Abstract
Polymerase chain reaction with confronting two-pair primers (PCR-CTPP) is an inexpensive, time-saving genotyping method that is applicable for most single nucleotide polymorphisms. To date, we have applied PCR-CTPP successfully for the genotyping of more than 30 polymorphisms. This paper demonstrates the differences in DNA amplification among different annealing temperatures of PCR-CTPP with given melting temperatures for four primers. The NQO1 C609T (Pro187Ser) polymorphism was used as an example. Two sets of four primers were applied for PCR-CTPP; the first set with different melting temperatures (Tms), and the second with similar Tms. The comparisons with one-pair primer PCR (allele-specific PCR) revealed that PCR-CTPP amplified DNA more specifically than allele-specific PCR. The primers with different Tms caused competitive DNA amplification for heterozygous genotype. Four primers with similar Tms amplified both alleles unspecifically at a lower annealing temperature, while the same DNA samples were correctly genotyped under an optimal annealing temperature. These findings are unique for PCR-CTPP, and important characteristics when the primers and annealing temperatures in PCR-CTPP are designed. The knowledge of these characteristics will extend the applicability of PCR-CTPP for polymorphism genotyping.
Polymerase chain reaction (PCR) is the most common technique used for genotyping, which has promoted studies on the associations of genotypes with disease risk and prognosis. 1, 2, 3, 4 Several PCR methods are available and a method suitable to the nature of each study is adopted for genotyping. Type of polymorphism, accuracy of genotyping, number of samples, and available PCR equipment are factors to be taken into account when a PCR method is chosen for a given study.
Genotyping for variable number of tandem repeat (VNTR) polymorphisms can be conducted simply by PCR followed by electrophoresis, because DNA products with different sizes specific to each allele are amplified by PCR. PCR-RFLP (restriction fragment length polymorphism) requires another step for incubation with a restriction enzyme before electrophoresis. 5 Selected restriction enzyme digests DNA products with a particular base sequence, which enables it to distinguish the allele with the particular sequence from the alleles without it. It has been estimated that half of single nucleotide polymorphisms (SNPs) occur in natural restriction sites. 6 PCR-SSCP (single-strand conformation polymorphism) is a genotyping technique to detect differences in the conformation of single strand DNA, 7 which requires electrophoresis equipment to keep a constant temperature. Allele-specific PCR is an alternative for genotyping, 8 whose PCR condition is set to avoid unspecific DNA products. The PCR method is conducted for each allele, that is, for different alleles in different tubes.
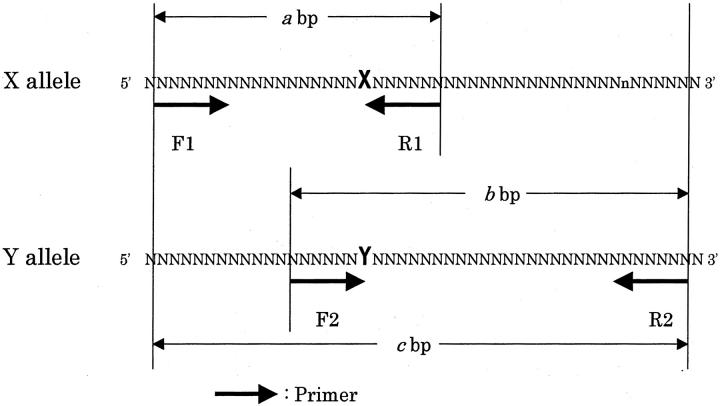
Polymerase chain reaction with confronting two-pair primers (PCR-CTPP) is a new genotyping method invented independently, 9, 10 which we recently found to be based on the same logic as bi-directional PCR amplification of specific alleles (Bi-PASA) whose real applications for genotyping have been infrequently reported. 11 Both methods for single nucleotide polymorphisms produce allele-specific DNA bands with different lengths by adding four designed primers into one tube containing an ordinarily prepared PCR mixture. The amplification of allele-specific bands with different lengths makes genotyping possible by electrophoresis without other steps. As shown in Figure 1 , the four primers consist of F1 and R1 for X allele amplifying a a-bp DNA and F2 and R2 for Y allele producing a b-bp DNA. F1 and R2 produce a c-bp common band. R1 and F2 confront each other at the 3′ end with the base specific to the allele. The difference between the two methods is only in the sequence design for the inner primers R1 and F2. Bi-PASA uses a G+C-rich tail at the 5′ end of the inner primers, 11, 12 while our method does not.
Figure 1.
Logic of polymerase chain reaction with confronting two-pair primers. At the 3′ end of inner primers R1 and F2, the base specific to each allele is designed. The difference between a-bp and b-bp should be large enough to be distinguishable by electrophoresis.
There is no doubt that, if applicable, PCR-CTPP is an inexpensive, time-saving method compared with PCR-RFLP and PCR-SSCP, although the research groups applying this method for genotyping are still limited. We have succeeded in the genotyping of more than 30 polymorphisms. 13 This paper demonstrates the effects of primer melting temperature (Tm) and PCR annealing temperature on the DNA amplification. The effects are exemplified by NAD(P)H:quinone oxidoreductase gene (NQO1) C609T(Pro187Ser) polymorphism. The enzyme detoxifies quinones and reduces oxidative stress. The T/T(Ser/Ser) genotype causes complete loss of enzyme activity, which was reportedly associated with acute myeloid leukemia, leukemia/myelodysplastic syndrome, 14, 15, 16, 17 colorectal cancer, 18 and lung cancer for smokers. 19
Materials and Methods
Genotyping
DNA was extracted from 200 μl of buffy coat preserved at −40°C by QIAamp DNA Blood mini kit (Qiagen Inc., Valencia, CA). PCR-CTPP for the NQO1 polymorphism was conducted using two sets of four primers. The first set of primers were as follows; F1: 5′-TAT CAG AGT GTC TTA CTG AGA (Tm at primer concentration 50 nmol/L and salt concentration 50 mmol/L, 46.4°C) and R1: 5′- AAT GCT ATA TGT CAG TTG AGG (51.6°C) for C allele amplifying a 161-bp band, F2: 5′-GTG GCT TCC AAG TCT TAG AAT (54.9°C) and R2: TTT CTA GCT TTG ATC TGG TTG (54.5°C) for T allele amplifying a 283-bp band. A 403-bp band was designed to be amplified between primers F1 and R2. Although all of the primers were 21 mers with 8 GC bases, the Tms estimated by a Tm prediction program 20 were different, as shown in the above parentheses. The Tms for F2 and R2 were higher than those for F1 and R1. Four primers of the second set had similar melting temperatures; F1′: 5′-CCT TAT CAG AGT GTC TTA CTG AGA (54.4°C) and R1′: 5′-CAA TGC TAT ATG TCA GTT GAG G (54.7°C), as well as F2 (54.9°C) and R2 (54.5°C). F1′ was three bases (CCT) longer than F1, and R1′ one base (C) longer than R1. Accordingly, the DNA amplified by the second set was 165 bp between F1′ and R1′ and 406 bp between F1′ and R2.
Genomic DNA (30 ng to 100 ng) was used in a volume of 25 μl with 0.18 mmol/L dNTPs, 12.5 pmol of each primer, 0.5 units of AmpliTaq Gold (Perkin-Elmer Corp., Foster City, CA), and 2.5 μl of 10X PCR buffer including 15 mmol/L MgCl2. GeneAmp PCR System 9700 (PE Biosystems, Foster City, CA) was used for PCR. The amplification was conducted by 10 minutes of initial denaturation at 95°C, followed by 35 cycles of 1 minute at 95°C, 1 minute at X°C, and 1 minute at 72°C, and a 5 minute final extension at 72°C, where annealing temperature, X, was a variable for this study. All PCR products were visualized on a 2% agarose gel containing a 2 μl/100 ml of ethidium bromide.
The results of PCR-CTPP genotyping were confirmed by PCR-RFLP with HinF1 enzyme, which produces 188-bp and 85-bp bands for C allele and 151-bp and 85-bp bands for T allele using primers F: 5′-AGT GGC ATT CTG CAT TTC TGT G, and R: 5′-GAT GGA CTT GCC CAA GTG ATG. 16 Forty-seven samples (18 for C/C, 20 for C/T, and 9 for T/T) were successfully genotyped, providing the same results as PCR-CTPP.
Results
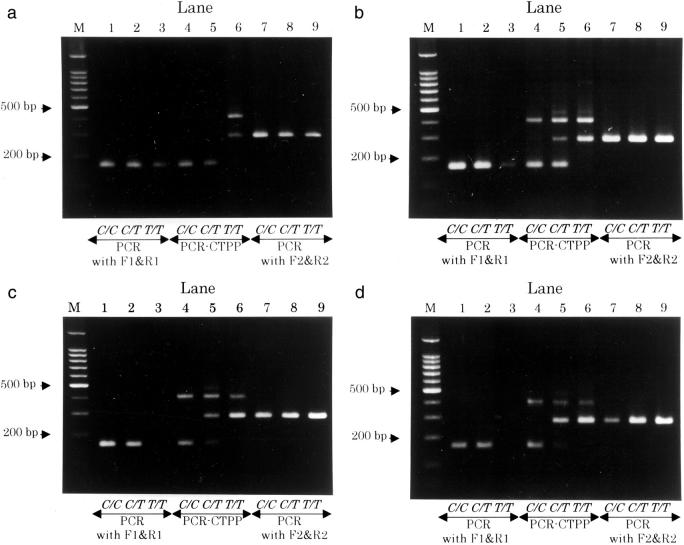
Figure 2 shows the gels for the first set of primers (F1, R1, F2, and R2) according to annealing temperature. DNA from three individuals with a different genotype of NQO1 C609T was used for this demonstration. At annealing temperature 59°C, one-pair PCR (allele-specific PCR) with F1&R1 or F2&R2 amplified DNA for all of the genotypes unspecifically, as shown in Figure 2 , lanes 1 to 3 and 7 to 9, respectively. PCR-CTPP produced a specific band for the homozygous genotypes; a 161-bp band for C/C genotype (lane 4) and a 283-bp band for T/T genotype (lane 6). For C/T genotype, two bands were amplified, but the strength of the amplified bands was competitive, depending on annealing temperature. At 59°C (Figure 2a) the 161-bp band for C allele was stronger, while at 64°C (Figure 2d) the 283-bp band for T allele was stronger. Figure 2b at 61°C and Figure 2c at 62°C was between them. Both allowed correct typing for the three genotypes, but not at the other temperatures. In allele-specific PCR, the allele specificity was observed between 61°C and 64°C for F1 and R1, but not for F2 and R2.
Figure 2.
Gel showing the genotypes for NAD(P)H:quinone oxidoreductase gene (NQO1), C609T(Pro187Ser) polymorphism by primers with different melting temperatures. Lane M contains a 100-bp DNA ladder, lanes 1 to 3 for C allele primers (F1 and R1), lanes 4 to 6 for four primers (F1, R1, F2, and R2), and lanes 7 to 9 for T allele primers (F2 and R2). PCR was conducted for template DNA from three individuals with C/C, C/T, or T/T genotype, from left to right. The annealing temperatures were 59°C (a), 61°C (b), 62°C (c), and 64°C (d).
For the second set of primers with similar Tms (F1′, R1′, F2, and R2), results performed at annealing temperatures 58°C and 63°C are shown. At 58°C (Figure 3a) , PCR-CTPP amplified both alleles (lanes 4 to 6), which was useless for genotyping, but PCR-CTPP at 63°C (Figure 3b) produced an acceptable specificity for the genotyping (lanes 4 to 6). Competitive DNA amplification for C/T genotype demonstrated by the first set of primers was not observed for the second set. When the annealing temperature was set at 66°C, no observable bands were amplified. Correct genotyping was possible at an annealing temperature between 62°C and 63°C.
Figure 3.
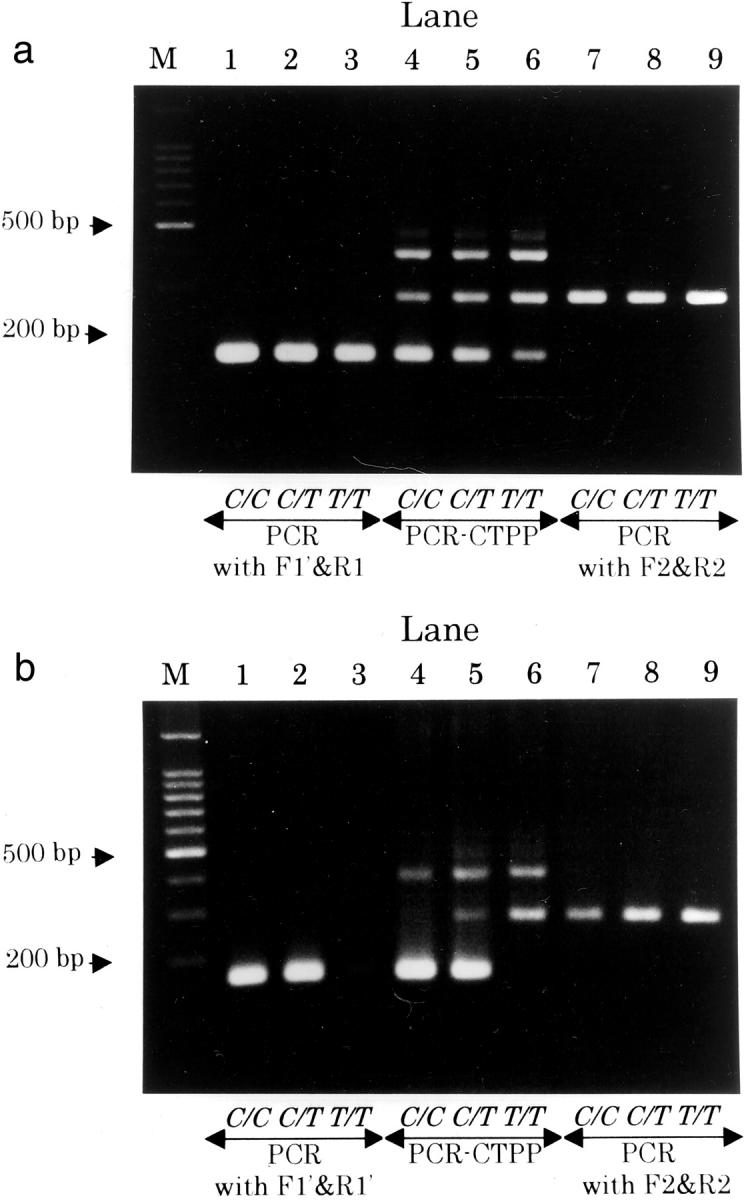
Gel showing the genotypes for NAD(P)H:quinone oxidoreductase gene (NQO1), C609T(Pro187Ser) polymorphism by primers with a similar melting temperatures. Lane M contains a 100-bp DNA ladder, lanes 1 to 3 for C allele primers (F1′ and R1′), lanes 4 to 6 for four primers (F1′, R1′, F2, and R2), and lanes 7 to 9 for T allele primers (F2 and R2). PCR was conducted for template DNA from three individuals with C/C, C/T, or T/T genotype, from left to right. The annealing temperatures were 58°C (a) and 63°C (b).
Discussion
This paper has reported technical points for PCR-CTPP, which is used only by a few research teams at present. However, we have successfully designed PCR-CTPP conditions for more than 30 polymorphisms. These genotyping conditions will be reported in our case-control studies elsewhere. There is no doubt that this method is applicable to most single nucleotide polymorphisms.
The accuracy of PCR-CTPP for other polymorphisms has been confirmed for IL-1B, 21 ALDH2, 22 FUT2, XRCC1, DRD2, and L-myc by PCR-RFLP, and for IL-1B, ALDH2, and FUT2 by DNA direct sequence. The DNA with a length expected from the sequence itself indicates that the PCR-CTPP amplified the part of polymorphism under examination, unless the highly homologous genes exist. The problem of highly homologous genes relates not only to PCR-CTPP, but also to PCR-RFLP and PCR-SSCP.
Through the process of finding a suitable condition, we learned that the annealing temperature is more important in PCR-CTPP than in PCR-RFLP. While PCR-RFLP requires a DNA amount enough for a digestion step, PCR-CTPP needs balanced amplification between the two allele-specific DNA products. If the actual Tm differs between the two-pair primers, DNA is amplified competitively as seen for the first set of four primers. When the Tm is similar among the four primers, the DNA products are balanced. In such cases, a low annealing temperature causes the unspecific amplification, which may be interpreted as erroneous heterozygous genotype for all samples. In either case for the Tm, an appropriate annealing temperature needs to be found by changing the estimated Tm. Since we have found that, in many instances, different Tms did not provide an optimal annealing temperature, we prefer similar Tms for the four primers. The range of annealing temperatures producing correct genotyping might be wider for primers with similar Tms than for those with different Tms.
Until recently we used a Tm value calculated by a method based solely on the number of bases and GC percentage for PCR-CTPP 23 to design primers with similar Tms. At that time it usually took more than 10 pilot PCRs to find a PCR-CTPP condition for clear genotyping. When a nearest-neighbor algorithm based on a base sequence 20 was used for the Tm estimation, we found that there was a substantial difference in the Tm for some primers between the two methods. All of the primer sets by which PCR-CTPP failed accurate genotyping had different Tm values, about 10°C of difference, even with the same number of bases and GC percentage. Roughly 50% of the designed PCR-CTPP had been aborted before establishing the condition; currently, 75% of the designed primers produce an acceptable level of accurate genotyping. Although the actual Tm may still differ from the estimate by the algorithm, 24 the estimated Tm based on base sequence is applicable to the primer design for PCR-CTPP. Since the Tm is important for this method, further basic studies will be desirable to establish a more comprehensive automated PCR-CTPP genotyping system.
Multiplex PCR is one of the promising applications for PCR-CTPP. We established two sets of duplex PCR-CTPP; independent duplex PCR-CTPP for IL-1B C-31T and IL-1RN VNTR polymorphisms, and related duplex PCR-CTPP for the FUT2 gene. 10 There are sets of polymorphisms to be genotyped at the same time for predicting disease susceptibility, eg, cytochrome p450s and glutathione S-transferases. 25 The primer design taking the Tm into account may produce a stable condition for multiplex PCR-CTPP, which will accelerate studies of genetic epidemiology. 3
In conclusion, the advantages and disadvantages of PCR-CTPP have become clear as the application increases. The competitive or unspecific amplification is quite an interesting finding unique to PCR-CTPP, which is one of the problems to disturb correct genotyping by PCR-CTPP. The problem is simply resolved by choosing four primers with similar Tms and by changing the annealing temperature. This method should quickly replace PCR-RFLP, because the digestion step can be skipped, resulting in lower costs and shorter genotyping times. For the researchers who wish to design the PCR-CTPP conditions, the findings reported in this paper are useful to avoid erroneous genotyping.
Acknowledgments
We thank Ms. Naomi Takeuchi for technical assistance.
Address reprint requests to Nobuyuki Hamajima, Division of Epidemiology and Prevention, Aichi Cancer Center Research Institute, 1–1 Kanokoden, Chikusa-ku, Nagoya 464-8681, Japan. E-mail: nhamajim@aichi-cc.jp.
Footnotes
Supported in part by a Grant-in-Aid for Scientific Research from the Ministry of Education, Science, Sports, Culture, and Technology of Japan.
References
- 1.Hussain SP, Harris CC: Molecular epidemiology of human cancer: contribution of mutation spectra studies of tumor suppressor genes. Cancer Res 1998, 58:4023-4037 [PubMed] [Google Scholar]
- 2.Perera FP: Molecular epidemiology: on the path to prevention? J Natl Cancer Inst 2000, 92:602-612 [DOI] [PubMed] [Google Scholar]
- 3.Schork NJ, Fallin D, Lanchbury JS: Single nucleotide polymorphisms and the future of genetic epidemiology. Clin Genet 2000, 58:250-264 [DOI] [PubMed] [Google Scholar]
- 4.Rock CL, Lampe JW, Patterson RE: Nutrition, genetics, and risk of cancer. Annu Rev Public Health 2000, 21:47-64 [DOI] [PubMed] [Google Scholar]
- 5.Erlich HA, Gelfand D, Sninsky JJ: Recent advances in the polymerase chain reaction. Science 1991, 252:1643-1651 [DOI] [PubMed] [Google Scholar]
- 6.Landegren U, Kaiser R, Sanders J, Hood L: A ligase-mediated gene detection technique. Science 1988, 241:1077-1080 [DOI] [PubMed] [Google Scholar]
- 7.Orita M, Iwahana H, Kanazawa H, Hayashi K, Sekiya T: Detection of polymorphisms of human DNA by gel electrophoresis as single-strand conformation polymorphisms. Proc Natl Acad Sci USA 1989, 86:2766-2770 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Takeda S, Ichii S, Nakamura Y: Detection of K-ras mutation in sputum by mutant-allele-specific amplification (MASA). Hum Mutat 1993, 2:112-117 [DOI] [PubMed] [Google Scholar]
- 9.Hamajima N, Saito T, Matsuo K, Kozaki K, Takahashi T, Tajima K: Polymerase chain reaction with confronting two-pair primers for polymorphism genotyping. Jpn J Cancer Res 2000, 91:865-868 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hamajima N: PCR-CTPP: a new genotyping technique in the era of genetic epidemiology. Exp Rev Mol Diagn 2001, 1:119-123 [DOI] [PubMed] [Google Scholar]
- 11.Liu Q, Thorland EC, Heit JA, Sommer SS: Overlapping PCR for bidirectional PCR amplification of specific alleles: a rapid one-tube method for simultaneously differentiating homozygotes and heterozygotes. Genome Res 1997, 7:389-398 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ye S, Dhillon S, Ke X, Collins AR, Day INM: An efficient procedure for genotyping single nucleotide polymorphisms. Nucleic Acids Res 2001, 29:e88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hamajima N, Matsuo K, Saito T, Hirose K, Inoue M, Takezaki T, Kuroishi T, Tajima K: Gene-environment interactions and polymorphisms studies of cancer risk in the Hospital-based Epidemiologic Research Program at Aichi Cancer Center II (HERPACC-II). Asian Pacific J Cancer Prev 2001, 2:99-107 [PubMed] [Google Scholar]
- 14.Wiemels JL, Pagnamenta A, Taylor GM, Eden GM, Alexander OB, Greaves MF, : United Kingdom Childhood Cancer Study Investigators: A lack of a functional NAD(P)H: quinone oxidoreductase allele is selectively associated with pediatric leukemias that have MLL fusions. Cancer Res 1999, 59:4095-4099 [PubMed] [Google Scholar]
- 15.Larson RA, Wang Y, Banerjee M, Wiemels J, Hartford C, Le Beau MM, Smith MT: Prevalence of the inactivating 609C->T polymorphism in the NAD(P)H: quinone oxidoreductase (NQO1) gene in patients with primary and therapy-related myeloid leukemia. Blood 1999, 94:803-807 [PubMed] [Google Scholar]
- 16.Naoe T, Takayama K, Yokozawa T, Kiyoi H, Seto M, Uike N, Ino T, Utsunomiya A, Maruta A, Jin-nai I, Kamada N, Kubota Y, Nakamura H, Shimazaki C, Horiike S, Kodera Y, Saito H, Ueda R, Wiemels J, Ohno R: Analysis of genetic polymorphism in NQO1, GST-M1, GST-T1, and CYP3A4 in 469 Japanese patients with therapy-related leukemia/myelodysplastic syndrome and de novo acute myeloid leukemia. Clin Cancer Res 2000, 6:4091-4095 [PubMed] [Google Scholar]
- 17.Smith MT, Wang Y, Kane E, Rollinson S, Wiemels JL, Roman E, Roddam P, Cartwright R, Morgan G: Low NAD(P)H: quinone oxidoreductase 1 activity is associated with increased risk of acute leukemia in adults. Blood 2001, 97:1422-1426 [DOI] [PubMed] [Google Scholar]
- 18.Lafuente MJ, Casterad X, Trias M, Ascaso C, Molina R, Ballesta A, Zheng S, Wiencke JK, Lafuente A: NAD(P)H: quinone oxidoreductase-dependent risk for colorectal cancer and its association with the presence of K-ras mutations in tumors. Carcinogenesis 2000, 21:1813-1819 [DOI] [PubMed] [Google Scholar]
- 19.Xu LL, Wain JC, Miller DP, Thurston SW, Su L, Lynch TJ, Christiani DC: The NAD(P)H: quinone oxidoreductase 1 gene polymorphism and lung cancer: differential susceptibility based on smoking behavior. Cancer Epidemiol Biomark Prev 2001, 10:303-309 [PubMed] [Google Scholar]
- 20.Breslauer KJ, Frank R, Blocker H, Marky LA: Predicting DNA duplex stability from the base sequence. Proc Natl Acad Sci USA 986, 83:3746–3750 [DOI] [PMC free article] [PubMed]
- 21.Hamajima N, Matsuo K, Saito T, Tajima K, Okuma K, Yamao K, Tominaga S: Interleukin 1 polymorphisms, lifestyle factors, and Helicobacter pylori infection. Jpn J Cancer Res 2001, 92:383-389 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Matsuo K, Hamajima N, Shinoda M, Hatooka S, Inoue M, Takezaki T, Tajima K: Gene-environment interaction between aldehyde dehydrogenase-2 (ALDH2) polymorphism and alcohol consumption for the risk of esophageal cancer. Carcinogenesis 2001, 22:913-916 [DOI] [PubMed] [Google Scholar]
- 23.Rychlik W, Spencer WJ, Rhoads RE: Optimization of the annealing temperature for DNA amplification in vitro. Nucleic Acids Res 1990, 18:6409-6412 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Akey JM, Sosnoski D, Parra E, Dios S, Hiester K, Su B, Bonilla C, Jin L, Shriver MD: Melting curve analysis of SNPs (McSNP): a gel-free and inexpensive approach for SNP genotyping. BioTechniques 2001, 30:358-367 [DOI] [PubMed] [Google Scholar]
- 25.Bartsch H, Nair U, Risch A, Rojas M, Wikman H, Alexandrov K: Genetic polymorphism of CYP genes, alone or in combination, as a risk modifier of tobacco-related cancers. Cancer Epidemiol Biomarkers Prev 2000, 9:3-28 [PubMed] [Google Scholar]