP fusion product has different activity from other types of CHOP-associated fusion products found in MLS/RCLS.
The relationship between histopathological features and the type of fusion genes is an another intriguing matter. For example, the presence of epithelial features in synovial sarcoma was found to be associated with the SYT-SSX1 fusion genes. 9, 11 MLS/RCLS usually shows a histological transition from a typical paucicellular area of MLS with scattered lipoblasts in a faintly bluish myxoid background partitioned by a plexiform capillary network to a dense cellular area of RCLS. The latter cellular area, mainly composed of lipoblastic cells, tends to be located in the periphery of the lobular arrangement of the MLS/RCLS. 24 However, these classical features were not prominent in the two tumors with the type 2 EWS-CHOP gene. These two tumors shared common histopathological features in that small cells with primitive or undifferentiated mesenchymal qualities proliferated with a moderate degree of intercellular edema or myxoid change. These tumors may even simulate, in some respects, the primitive or undifferentiated cellular area of embryonal rhabdomyosarcoma. 24 Rhabdomyoblasts, however, were not identified. Although these features can be regarded as indicative of a variant of MLS/RCLS, a diagnosis of MLS/RCLS might not be appropriate especially in the case of KS685, where no typical lipoblasts were found. The diagnosis made by an external consultant was a high-grade myxoid liposarcoma for KS682 and an undifferentiated spindle myxoid sarcoma for KS685. To determine whether the novel type of EWS-CHOP fusion gene described in this study relates to a specific histology or not, a molecular investigation of tumors with histopathological features comparable to our two cases should be carried out. Such tumors could have been diagnosed as, for example, cellular MLS/RCLS, 32 atypical MLS/RCLS, or in some cases, even primitive or unclassified mesenchymal tumor.
Figure 2.
Partial sequences of the type 2 EWS-CHOP fusion transcript (a). The exon 2 sequence of the CHOP gene was found at the end of exon 10 of the EWS gene (b).
Acknowledgments
We thank Drs. Shigeaki Wakita, Tsugumaro Cho, Hitoshi Murakami, and Yoshimichi Ueda for their kind cooperation in this study, and Dr. Christopher D. M. Fletcher for his valuable comments on the histological diagnoses.
Address reprint requests to Junya Toguchida, Institute for Frontier Medical Sciences, Kyoto University, 53 Kawahara-cho, Shogoin, Sakyo-ku, Kyoto 606-8507, Japan. E-mail: togjun@frontier.kyoto-u.ac.jp.
Footnotes
Supported in part by a grant from the Ministry of Education, Culture, Sports, Science, and Technology, and from the Ministry of Health, Labor, and Welfare of Japan.
References
- 1.Aman P: Fusion genes in solid tumors. Semin Cancer Biol 1999, 9:303-318 [DOI] [PubMed] [Google Scholar]
- 2.Delattre O, Zucman J, Plougastel B, Desmaze C, Melot T, Peter M, Kovar H, Joubert I, de Jong P, Rouleau G, Aurias A, Thomas G: Gene fusion with an ETS DNA-binding domain caused by chromosome translocation in human tumours. Nature 1992, 359:162-165 [DOI] [PubMed] [Google Scholar]
- 3.Sorensen PH, Lessnick SL, Lopez-Terrada D, Liu XF, Triche TJ, Denny CT: A second Ewing’s sarcoma translocation, t(21;22), fuses the EWS gene to another ETS-family transcription factor, ERG. Nat Genet 1994, 6:146-151 [DOI] [PubMed] [Google Scholar]
- 4.Jeon IS, Davis JN, Braun BS, Sublett JE, Roussel MF, Denny CT, Shapiro DN: A variant Ewing’s sarcoma translocation (7;22) fuses the EWS gene to the ETS gene ETV1. Oncogene 1995, 10:1229-1234 [PubMed] [Google Scholar]
- 5.Kaneko Y, Yoshida K, Handa M, Toyoda Y, Nishihira H, Tanaka Y, Sasaki Y, Ishida S, Higashino F, Fujinaga K: Fusion of an ETS-family gene, EIAF, to EWS by t(17;22)(q12;q12) chromosome translocation in an undifferentiated sarcoma of infancy. Genes Chromosomes Cancer 1996, 15:115-121 [DOI] [PubMed] [Google Scholar]
- 6.Peter M, Couturier J, Pacquement H, Michon J, Thomas G, Magdelenat H, Delattre O: A new member of the ETS family fused to EWS in Ewing tumors. Oncogene 1997, 14:1159-1164 [DOI] [PubMed] [Google Scholar]
- 7.Zucman J, Melot T, Desmaze C, Ghysdael J, Plougastel B, Peter M, Zucker JM, Triche TJ, Sheer D, Turc-Carel C, Ambros P, Combaret V, Lenoir G, Aurias A, Thomas G, Delattre O: Combinatorial generation of variable fusion proteins in the Ewing family of tumours. EMBO J 1993, 12:4481-4487 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.de Alava E, Kawai A, Healey JH, Fligman I, Meyers PA, Huvos AG, Gerald WL, Jhanwar SC, Argani P, Antonescu CR, Pardo-Mindan FJ, Ginsberg J, Womer R, Lawlor ER, Wunder J, Andrulis I, Sorensen PH, Barr FG, Ladanyi M: EWS-FLI1 fusion transcript structure is an independent determinant of prognosis in Ewing’s sarcoma. J Clin Oncol 1998, 16:1248-1255 [DOI] [PubMed] [Google Scholar]
- 9.Kawai A, Woodruff J, Healey JH, Brennan MF, Antonescu CR, Ladanyi M: SYT-SSX gene fusion as a determinant of morphology and prognosis in synovial sarcoma. N Engl J Med 1998, 338:153-160 [DOI] [PubMed] [Google Scholar]
- 10.Kelly KM, Womer RB, Sorensen PH, Xiong QB, Barr FG: Common and variant gene fusions predict distinct clinical phenotypes in rhabdomyosarcoma. J Clin Oncol 1997, 15:1831-1836 [DOI] [PubMed] [Google Scholar]
- 11.Nilsson G, Skytting B, Xie Y, Brodin B, Perfekt R, Mandahl N, Lundeberg J, Uhlen M, Larsson O: The SYT-SSX1 variant of synovial sarcoma is associated with a high rate of tumor cell proliferation and poor clinical outcome. Cancer Res 1999, 59:3180-3184 [PubMed] [Google Scholar]
- 12.Zoubek A, Dockhorn-Dworniczak B, Delattre O, Christiansen H, Niggli F, Gatterer-Menz I, Smith TL, Jurgens H, Gadner H, Kovar H: Does expression of different EWS chimeric transcripts define clinically distinct risk groups of Ewing tumor patients? J Clin Oncol 1996, 14:1245-1251 [DOI] [PubMed] [Google Scholar]
- 13.Crozat A, Aman P, Mandahl N, Ron D: Fusion of CHOP to a novel RNA-binding protein in human myxoid liposarcoma. Nature 1993, 363:640-644 [DOI] [PubMed] [Google Scholar]
- 14.Rabbitts TH, Forster A, Larson R, Nathan P: Fusion of the dominant negative transcription regulator CHOP with a novel gene FUS by translocation t(12;16) in malignant liposarcoma. Nat Genet 1993, 4:175-180 [DOI] [PubMed] [Google Scholar]
- 15.Panagopoulos I, Hoglund M, Mertens F, Mandahl N, Mitelman F, Aman P: Fusion of the EWS and CHOP genes in myxoid liposarcoma. Oncogene 1996, 12:489-494 [PubMed] [Google Scholar]
- 16.Antonescu CR, Elahi A, Humphrey M, Lui MY, Healey JH, Brennan MF, Woodruff JM, Jhanwar SC, Ladanyi M: Specificity of TLS-CHOP rearrangement for classic myxoid/round cell liposarcoma: absence in predominantly myxoid well-differentiated liposarcomas. J Mol Diagn 2000, 2:132-138 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hisaoka M, Tsuji S, Morimitsu Y, Hashimoto H, Shimajiri S, Komiya S, Ushijima M: Detection of TLS/FUS-CHOP fusion transcripts in myxoid and round cell liposarcomas by nested reverse transcription-polymerase chain reaction using archival paraffin-embedded tissues. Diagn Mol Pathol 1998, 7:96-101 [DOI] [PubMed] [Google Scholar]
- 18.Panagopoulos I, Aman P, Mertens F, Mandahl N, Rydholm A, Bauer HF, Mitelman F: Genomic PCR detects tumor cells in peripheral blood from patients with myxoid liposarcoma. Genes Chromosomes Cancer 1996, 17:102-107 [DOI] [PubMed] [Google Scholar]
- 19.Panagopoulos I, Mertens F, Isaksson M, Mandahl N: A novel FUS/CHOP chimera in myxoid liposarcoma. Biochem Biophys Res Commun 2000, 279:838-845 [DOI] [PubMed] [Google Scholar]
- 20.Antonescu CR, Elahi A, Healey JH, Brennan MF, Lui MY, Lewis J, Jhanwar SC, Woodruff JM, Ladanyi M: Monoclonality of multifocal myxoid liposarcoma: confirmation by analysis of TLS-CHOP or EWS-CHOP rearrangements. Clin Cancer Res 2000, 6:2788-2793 [PubMed] [Google Scholar]
- 21.Dal Cin P, Sciot R, Panagopoulos I, Aman P, Samson I, Mandahl N, Mitelman F, Van den Berghe H, Fletcher CD: Additional evidence of a variant translocation t(12;22) with EWS/CHOP fusion in myxoid liposarcoma: clinicopathological features. J Pathol 1997, 182:437-4419306965 [Google Scholar]
- 22.Kanoe H, Nakayama T, Hosaka T, Murakami H, Yamamoto H, Nakashima Y, Tsuboyama T, Nakamura T, Ron D, Sasaki MS, Toguchida J: Characteristics of genomic breakpoints in TLS-CHOP translocations in liposarcomas suggest the involvement of translin and topoisomerase II in the process of translocation. Oncogene 1999, 18:721-729 [DOI] [PubMed] [Google Scholar]
- 23.Hosaka T, Kanoe H, Nakayama T, Murakami H, Yamamoto H, Nakamata T, Tsuboyama T, Oka M, Kasai M, Sasaki MS, Nakamura T, Toguchida J: Translin binds to the sequences adjacent to the breakpoints of the TLS and CHOP genes in liposarcomas with translocation t(12;16). Oncogene 2000, 19:5821-5825 [DOI] [PubMed] [Google Scholar]
- 24.Weiss SW, Goldblum JR: Enzinger and Weiss’s Soft Tissue Tumors ed 5. 2001. Mosby St. Louis, MO
- 25.Dei Tos AP: Liposarcoma: new entities and evolving concepts. Ann Diagn Pathol 2000, 4:252-266 [DOI] [PubMed] [Google Scholar]
- 26.Knight JC, Renwick PJ, Cin PD, Van den Berghe H, Fletcher CD: Translocation t(12;16)(q13;p11) in myxoid liposarcoma and round cell liposarcoma: molecular and cytogenetic analysis. Cancer Res 1995, 55:24-27 [PubMed] [Google Scholar]
- 27.Naito N, Kawai A, Ouchida M, Dan’ura T, Morimoto Y, Ozaki T, Shimizu K, Inoue H: A reverse transcriptase-polymerase chain reaction assay in the diagnosis of soft tissue sarcomas. Cancer 2000, 89:1992-1998 [DOI] [PubMed] [Google Scholar]
- 28.Schneider-Stock R, Rys J, Walter H, Limon J, Iliszko M, Niezabitowski A, Roessner A: A rare chimeric TLS/FUS-CHOP transcript in a patient with multiple liposarcomas: a case report. Cancer Genet Cytogenet 1999, 111:130-133 [DOI] [PubMed] [Google Scholar]
- 29.Kuroda M, Ishida T, Horiuchi H, Kida N, Uozaki H, Takeuchi H, Tsuji K, Imamura T, Mori S, Machinami R, Watanabe T: Chimeric TLS/FUS-CHOP gene expression and the heterogeneity of its junction in human myxoid and round cell liposarcoma. Am J Pathol 1995, 147:1221-1227 [PMC free article] [PubMed] [Google Scholar]
- 30.Lin PP, Brody RI, Hamelin AC, Bradner JE, Healey JH, Ladanyi M: Differential transactivation by alternative EWS-FLI1 fusion proteins correlates with clinical heterogeneity in Ewing’s sarcoma. Cancer Res 1999, 59:1428-1432 [PubMed] [Google Scholar]
- 31.Ron D: TLS-CHOP and the role of RNA-binding proteins in oncogenic transformation. Curr Top Microbiol Immunol 1997, 220:131-142 [DOI] [PubMed] [Google Scholar]
- 32.Kempson RL, Fletcher CDM, Evans HL, Hendrickson MR, Sibley RK: Tumors of the Soft Tissues, Atlas of Tumor Pathology, 3rd series, Fascicle 30 2001. Armed Forces Institute of Pathology Washington, DC