Abstract
Specific assays capable of distinguishing normal and atypical cervical changes from pre-cancerous lesions are direly needed to improve screening for cervical cancer. Specific genes transcripts that are up-regulated in dysplastic and cancer cells can be exploited as new markers for cervical cancer screening provided that they can be detected in heterogeneous populations such as those collected for Papanicolaou tests. We hypothesized that expression of the HPV early region gene E7 might distinguish between normal samples (absent expression) and high-grade lesions (detectable E7 expression). Our goal was to detect and measure gene expression in cells scraped from the cervix using real time quantitative reverse transcription-polymerase chain reaction (TaqMan). We have optimized collection and extraction procedures to provide suitable RNA for TaqMan analysis in clinical samples collected for cervical cancer screening and have demonstrated efficient measurements of housekeeping genes in these samples. HPV 16 or 18 early gene E7 transcripts were detected in 47% of samples with a clinical diagnosis of high-grade SIL and in 0% of cytologically normal samples (P = 0.006). Our study demonstrates that the TaqMan assay can be reliably applied to samples collected for cervical cancer screening, and that presence of detectable HPV E7 transcripts can distinguish between normal and abnormal samples.
World-wide, cervical cancer is the leading cause of cancer morbidity in women. 1 Human cervical epithelium can undergo a series of progressive neoplastic changes known as cervical dysplasia, cervical intraepithelial neoplasia (CIN), or squamous intraepithelial lesions (SIL) of graded severity that, in certain instances, are precursors to invasive cervical cancer. Papanicolaou smear screening is a cytological test that successfully detects cervical cancer precursors and has resulted in a significant decrease in cervical cancer incidence in countries where it has been implemented. 1, 2, 3 However, this test also detects a high proportion of clinically insignificant minor abnormalities resulting in stressful and expensive medical intervention. 4 In addition, the Papanicolaou test requires highly trained specialists and expertise not available in most developing countries.
Humanpapilloma virus (HPV) is present in approximately 90% of high-grade dysplasias and cervical cancers. 5 While measurement of HPV DNA is sensitive, the high prevalence of HPV in asymptomatic populations may limit its usefulness as a screening test. 6, 7, 8 Therefore, the availability of specific diagnostic markers to discriminate patients with clinically benign atypical changes from those with cancer and high-grade dysplasia would be extremely useful. Measurement of HPV neoplastic activity such as expression of early region transcripts might provide such distinction. RNA levels can be measured in cultured cells and tissues using recent technological advances in reverse transcription-polymerase chain reaction (RT-PCR) that allow rapid and accurate quantitative analyses, 9, 10, 11 including applications suitable for samples with moderately or highly degraded RNA such as commonly found in clinical specimens. 12 The present study is the first demonstration of the feasibility of measuring RNA transcripts in samples collected for cervical cancer screening using real time quantitative RT-PCR (TaqMan). Sample collection and RNA extraction procedures were optimized for the TaqMan assay to allow reliable expression measurements of housekeeping genes in clinical samples. Expression of the HPV 16 or 18 E7 oncogene was detected in 47% of samples from women with high-grade SIL and 0% of samples from women with normal cytology, indicating that E7 expression is technically feasible in these samples and may have diagnostic significance.
Materials and Methods
Collection Methods
Cervical cells were collected from patients receiving Papanicolaou smears at the University of California at San Francisco (UCSF). The UCSF Committee on Human Research approved the protocol for enrolling subjects and collecting samples. In the gynecology clinic, where conventional Papanicolaou smears are made, cells remaining on the collection devices after preparation of the clinical sample were collected for study purposes. In the dysplasia clinic, where liquid-based Papanicolaou tests are used for clinical diagnosis, a second cervical scrape with a spatula and a brush was obtained for study purposes. The collection devices containing the study sample were immediately agitated in a 50-ml centrifugation tube containing either 3 ml of Trizol reagent (Gibco, Life Technologies), or 12 ml of Dulbecco’s modified Eagle’s medium (DME), or 95% ethanol. Eight to 10 clinical samples were collected into each of the different collection media for comparison. Trizol suspensions were immediately snap-frozen in liquid nitrogen. Cell suspensions in DME or ethanol were centrifuged at 1200 rpm for 10 minutes. The supernatant was decanted and the cell pellet snap-frozen in liquid nitrogen. Samples were frozen within 20 minutes of collection and stored at −80°C until RNA extraction. Diagnoses of samples in Table 2 are from cytology reports of the corresponding clinical Papanicolaou smear samples signed out by cytopathologists at UCSF.
Table 2.
Detection of E7 RNA Expression by TaqMan RT-PCR in Cytologically Normal and Abnormal Samples
| HPV E7 type | Number (%) of normal samples with E7 mRNA | Number (%) of HGSIL samples with E7 mRNA |
|---|---|---|
| 16 alone | 0 | 2 (13) |
| 18 alone | 0 | 2 (13) |
| 16 and 18 | 0 | 3 (20) |
A total of 7 patients with HGSIL had E7 transcripts detected. Five samples contained type 16 E7 transcripts and 5 samples contained type 18 E7 transcripts as follows: 2 each with type 16 and type 18 alone, and 3 with both type 16 and 18.
Extraction Methods
Different RNA extraction procedures were tested for their ability to provide RNA that would be suitable for the TaqMan assay. Frozen cell pellets were thawed on ice for 5 minutes, centrifuged at 1600 rpm, and the remaining supernatant was removed.
RNeasy Extraction
Cell pellets were resuspended in 350 μl of RNeasy (RLT) lysis buffer from the RNeasy kit (Qiagen Inc, Valencia, CA) and homogenized through Qiashredder columns (Qiagen). RNA extraction was performed through RNeasy columns according to manufacturer’s instructions. Extracted RNA was eluted with 45 μl of diethylpyrocarbonate (DEPC) water.
Trizol Extraction
One ml of Trizol solution was added to cell pellets. The samples were homogenized by aspirating approximately 10 times through a 23-gauge needle before chloroform extraction and ethanol precipitation following manufacturer’s instructions. In some cases, the upper phase after chloroform extraction was collected and re-extracted with acidic phenol and chloroform before ethanol precipitation. RNA precipitates were resuspended in 45 μl of DEPC water.
The purity and quantity of RNA obtained by these extraction methods was measured by optical density reading (OD) at 260 nm and 280 nm. Integrity of RNA samples was assessed by Agilent biosizing gel separation. Agilent technology was not used to quantify RNA.
RT-PCR
The reverse transcription reaction was performed in 100-μl-volume consisting of the following mixture: 5 μl of RNA template (100 to 300 ng as measured by OD), 1 mmol/L each dNTP (Roche Molecular Systems, Alameda, CA, USA), 5 μmol/L random hexamers (Invitrogen, Carlsbad, CA, USA), 250 units of Moloney murine leukemia virus (MMLV) reverse transcriptase (RT) (Invitrogen), 40 units of RNase inhibitor (Roche Molecular Systems), 7.5 mmol/L MgCl2, and 1X PCR buffer (Applied Biosystems, Foster City, CA, USA). The reaction was carried out in RNase-free condition at 25°C for 10 minutes, 48°C for 40 minutes, and 95°C for 5 minutes in a Perkin Elmer GenAmp PCR cycler. Each RT reaction was accompanied by a “no RT” control in which the reverse transcriptase was replaced by DEPC water. The cDNA was stored at −20°C until use in the TaqMan PCR reaction.
The PCR reaction was carried out in 50-μl volumes containing 10 μl of cDNA template from the RT reaction, 200 to 500 nmol/L of each primer, 100 to 200 nmol/L of probe, 200 μmol/L of each dNTPs (Roche Molecular Systems), 1X TaqMan PCR buffer A (Applied Biosystems), 0.025U/μl Taq Gold polymerase (Applied Biosystems) and 5.5 mmol/L MgCl2. PCR was performed as follows: 95°C for 12 minutes to activate the Taq Gold, 45 cycles of 95°C for 15 seconds, and 60°C for 1 minute, consecutively. Each amplification experiment was performed in a 96-well PCR plate covered with optical caps in the ABI 7700 real-time PCR instrument.
Primers and Probes
All TaqMan PCR primers and probes were designed by the Genome Core Facility at the Cancer Research Institute of UCSF using Primer Express Software (Applied Biosystems) and synthesized by Integrated DNA Technologies (Coralville, IA). Fluorescent probes were labeled at the 5′ end with 6-carboxy fluorescein (FAM) and the 3′ end with 6-carboxy tetramethyl rhodamine (TAMRA). β-glucuronidase (GUS) intron spanning primers and probes amplifying an 81-bp fragment, and glyceraldehyde-3-phosphate dehydrogenase (GAPDH) primers amplifying a 72-bp sequence, have been previously published. 12 HPV type-specific primers for E7 were synthesized to amplify an 82-bp fragment for HPV 16 (forward primer 5′-CCGGACAGAGCCCATTACAAT-3′, reverse primer 5′-ACGTGTGTGCTTTGTACGCAC-3′, probe 5′-(FAM)-TGTTGCAAGTGTGACTCTACGCTTCGGT-(TAMRA)-3′, and an 81-bpfragment for HPV 18 (forward primer 5′-GACTCAGAGGAAGAAAACGATGAAA-3′, reverse primer 5′-GTGACGTTGTGGTTCGGCT-3′, probe 5′-(FAM)-TGGAGTTAATCATCAACATTTACCA-(TAMRA)-3′. E7 primers were selected to amplify regions of sequence diversity between HPV 16 or 18 to limit cross-reactivity with other HPV types. Alignment of the 28 bases of the HPV 16 TaqMan probe with homologous sequences in HPV 18, 6, and 11 revealed 10, 15, and 14 shared bases respectively (35, 54, and 50% identity). Alignment of the 30 bases of the HPV 18 TaqMan probe with HPV 16, 6 and 11 sequences revealed 9, 9, and 8 shared bases respectively (30, 30, and 27% identity). The low degree of homology makes cross-reactivity of the probes with low risk viral types extremely unlikely.
Measurements
Data from the ABI Prism 7700 sequence detection instrument were analyzed with Sequence Detector software version 1.6.3 and 1.7. 12 The numeric data were then imported to an MS Excel spreadsheet for further analysis. Relative expression of each mRNA species was expressed as the cycle threshold (CT) value reached during the amplification reaction. The CT represents the cycle number at which the sample amplification plot reaches a fluorescence threshold above background. Higher expression of a transcript is reflected by lower CT values, since the amplification products reach the critical threshold sooner than less abundant transcripts. For E7 measurements, we compared the CT value of the “no-RT” control to that of the “plus RT” measurement. Due to the exponential nature of the PCR reaction, a CT value of the “no-RT” control that is four cycles higher than that of the RT sample indicates that 2−4 ×100 (6.2%) of the “plus RT” signal generated is the result of contaminating DNA. Therefore, for our data analysis of E7 expression, we considered a CT value significant when it was at least four cycles lower than the CT value of the corresponding “no RT” control.
RT linearity was assessed in our clinical samples by using increasing amount of RNA in the RT reaction and performing the PCR amplification with constant volume input of cDNA. Efficiency was determined using the equation 10∧(1/−slope)−1.
Controls
In each amplification reaction, a “no RT” control was run concurrently with the test samples, to determine possible contribution of genomic DNA or cDNA contaminants. To establish reproducibility, PCR reactions were always performed in duplicate or triplicate for all of the samples depending on the amount of RNA available. Human cervical cancer cell lines were used as a source of HPV E7 RNA (CaSki for HPV 16 and HeLa for HPV 18).
Statistical Analysis
Fisher’s exact test was used to compare proportions of samples with measurable E7 expression in different clinical categories. Two-sided Student’s t-test was used to compare RNA yield and absorbance ratios for the different RNA extraction procedures.
Results
Cervical cells scraped from the cervix exhibited a high proportion of degraded RNA despite immediate freezing in liquid nitrogen after collection into Trizol, cell culture medium, or ethanol (Figure 1) . There were no significant differences in RNA yield or size based on comparisons of 8 to10 samples in each of the collection media. Trizol was rejected as a collection solution due to concerns about exposure of patients and providers to its toxicity. RNA extracted from samples collected in ethanol contained variable amounts of genomic DNA (data not shown). Therefore, we chose to collect cervical samples into DME cell culture medium.
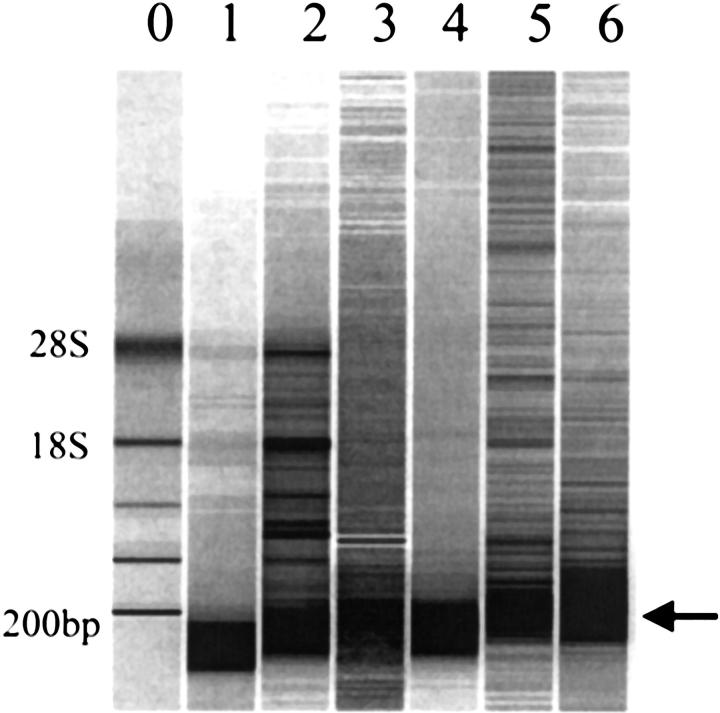
Figure 1.
RNA from cells scraped from the cervix demonstrates variable degrees of degradation regardless of collection method. Cells scraped from the cervix were collected directly into Trizol (lanes 1 and 2), into DME (lanes 3 and 4) or into 95% ethanol (lanes 5 and 6) and immediately snap-frozen in liquid nitrogen. RNA was extracted with Trizol and analyzed by Agilent biosizing gel electrophoresis. RNA quantity was calculated from the absorbance at 260 nm, and 1 μl (approx 100 ng) was loaded per lane. Ribosomal RNA was used as the molecular weight maker (lane 0); positions of 28S and 18S rRNA are indicated on the left. Arrow (right) indicates 200 bp.
RNeasy versus Trizol RNA Extraction
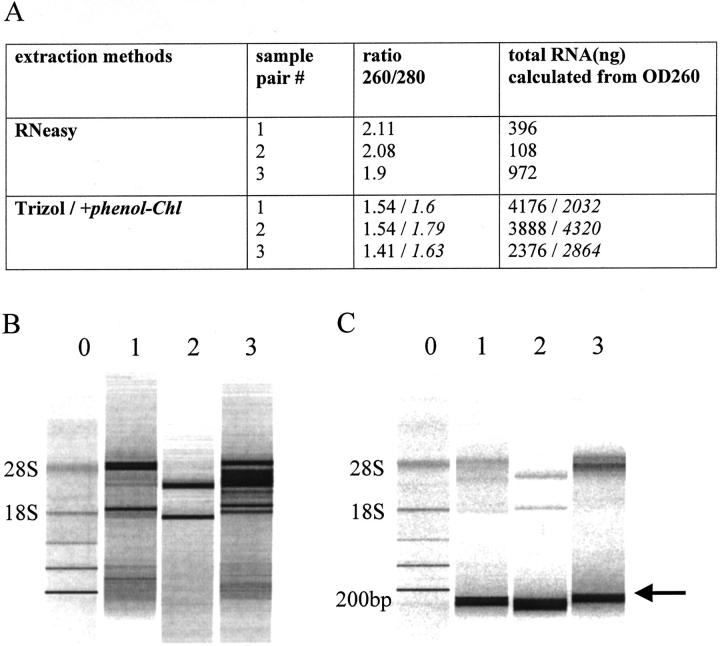
Direct comparison of RNA extracted by different methods from the same sample is shown in Figure 2 , in which pairs of clinical samples were pooled, divided equally and processed in parallel for RNA extraction by RNeasy and Trizol protocols. Yield and purity of RNA extracted by different methods from unmatched samples are compared in Table 1 . The average total yield of RNA obtained with the RNeasy columns was much lower (mean, 361 ng) than the amount obtained with the other methods, but RNeasy produced RNA of higher purity as indicated by the higher 260/280 absorbance ratio (Table 1) . The lower yields obtained with RNeasy columns reflect the fact that RNA from cervical samples is partially degraded, and RNeasy extraction excludes small RNA molecules (compare Figure 2B and Figure 2C for each parallel sample nos. 1 to 3). The average yield of RNA extracted with RNeasy is insufficient for expression measurements of several genes in replicates by TaqMan RT-PCR. On the other hand, Trizol extraction recovers both small and large RNA fragments that are more representative of the clinical samples. To improve the quality and yield of RNA extracted with Trizol, re-extraction with acidic phenol-chloroform was performed and resulted in a significant increase in RNA yield (3.6 μg versus 5.7 μg, P = 0.02). However, the difference in the mean 260/280 absorbance ratios (1.5 vs. 1.6) was not significant (Table 1 and Figure 2A ).
Figure 2.
Direct comparisons of RNA yield, purity, and integrity using different RNA extraction procedures. Pairs of clinical samples were pooled, divided into two equal aliquots, and extracted in parallel with RNeasy or Trizol. RNA concentration and yield were calculated from absorbance at 260 nm. In A, values in italics were obtained after an additional acidic phenol-chloroform extraction of the Trizol-extracted RNA. RNA size was assessed by Agilent gel electrophoresis of samples extracted with RNeasy (B), and Trizol (C). One ul was added to each lane, corresponding to approximately 25 ng (B) and 100 ng (C). Numbers 1 to 3 above each lane in B and C correspond to the samples number in A; matched numbers represent identical aliquots extracted in parallel. Ribosomal RNA was used as the molecular weight maker (lane 0); positions of 28S and 18S rRNA are indicated on the left. Arrow (right) indicates 200 bp.
Table 1.
Comparison of Three Methods of RNA Extraction of Cells Scraped from the Cervix
| RNA extraction method | |||
|---|---|---|---|
| RNeasy (n = 5) | Trizol (n = 8) | Trizol followed by acidic phenol-chloroform re-extraction (n = 21) | |
| Mean yield* (ng)± SD | 361 ± 313 | 3650 ± 1827 | 5730 ± 2043 |
| Range of RNA yields | 96–874 | 2144–7956 | 2556–8719 |
| Mean OD ratio (260/280)± SD | 2.0 ± 0.1 | 1.5 ± 0.08 | 1.6 ± 0.1 |
Yield was calculated from the absorbance at 260 nm.
The difference in yield between RNeasy and the other extraction methods is statistically significant (p < 0.01). The difference in yield between Trizol versus Trizol/acidic phenol-chloroform is also statistically significant (p = 0.02).
The difference in absorbance ratio between RNeasy and the other extraction methods is statistically significant (p < 0.0001). The difference in absorbance ratio between Trizol versus Trizol/acidic phenol-chloroform is not statistically significant (p = 0.3).
TaqMan Expression Measurements from Cells Scraped from the Cervix
Measurement of Housekeeping Genes
Godfrey et al 12 have shown that sensitive and linear RNA transcript measurements can be obtained from formalin-fixed paraffin embedded clinical samples despite significant RNA degradation, as long as the amplicon size is <130 bases.We chose to validate performance of the TaqMan assay in our clinical application (cells scraped from the cervix) using β-GUS and GAPDH genes, as they are well accepted as internal standards 13, 14, 15 and have been optimized for this assay. 12 To test the effect of degraded RNA on the RT efficiency, GAPDH was amplified from a pooled clinical sample equally divided and extracted in parallel by RNeasy and Trizol/phenol-chloroform procedures. Although RNA extracted with Trizol/phenol-chloroform was more degraded than RNA obtained with RNeasy extraction, it still reverse transcribed linearly and with slightly higher efficiency than RNA extracted with RNeasy (83% versus 62%, Figure 3A ). As a result, all subsequent measurements were performed using RNA extracted with Trizol/phenol-chloroform.
Figure 3.
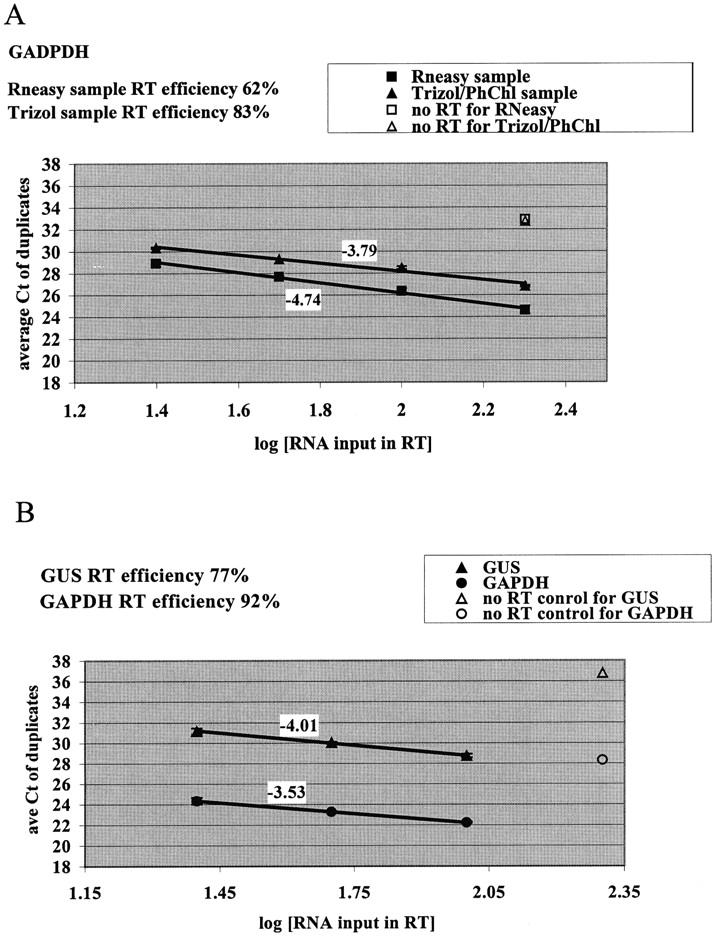
Efficient and linear measurements of housekeeping genes in clinical samples collected for cervical cancer screening by TaqMan. A: Increasing amounts of RNA in the RT reaction were used to measure GAPDH amplification using RNA extracted by RNeasy or Trizol/phenol-chloroform. The “no RT” controls contained the highest amount of RNA that was used in the RT reaction, but the RT enzyme was omitted. Each data point indicates average of duplicate determinations with error bars corresponding to the SD. Numbers next to the lines indicate the slopes. B: Simultaneous amplification of GAPDH (circles) and GUS (triangles) using RNA extracted with Trizol/phenol-chloroform from a single clinical sample. The “no RT” control contained the highest amount of RNA that was used in the RT reaction, but the RT enzyme was omitted. Each data point indicates average of duplicate determinations with error bars corresponding to the SD. Numbers next to the lines indicate the slopes.
RNA from clinical samples prepared with Trizol/phenol-chloroform demonstrated linear TaqMan measurements of GUS and GAPDH (Figure 3B) . The efficiency was calculated from the slope and was 77% for GUS and 92% for GAPDH. These data demonstrate reliable performance in the TaqMan assay for two common housekeeping genes using RNA from clinical Pap samples.
HPV Early Gene E7 Expression in Cervical Samples with High-Grade Dysplasia
HPV 16 and 18 are the most common HPV types present in high-grade dysplasia and cancer, 5 and were therefore selected as initial HPV types to be tested for E7 expression with TaqMan technology. HPV 16 E7 and HPV 18 E7 TaqMan PCR primers and probes were designed to produce short amplicons (82 and 81 bases, respectively) and were tested on cDNA from CaSki and HeLa human cervical cancer cell lines. CaSki (HPV 16) and HeLa (HPV 18) cervical cancer cell lines were used to measure the ability of HPV 16 and 18 E7 primer pairs to detect the cognate E7 mRNA (Figure 4) . While HPV 16 E7 primers were restricted to measuring E7 in cDNA from CaSki cells and did not measure HPV 18 E7 in cDNA from HeLa cells, HPV 18 E7 primers showed a low level of detection in cDNA from in CaSki cells. However the difference in Ct values for HPV 18 E7 between HeLa and CaSki cells was >10 cycles, indicating that the relative contribution of mismatched E7 primer-probes in clinical samples would be vanishingly small (>2−10).
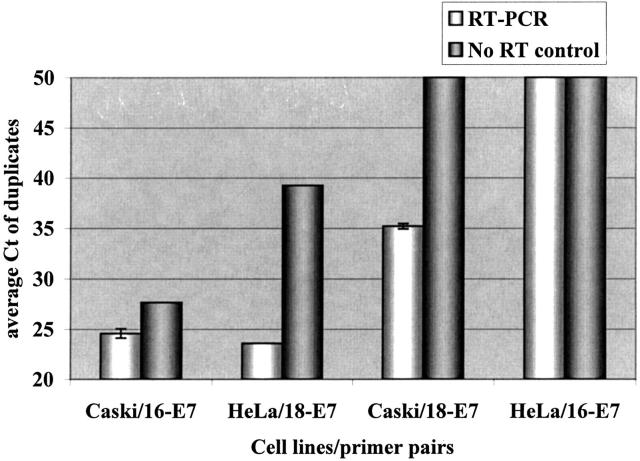
Figure 4.
Measurement of HPV 16 and 18 E7 in cervical cancer cell lines by TaqMan. The specificity of E7 primers for their respective HPV types were tested using RNA extracted from both Caski (HPV 16) and HeLa (HPV 18) cells (white bars). “No RT” controls (dark gray bars) were corresponding reactions in which the RT enzyme was omitted. Each data point indicates average of duplicate determinations with error bars corresponding to the SD.
HPV 16 and 18 E7 transcripts were measured in cervical samples collected from women with corresponding normal and high grade Papanicolaou smears (Table 2) . A sample was scored as positive for E7 expression when the CT value was at least four cycles lower than the CT value of the corresponding “no RT” control, representing a signal that was due to the presence of RNA and not contaminating DNA. Using this criterion, E7 RNA transcripts from HPV 16 or HPV 18 were detected in cells scraped from the cervix of 7 of 15 women with cytologically diagnosed high-grade dysplasia. Five high-grade samples demonstrated expression of either HPV 16 or 18 E7 RNA, and three high-grade samples demonstrated expression of E7 from both HPV 16 and 18 (Table 2) . None of the samples from women with cytologically normal Papanicolaou smears had detectable HPV 16 or 18 E7 RNA expression (Table 2) . The difference in E7 detection between dysplastic versus normal samples is statistically significant (P = 0.006).
Discussion
This report is the first demonstration that TaqMan RT-PCR can reliably measure gene expression in RNA extracted from samples collected for cervical cancer screening. The experimental use of cells scraped from the cervix to detect RNA transcripts has been previously exploited only in a limited number of studies and has required the use of two rounds of PCR amplification. 16, 17, 18 A large proportion of RNA extracted from the cervical cell samples was approximately 200 bp in length. Partial RNA degradation has previously been reported in cells from Papanicolaou smear samples and has been attributed to autolysis that occurs during the normal processes of epithelial cell maturation and senescence. 19 The presence of partially degraded RNA in cervical samples does not preclude efficient measurement of RNA transcripts as long as the amplicon size is kept small. 12 In our samples, measurements of reference gene expression were linear with respect to input RNA levels regardless of RNA extraction technique used on the clinical samples. As an automated, high throughput method, TaqMan is a suitable scientific platform for validation of potential cervical cancer screening markers.
Our data demonstrate that TaqMan detects HPV 16 or 18 E7 transcripts in 47% of clinical samples from women with high-grade dysplasia. HPV 16 or 18 are typically present in 47 and 5% of high-grade lesions respectively, 5 therefore, absence of detectable E7 transcripts in 53% of dysplastic samples is probably due to the presence of other HPV types in these samples not detected with our primer/probe combinations. Detection of HPV early gene transcripts in cervical dysplasia has been reported in a few studies. 16, 17, 20 The methods for E7 RNA detection described here are based on comparing the CT value with the corresponding “no RT” control, to measure the presence of E7 transcripts rather than the presence of viral DNA. HPV DNA is commonly detected in cytologically normal 6, 7, 8 and equivocal Pap smears such as low grade SIL and atypia, 21, 22 but the status of E7 transcription is unknown. High grade SIL and cancer have high viral loads and high rates of viral integration into the host cell genome, resulting in up-regulation of genes in the early region. 23, 24, 25 Therefore the detection of E7 RNA transcripts may be more specific than the detection of HPV DNA in the triage of women with minimally abnormal Pap smears. This work sets the stage for a clinical cohort study to measure the correlation between the presence of measurable E7 transcripts, HPV DNA, and disease status.
Acknowledgments
We thank Mamie Fung and Marielena Chavira for technical assistance with the TaqMan experiments, Dr. Supriya Shivakumar for sharing her expertise in molecular biology, and Dr. Vivian Weinberg for statistical analysis.
Address reprint requests to K. Smith-McCune, 2340 Sutter St, Room S-371, UCSF, San Francisco, CA 94143-0128. E-mail: kmccune@cc.ucsf.edu.
Footnotes
Supported by grants from the National Cancer Institute (CA80780), the Research Evaluation and Allocation Committee of UCSF School of Medicine, and Mount Zion Health Systems.
References
- 1.Parkin D, Pisani P, Ferlay J: Estimates of the worldwide incidence of eighteen major cancers in 1985. Int J Cancer 1993, 54:594-606 [DOI] [PubMed] [Google Scholar]
- 2.Papanicolaou G, Traut H: Diagnosis of uterine cancer by the vaginal smear. 1943. Oxford University Press, New York
- 3.Eddy D: Screening for cervical cancer. Ann Intern Med 1990, 113:214-226 [DOI] [PubMed] [Google Scholar]
- 4.Kurman R, Henson D, Herbst A, Noller K, Schiffman M: Interim guidelines for management of abnormal cervical cytology. JAMA 1994, 271:1866-1869 [PubMed] [Google Scholar]
- 5.Lorincz AT, Reid R, Jensen AB, Greenberg MD, Lancaster W, Kurman RJ: Human papillomavirus infection of the cervix: relative risk associations of 15 common anogenital types. Obstet Gynecol 1992, 79:328-337 [DOI] [PubMed] [Google Scholar]
- 6.Wheeler C, Parmenter C, Hunt W, Becker T, Greer C, Hildesheim A, Manos M: Determinants of genital human papillomavirus infection among cytologically normal women attending the University of New Mexico student health center. Sex Transm Dis 1993, 20:286-299 [DOI] [PubMed] [Google Scholar]
- 7.Bauer H, Hildesheim A, Schiffman M, Glass A, Rush B, Scott D, Cadell D, Kurman R, Manos M: Determinants of genital human papillomavirus infection in low-risk women in Portland, Oregon. Sex Transm Dis 1993, 20:274-278 [DOI] [PubMed] [Google Scholar]
- 8.Denny L, Kuhn L, Pollack A, Wainwright H, Wright TC, Jr: Evaluation of alternative methods of cervical cancer screening for resource-poor settings. Cancer 2000, 89:826-833 [DOI] [PubMed] [Google Scholar]
- 9.Lie YS, Petropoulos CJ: Advances in quantitative PCR technology: 5′ nuclease assays. Curr Opin Biotechnol 1998, 9:43-48 [DOI] [PubMed] [Google Scholar]
- 10.Heid CA, Stevens J, Livak KJ, Williams PM: Real time quantitative PCR. Genome Res 1996, 6:986-994 [DOI] [PubMed] [Google Scholar]
- 11.Gibson UE, Heid CA, Williams PM: A novel method for real time quantitative RT-PCR. Genome Res 1996, 6:995-1001 [DOI] [PubMed] [Google Scholar]
- 12.Godfrey TE, Kim SH, Chavira M, Ruff DW, Warren RS, Gray JW, Jensen RH: Quantitative mRNA expression analysis from formalin-fixed, paraffin- embedded tissues using 5′ nuclease quantitative reverse transcription-polymerase chain reaction. J Mol Diagn 2000, 2:84-91 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Maeda I, Takano T, Matsuzuka F, Maruyama T, Higashiyama T, Liu G, Kuma K, Amino N: Rapid screening of specific changes in mRNA in thyroid carcinomas by sequence specific-differential display: decreased expression of acid ceramidase mRNA in malignant and benign thyroid tumors. Int J Cancer 1999, 81:700-704 [DOI] [PubMed] [Google Scholar]
- 14.Leutenegger CM, Mislin CN, Sigrist B, Ehrengruber MU, Hofmann-Lehmann R, Lutz H: Quantitative real-time PCR for the measurement of feline cytokine mRNA. Vet Immunol Immunopathol 1999, 71:291-305 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Taubert H, Koehler T, Meye A, Bartel F, Lautenschlager C, Borchert S, Bache M, Schmidt H, Wurl P: mdm2 mRNA level is a prognostic factor in soft tissue sarcoma. Mol Med 2000, 6:50-59 [PMC free article] [PubMed] [Google Scholar]
- 16.Fujii T, Tsukazaki K, Kiguchi K, Kubushiro K, Yajima M, Nozawa S: The major E6/E7 transcript of HPV-16 in exfoliated cells from cervical neoplasia patients. Gynecol Oncol 1995, 58:210-215 [DOI] [PubMed] [Google Scholar]
- 17.Sotlar K, Selinka HC, Menton M, Kandolf R, Bultmann B: Detection of human papillomavirus type 16 E6/E7 oncogene transcripts in dysplastic and nondysplastic cervical scrapes by nested RT-PCR. Gynecol Oncol 1998, 69:114-121 [DOI] [PubMed] [Google Scholar]
- 18.Lin WM, Ashfaq R, Michalopulos EA, Maitra A, Gazdar AF, Muller CY: Molecular Papanicolaou tests in the twenty-first century: molecular analyses with fluid-based Papanicolaou technology. Am J Obstet Gynecol 2000, 183:39-45 [DOI] [PubMed] [Google Scholar]
- 19.Bhattacharya PK, Pappelis AJ: Changes in nucleic acid and protein content in nuclei of human cervical cells. Mech Ageing Dev 1984, 27:135-142 [DOI] [PubMed] [Google Scholar]
- 20.Durst M, Glitz D, Schneider A, zur Hausen H: Human papillomavirus type 16 (HPV 16) gene expression and DNA replication in cervical neoplasia: analysis by in situ hybridization. Virology 1992, 189:132-140 [DOI] [PubMed] [Google Scholar]
- 21.Koutsky L: Human Papillomavirus testing for triage of women with cytologic evidence of low-grade squamous intrapithelial lesions: baseline data from a randomized trial. J Natl Cancer Inst 2000, 92:397-402 [DOI] [PubMed] [Google Scholar]
- 22.Solomon D, Schiffman M, Tarone R: Comparison of three management strategies for patients with atypical squamous cells of undetermined significance: baseline results from a randomized trial. J Natl Cancer Inst 2001, 93:293-299 [DOI] [PubMed] [Google Scholar]
- 23.Stoler MH, Rhodes CR, Whitbeck A, Wolinsky SM, Chow LT, Broker TR: Human papillomavirus type 16 and 18 gene expression in cervical neoplasias. Hum Pathol 1992, 23:117-128 [DOI] [PubMed] [Google Scholar]
- 24.Vormwald-Dogan V, Fischer B, Bludau H, Freese UK, Gissmann L, Glitz D, Schwartz E, Durst M: Sense and antisense transcripts of human papillomavirus type 16 in cervical cancers. J Gen Virol 1992, 73:1833-1838 [DOI] [PubMed] [Google Scholar]
- 25.Daniel B, Rangarajan A, Mukherjee G, Vallikad E, Krishna S: The link between integration and expression of human papillomavirus type 16 genomes and cellular changes in the evolution of cervical intraepithelial neoplastic lesions. J Gen Virol 1997, 78:1095-1101 [DOI] [PubMed] [Google Scholar]