Abstract
Submarine mud volcanoes are formed by expulsions of mud, fluids, and gases from deeply buried subsurface sources. They are highly reduced benthic habitats and often associated with intensive methane seepage. In this study, the microbial diversity and community structure in methane-rich sediments of the Haakon Mosby Mud Volcano (HMMV) were investigated by comparative sequence analysis of 16S rRNA genes and fluorescence in situ hybridization. In the active volcano center, which has a diameter of about 500 m, the main methane-consuming process was bacterial aerobic oxidation. In this zone, aerobic methanotrophs belonging to three bacterial clades closely affiliated with Methylobacter and Methylophaga species accounted for 56% ± 8% of total cells. In sediments below Beggiatoa mats encircling the center of the HMMV, methanotrophic archaea of the ANME-3 clade dominated the zone of anaerobic methane oxidation. ANME-3 archaea form cell aggregates mostly associated with sulfate-reducing bacteria of the Desulfobulbus (DBB) branch. These ANME-3/DBB aggregates were highly abundant and accounted for up to 94% ± 2% of total microbial biomass at 2 to 3 cm below the surface. ANME-3/DBB aggregates could be further enriched by flow cytometry to identify their phylogenetic relationships. At the outer rim of the mud volcano, the seafloor was colonized by tubeworms (Siboglinidae, formerly known as Pogonophora). Here, both aerobic and anaerobic methane oxidizers were found, however, in lower abundances. The level of microbial diversity at this site was higher than that at the central and Beggiatoa species-covered part of the HMMV. Analysis of methyl-coenzyme M-reductase alpha subunit (mcrA) genes showed a strong dominance of a novel lineage, mcrA group f, which could be assigned to ANME-3 archaea. Our results further support the hypothesis of Niemann et al. (54), that high methane availability and different fluid flow regimens at the HMMV provide distinct niches for aerobic and anaerobic methanotrophs.
Large amounts of methane are stored in the subsurface ocean as crystalline gas hydrate, dissolved in porewater, and as free gas. Most of the methane which rises from subsurface reservoirs is consumed by anaerobic microorganisms inhabiting sulfate-penetrated sediment layers before it reaches the seafloor (65). This microbially mediated anaerobic oxidation of methane (AOM) controls the emission of the greenhouse gas methane from the ocean to the atmosphere (see reference 23 and references therein).
In marine habitats, the metabolic process of AOM is proposed to be a reversed methanogenesis coupled to the reduction of sulfate involving methanotrophic archaea (ANME archaea) and sulfate-reducing bacteria (SRB). ANME archaea and SRB are assumed to interact syntrophically (26) and form microbial consortia which oxidize methane with equimolar amounts of sulfate, yielding bicarbonate and sulfide (26, 51, 52). Sulfide produced as a by-product of AOM at cold seeps often supports chemosynthetic communities which derive energy from sulfide oxidation. Three archaeal clades (ANME-1, ANME-2, and ANME-3) oxidize methane under anoxic marine conditions (5, 54, 58, 59). ANME-1 archaea, distantly related to Methanosarcinales and Methanomicrobiales, occur in sediments mostly as single cells (35, 53, 58) but also aggregated (56, 59) or associated with SRB in microbial mats (mat-type consortia [35, 46]). ANME-2 archaea belong to the order Methanosarcinales and are usually associated with SRB of the Desulfosarcina-Desulfococcus (DSS) branch, forming structured consortia (shell-type and mixed-type ANME-2/DSS aggregates [5, 35]). Very recently, we identified ANME-3 archaea related to the genera Methanococcoides and Methanolobus which live syntrophically with SRB of the Desulfobulbus (DBB) branch (ANME-3/DBB aggregates) (54). Today, neither members of the ANME groups nor their sulfate-reducing bacterial partners have been obtained in pure culture. The enzymes and biochemical pathways of AOM remain largely unknown. However, recent metagenomic studies have identified a modified methyl-coenzyme M-reductase (“methanase”) which may catalyze the activation of methane under anoxic conditions (20, 21, 36).
In contrast to AOM, the microbially mediated aerobic oxidation of methane appears to be only a minor biological sink of methane in the ocean. Most known aerobic methanotrophs belong to the classes Alphaproteobacteria and Gammaproteobacteria and depend on the availability of oxygen (22). Hence, in marine habitats aerobic methanotrophs are restricted to the oxic water column and sediment-water interface or exist as symbionts of mussels, clams, and tubeworms.
This study provides for the first time a detailed quantitative analysis of the composition and distribution of methanotrophic guilds at an active methane-seeping mud volcano. The Haakon Mosby Mud Volcano (HMMV) is a submarine mud volcano and is the only one in a polar region which has been studied in great detail (8, 16, 18, 25, 38, 47, 61, 62, 68, 80, 81). Very recently, we showed that the HMMV hosts three key microbial communities (Fig. 1): aerobic methanotrophic bacteria at the volcano center, anaerobic methanotrophic archaea of the ANME-2 group below siboglinid tubeworms, and the new clade of ANME-3 archaea associated with Beggiatoa mats (54). These populations are highly influenced by the upward flow of sulfate- and oxygen-free fluids which restrict the availability of these electron acceptors for methane oxidation. The biogeochemistry of the HMMV is discussed in detail by De Beer et al. (9) and Niemann et al. (54). In this study, we further investigated the diversity and distribution of methanotrophic guilds at the HMMV and present detailed phylogenetic, microscopic, and quantitative analysis of the novel ANME-3 consortia as well as other bacteria and archaea at the HMMV. Furthermore, we analyzed the diversity of genes encoding the alpha subunit of methyl-coenzyme M-reductase (mcrA), which is the key enzyme of methanogenesis and identified a new lineage of mcrA. This mcrA type could be retrieved from sorted ANME-3 aggregates and thereby assigned to ANME-3 archaea.
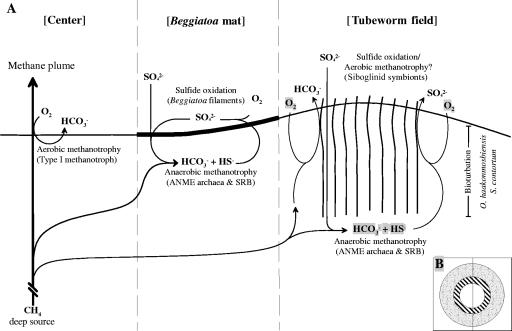
FIG. 1.
Schematic diagram of the HMMV. (A) Dominant methane-consuming processes in sediments of the different sampling sites. (B) Circular zonation of the mud volcano. The inner circle represents uncovered sediments of the thermal center, the surrounding area is covered with Beggiatoa mats, and the rim of the crater is populated by siboglinid tubeworms.
MATERIALS AND METHODS
Site description and sample collection.
The HMMV is an active cold seep situated at the Norwegian-Barents-Spitzbergen continental margin (72°00.25′N, 14°43.50′E) in a water depth of 1,250 m. Sediment samples were collected in August 2001 and June/July 2003 during joint cruises of the Alfred Wegener Institute and IFREMER with research vessels L'Atalante and Polarstern, respectively, and the remotely operated vehicle (ROV) VICTOR 6000 (IFREMER) in the framework of a German/French collaborative program (33). During dives with VICTOR 6000, signs of recent mud expulsion were observed in the northern part of the HMMV. The center is a flat circular area of about 500 m in diameter (0.11 km2). It is encircled by sulfidic sediments covered with dense Beggiatoa mats in a stretch of about 40 to 120 m width (0.22 km2). The outer rim of HMMV is composed of hill and trench sections, elevated about 1 to 8 m higher than the center, and covered by dense accumulations of thread-like tubeworms (Siboglinidae) for a zone of 60 to 200 m (0.41 km2). The two dominant species have been identified as Sclerolinum contortum and Oligobrachia haakonmosbiensis (70). The tubes of the worms have a diameter of a few millimeters and reach lengths of up to 60 cm. Only a small part of the tube extends up to 5 cm into the water column, and the main part is buried in the sediments. Siboglinid tubeworms are known to harbor endosymbionts, which can be either thiotrophic or methanotrophic (66, 72).
Sediment samples were recovered by video-guided multiple corer (MUC) or ROV-operated push cores at different zones of HMMV as follows: two cores as replicates from center sediments (station ATL25, dive 5, site A, 72°00.22′N, 14°43.50′E; station PS64-312, MUC core, 72°00.25′N, 14°43.49′E), three cores as replicates from the Beggiatoa mat site (station ATL19, dive 4, 72°00.19′N, 14°43.67′; station PS317-2, dive 219, PC14/17, 72°00.16′N, 14°43.88′E; station PS322, MUC core, 72°00.18′N, 14°43.85′E), and three cores as replicates of sediments from the tubeworm field (station ATL22, MUC core, 72°00.08′N, 14°43.39′E; station PS64-326, dive 220, PC2, 72°00.06′N, 14°42.13′E; PS64-356, MUC core, 72°00.05′N, 14°44.18′E). The sediment biogeochemistries of the sampled sites are further described by De Beer et al. (9) and Niemann et al. (54).
DNA extraction, PCR amplification, and clone library construction.
We constructed one bacterial and one archaeal 16S rRNA gene library from each habitat to gain first insights into the diversity. For DNA extraction, cores were sectioned in 1-cm-thick layers and frozen at −20°C. Total community DNA was directly extracted from 2 g of wet sediments (center station ATL25) and Beggiatoa mats (station ATL19) at 0- to 5-cm depths and from the tubeworm field (station ATL22) at 6- to 17-cm depths by using the method of Zhou et al. (85). Crude DNA was purified with the Wizard DNA clean-up kit (Promega, Madison, WI). Domain-specific primers were used to amplify almost-full-length 16S rRNA genes from the extracted chromosomal DNAs by PCR: for Bacteria, primers GM3F (Escherichia coli 16S rRNA position 0008) (50) and EUB1492 (32) were used, and for Archaea, primers 20f (44) and Uni1392R (37) or 20f and Arch958R (10) were used. In addition to 16S rRNA genes, fragments of the gene coding for the alpha subunit of methyl-coenzyme M-reductase (mcrA) were amplified using the primers ME1 and ME2 (19). PCRs were performed and products purified as described previously (64). DNA was ligated in the pGEM T-Easy vector (Promega, Madison, WI) or the pCR4 TOPO vector (Invitrogen, Carlsbad, CA) and transformed into E. coli TOP10 cells (Invitrogen, Carlsbad, CA) according to the manufacturer's recommendations.
Sequencing and phylogenetic analysis.
Sequencing was performed by Taq cycle sequencing with a model ABI377 sequencer (Applied Biosystems). The presence of chimeric sequences in the clone libraries was determined with the CHIMERA_CHECK program of the Ribosomal Database Project II (Center for Microbial Ecology, Michigan State University [http://wdcm.nig.ac.jp/RDP/html/analyses.html]). Potential chimeras were eliminated before phylogenetic trees were constructed. Sequence data were analyzed with the ARB software package (41). Phylogenetic trees of 16S rRNA gene sequences were calculated by parsimony, neighbor-joining, and maximum-likelihood analysis with different sets of filters. For tree calculation, only almost-full-length sequences were considered. Partial sequences were inserted into the reconstructed tree by parsimony criteria without allowing changes in the overall tree topology. The phylogenetic tree of the protein-encoding gene mcrA was generated from deduced amino acid sequences by neighbor-joining analysis with a 30% amino acid frequency filter.
Fluorescence in situ hybridization (FISH).
Subsamples of sediment cores were sliced into 1-cm intervals, fixed with formaldehyde, and treated by sonication as described previously (63). Hybridization with monolabeled probes and microscopy counts of hybridized and DAPI (4′,6′-diamidino-2-phenylindole)-stained cells were performed as described previously (71). In situ hybridizations with horseradish peroxidase (HRP)-labeled probes followed by tyramide signal amplification (also known as catalyzed reporter deposition [CARD]) were carried out as described by Pernthaler et al. (60), except for cell wall permeabilization. For permeabilization of rigid archaeal cell walls, filters were incubated in 0.01 M HCl for 10 min at room temperature, subsequently incubated in 0.1 M HCl (1 min) or 1% Triton X-100 (10 min), and finally washed in MilliQ water and dehydrated in absolute ethanol. Hybridized samples were examined with an epifluorescence microscope (Axiophot II; Carl Zeiss, Jena, Germany). For each probe and sample, 700 to 1,000 DAPI-stained cells in 10 to 20 independent microscopic fields were counted. Micrographs of microbial aggregates were taken with a confocal laser scanning microscope (LSM510; Carl Zeiss, Jena, Germany). Probes were purchased from biomers.net (Ulm, Germany). Probe sequences and formamide concentrations required for specific hybridization are given in Table 1. The specificity of new probes was evaluated against reference strains having one or more mismatches.
TABLE 1.
Oligonucleotide probes used in this study
| Probe | Specificity | Probe sequence (5′-3′) | Target sitea 16S rRNA positions | % FAb | Reference |
|---|---|---|---|---|---|
| NON 338c | Background control | ACTCCTACGGGAGGCAGC | 338-355 | 10 | 82 |
| GCTGCCTCCCGTAGGAGT | |||||
| EUB338 I-IIIc | Most Bacteria | GCAGCCACCCGTAGGTGT | 338-355 | 35 | 7 |
| GCTGCCACCCGTAGGTGT | |||||
| Arch915c | Most Archaea | GTGCTCCCCCGCCAATTCCT | 915-935 | 35 | 75 |
| DSS658c | Desulfosarcina spp., Desulfofaba spp., Desulfococcus spp., Desulfofrigus spp. | TCCACTTCCCTCTCCCAT | 658-685 | 60 | 43 |
| DSV698c | Desulfovibrio spp. | GTTCCTCCAGATATCTACGG | 698-717 | 35 | 43 |
| DSR651c | Desulforhopalus spp. | CCCCCTCCAGTACTCAAG | 651-668 | 35 | 43 |
| DSB985c | Desulfobacter spp., Desulfobacula spp. | CACAGGATGTCAAACCCAG | 985-1003 | 20 | 43 |
| 660c | Desulfobulbus spp. | GAATTCCACTTTCCCCTCTG | 660-679 | 60 | 11 |
| DBB305c | Desulfobulbus sp.-related sequences | AGTGCCAGTGTGACGGAT | 305-322 | 25 | 54 |
| DBBA655 | Desulfobulbus sp.-related sequences (ANME-3 partner group) | CACTTTCCCCTCTAGTAC | 655-672 | 35 | This study |
| CF319ac | Cytophaga/Flavobacterium | TGGTCCGTGTCTCAGTAC | 319-336 | 35 | 43 |
| MetI-444c | HMMV-MetI (Gammaproteobacteria) | CCTGCCTGTTTTCCTCCC | 444-461 | 60 | 54 |
| MetI-851 | HMMV-MetI (Gammaproteobacteria) | TACGTTAGCTCCGCCACT | 851-868 | 75 | This study |
| MetII-844c | HMMV-MetII (Gammaproteobacteria) | GCTCCACCACTAAGACCT | 844-861 | 75 | 54 |
| MetII-457 | HMMV-MetII (Gammaproteobacteria) | GCCTGATATTATCTCAGC | 457-474 | 20 | This study |
| MPH-732c | HMMV-MPH (Gammaproteobacteria) | GTAATGGCCCAGTGAGTC | 732-749 | 40 | 54 |
| ANME1-350c | ANME-1 archaea | AGTTTTCGCGCCTGATGC | 350-367 | 40 | 5 |
| EelMS932c | ANME-2 archaea | AGCTCCACCCGTTGTAGT | 932-949 | 40 | 5 |
| ANME3-397c,f | ANME-3 archaea | ATATGCTGGCACTCAGTG | 397-414 | 40 | This study |
| ANME3-1249c,d,e | ANME-3 archaea | TCGGAGTAGGGACCCATT | 1250-1267 | 20c/40d | 54 |
| ANME-3-1249H3 | Helper probe for ANME-3-1249 | GTC CCA ATC ATT GTA GCC GGC | 1229-1249 | This study | |
| ANME-3-1249H5 | Helper probe for ANME-3-1249 | TTA TGA GAT TAC CAT CTC CTT | 1268-1288 | This study | |
| Cren-444d | HMMV-Cren (Crenarchaeota) | CCCCCAGCTTTATACACT | 444-461 | 55 | This study |
E. coli numbering as in reference 5a.
Percent (vol/vol) formamide (FA) in hybridization buffer for hybridization.
Used as a monolabeled probe.
Used as an HRP-conjugated probe.
Monolabeled probe ANME-3-1249 used together with unlabeled helper probes ANME-3-1249H3 and ANME-3-1249H5.
Non-group hit (zero mismatches): Methanohalobium evestigatum.
Sorting of ANME-3 aggregates by flow cytometry.
For flow cytometric sorting, a fixed and sonicated subsample from an HMMV enrichment culture (culture conditions according to the method described in reference 51, incubated at 4°C) was hybridized in 800 μl buffer containing 0.9 M sodium chloride, 0.1% sodium dodecyl sulfate, 20 mM Tris-HCl (pH 7.5), 20% formamide (according to the ANME-3 probe requirements), 2.5 ng μl−1of Cy3-labeled oligonucleotide probe ANME3-1249, and 1.25 ng μl−1 of each helper probe (ANME-3-1249H3 and ANME-3-1249H5) at 46°C. After 4 h of incubation, hybridization was stopped by the addition of 3 ml of washing buffer containing 0.215 M sodium chloride, 0.1% sodium dodecyl sulfate, 20 mM Tris-HCl (pH 7.5), and 5 mM EDTA (pH 8.0).
Cell sorting was done with a MoFlow flow cytometer (Cytomation Inc., Fort Collins, CO) similar to the procedure described by Sekar et al. (67). For the excitation of Cy3-labeled cells, the laser was tuned to a wavelength of 514 nm (500 mW). Side-angle light scatter (SSC) was detected through a band-pass filter (530 ± 20 nm; Cytomation). Orange fluorescence from hybridized cells was detected by using a band-pass filter (570 ± 20 nm). The system threshold was set for SSC. The instrument was aligned by use of polychromatic 0.5-μm calibration beads (Polysciences, Warrington, PA). Online analysis, sort control, and postanalysis were done with Summit software, version 3.1 (Cytomation). Probe-positive cells were detected in a bivariant dot plot diagram of SSC versus Cy3. The instrument was sterilized by subsequent runs of 70% ethanol and autoclaved distilled water. It was kept under sterile conditions during sorting by use of an in-line filter (filter cartridge; pore size, 0.2 μm; Pall, Ilfracombe, United Kingdom) and autoclaved sheath fluid.
Bacterial and archaeal 16S rRNA genes as well as mcrA genes were amplified by PCR directly from the sorted fraction using the primer combinations described above.
Total cell counts and size measurements of ANME-3 aggregates.
Total counts of individual cells were done by epifluorescence microscopy after staining with acridine orange according to the method of Meyer-Reil (45). Aggregates were counted after in situ hybridization. Total cell counts were defined as the sums of the numbers of single cells and estimated numbers of aggregated cells present in ANME-3 consortia. Since single cells in an aggregate cannot be counted exactly, a semiquantitative method was applied. In total, 172 individual aggregates of the sediment horizon with the highest aggregate abundance at the Beggiatoa site (ATL19; 1 to 2 cm) were measured after staining with DAPI. Average volumes were 150 μm3 for spherical aggregates and 330 μm3 for nonspherical aggregates. Cell numbers in the aggregates were calculated based on the assumption that aggregates are either spheres or cylinders consisting of ANME-3 cells with a volume of 0.2 μm3. This estimate was corrected by the packing density (74%; http://mathworld.wolfram.com/SpherePacking.html) for the closest packing of spheres. Since most aggregates were associated with only very few SRB, SRB cell numbers were neglected during calculation.
Nucleotide sequence accession numbers.
The sequence data reported here will appear in the EMBL, GenBank, and DDBJ nucleotide sequence databases under the accession no. AJ579313 to AJ579330, AJ704632 to AJ704644, AJ704654 to AJ704724, AM404330 to AM404331, and AM407728 to AM407730.
RESULTS
Bacterial diversity in HMMV sediments.
In total, 254 bacterial clones (from center sediments, 72; from Beggiatoa mats, 132; from the tubeworm field, 50) were analyzed. The compositions of the bacterial 16S rRNA gene libraries for the different sampling sites differed remarkably. Many HMMV 16S rRNA gene clones showed high sequence similarity (>97%) to sequences of uncultivated microorganisms from other methane-rich habitats.
In oxic sediments of the volcano center, the least bacterial diversity was observed. Applying a <97% 16S rRNA sequence similarity criterion (27), we found 13 phylotypes, of which 10 were related to methylotrophic Gammaproteobacteria (Methylobacter and Methylophaga) (Fig. 2). Only few sequences grouped within two other phylogenetic lineages, namely the Bacteroidetes phylum and the WCHB1 cluster (sequences isolated from a hydrocarbon- and chlorinated-solvent-contaminated aquifer) (15).
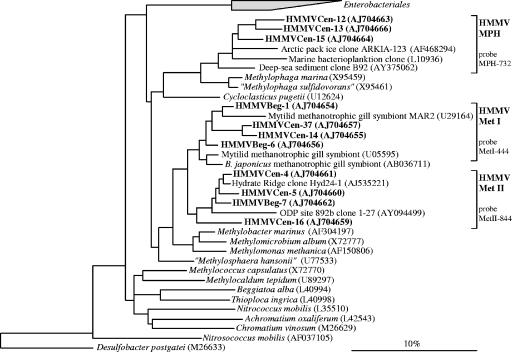
FIG. 2.
Phylogenetic tree showing the affiliations of HMMV 16S rRNA gene sequences to selected sequences of the Gammaproteobacteria. The tree was calculated with nearly full-length sequences (>1,450 bp) by maximum-likelihood analysis in combination with filters, which consider only 50% conserved regions of the 16S rRNA. Branching orders that were not supported by all calculation methods are shown as multifurcations. Partial sequences were subsequently inserted into the reconstructed consensus tree by parsimony criteria with global/local optimization, without allowing changes in the overall tree topology. Clone sequences from HMMV sediments are in boldface type. Clones with designations beginning HMMVCen are from center sediments; clones with designations beginning HMMVBeg are from the Beggiatoa site. The bar represents 10% estimated phylogenetic divergence. B. japonicus, Bathymodiolus japonicus.
In the clone library from anoxic sediments below Beggiatoa mats 25 phylotypes were detected. Most sequences were affiliated with Bacteroidetes phylum (five phylotypes, 59 sequences total). Additionally, aerobic methanotrophic Gammaproteobacteria (seven phylotypes, 37 sequences total) and Deltaproteobacteria related sequences (seven phylotypes, 22 sequences total) (Fig. 3) were abundant at this site.
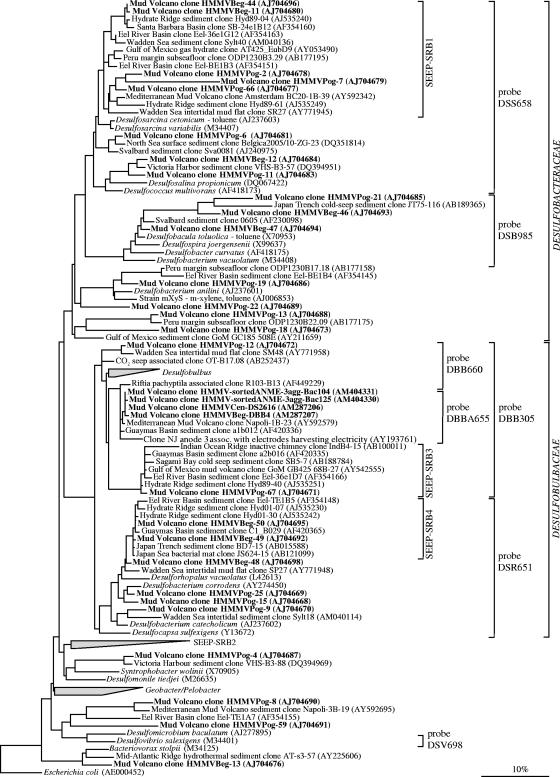
FIG. 3.
Phylogenetic tree showing the affiliations of HMMV 16S rRNA gene sequences to selected sequences of the Deltaproteobacteria. The tree was calculated with nearly full-length sequences (>1,450 bp) by maximum-likelihood analysis in combination with filters, which consider only 50% conserved regions of the 16S rRNA of Deltaproteobacteria. Branching orders that were not supported by all calculation methods are shown as multifurcations. Partial sequences were subsequently inserted into the reconstructed consensus tree by parsimony criteria, without allowing changes in the overall tree topology. Clone sequences from HMMV sediments are in boldface type. Clones with designations beginning HMMVCen are from center sediments, clones with designations beginning HMMVBeg are from the Beggiatoa site, clones with designations beginning HMMVPog are from the tubeworm site, and clones with designations beginning HMMV-sortedANME-3agg are from sorted ANME-3/DBB aggregates. The bar indicates 10% estimated phylogenetic divergence.
The level of bacterial diversity, in particular, the level of diversity of SRB within the class Deltaproteobacteria, was highest in the clone library from the tubeworm site with 37 phylotypes and 23 phylotypes, respectively (Fig. 3). Other phylogenetic lineages were only rarely retrieved (one to three sequences) and included the Actinobacteria (one phylotype), Acidobacterium/Holophaga (three phylotypes), Planctomycetes (two phylotypes), Verrucomicrobia (one phylotype), Betaproteobacteria (one phylotype), the Nitrospira lineage (one phylotype), and the uncultured OP8 lineages (one phylotype).
Methanotrophic and methylotrophic Gammaproteobacteria.
HMMV clone sequences related to aerobic methanotrophic Gammaproteobacteria were retrieved from surface sediments of the center (67 of 72 sequences) or from the adjacent Beggiatoa mat area (37 of 132 sequences) but not from the tubeworm site. Sediments at both sites were methane-rich and showed an in situ oxygen penetration depth of only a few millimeters or less (9). Three sequence clades could be defined: HMMV-MetI, HMMV-MetII, and HMMV-MPH (Fig. 2). HMMV-MetI (six phylotypes) and HMMV-MetII (one phylotype) were most abundant in the libraries and closely related to each other. The members of both groups belong to the order Methylococcales. HMMV-MetI 16S rRNA gene sequences were most closely related (94 to 97%) to sequences of methanotrophic mussel symbionts (13, 14). HMMV-MetII were most similar (96 to 99%) to clone sequence Hyd24-1 from gas hydrate-bearing sediments of Hydrate Ridge (34). Methylobacter marinus was the closest cultivated relative of both clusters (89 to 97%). The third gammaproteobacterial clade, HMMV-MPH, was related (87 to 94%) to the methylotrophic genus Methylophaga. The most similar sequences (92 to 100%) were obtained from marine bacterioplankton.
Deltaproteobacteria.
Sequences within the class Deltaproteobacteria were highly diverse (Fig. 3), and the highest diversity was found at the tubeworm site (23 phylotypes; 34 sequences total). Many known orders (Desulfovibrionales, Desulfobacterales, Synthrophobacterales, and Bdellovibrionales) were represented. Most clone sequences from sediments below Beggiatoa mats fell into the recently defined seep-specific clades of SRB (34), SEEP-SRB1 (Desulfosarcina/Desulfococcus-related), SEEP-SRB3 (Desulfobulbus-related), and SEEP-SRB4 (Desulforhopalus-related). Members of SEEP-SRB1 are supposed to be the sulfate-reducing partners of ANME-1 and ANME-2, while the ecological role of SEEP-SRB3 and SEEP-SRB4 is still unknown. Further HMMV sequences are related to Desulfobacterium anilii and its relatives. This group is known to include several cultivated as well as uncultivated hydrocarbon-degrading bacteria capable of complete oxidation of various aromatic hydrocarbons (84). Additionally, non-SRB sequences of the Deltaproteobacteria were also present in HMMV clone libraries (Fig. 3): a minor fraction of clone sequences (one phylotype) was related to Bacteriovorax stolpii (90 to 91%). Bacteriovorax spp. and its relatives are aerobic, obligately predatory bacteria that forage on a wide variety of susceptible gram-negative microorganisms (2) and thereby may control bacterial abundance. They have been found in a wide range of aquatic habitats, e.g., in freshwater and marine sediments and biofilms (31).
Archaeal diversity in HMMV sediments.
In total, 159 archaeal clones (from center sediments, 67; from Beggiatoa mats, 48; from the tubeworm field, 44) were analyzed. These were affiliated with only five different archaeal groups. The differences between the sampling sites were substantial. In the library of center sediments, only sequences belonging to the crenarchaeotal marine benthic group C (MBGC) were found (two phylotypes; 67 clone sequences total) (Fig. 4). This group has often been found at methane seeps (76, 79) but is not restricted to seeps (6, 39). Within MBGC, HMMV sequences form a separate branch (hereafter referred to as HMMV-Cren) of highly similar sequences often exceeding 99% similarity. In contrast, the Beggiatoa mat site contained only sequences affiliated with the novel euryarchaeotal clade named ANME-3 (35) (two phylotypes; 48 clone sequences total); HMMV data are shown in reference 54.
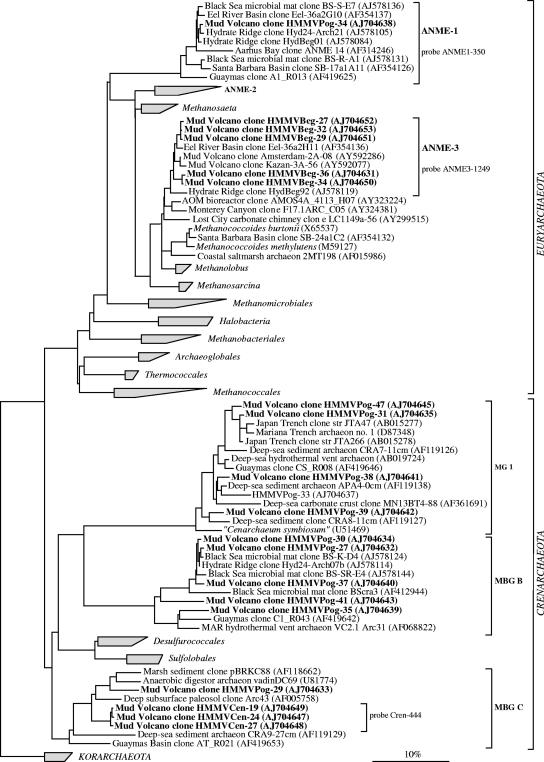
FIG. 4.
Phylogenetic tree showing the affiliations of HMMV 16S rRNA gene sequences to selected sequences of the domain Archaea. The tree was calculated with nearly full-length sequences (>1,300 bp) by maximum-likelihood analysis in combination with filters, which consider only 50% conserved regions of the 16S rRNA of Archaea. Branching orders that were not supported by all calculation methods are shown as multifurcations. Partial sequences were subsequently inserted into the reconstructed consensus tree by parsimony criteria, without allowing changes in the overall tree topology. Clone sequences from HMMV sediments are in boldface type. Clones with designations beginning HMMVCen are from center sediments, clones with designations beginning HMMVBeg are from the Beggiatoa site, and clones with designations beginning HMMVPog are from the tubeworm site. The bar indicates 10% estimated phylogenetic divergence. MBG, marine benthic group; MG, marine group.
The archaeal 16S rRNA gene library from the tubeworm site was again most diverse (13 phylotypes) and dominated by Crenarchaeota of the marine benthic group B (alternatively referred to as the deep-sea archaeal group, with five phylotypes and 40 clone sequences total). Marine benthic group B is generally associated with methane-rich surface and subsurface environments, but its function remains unknown (3, 29, 35). Other retrieved phylotypes belonged to euryarchaeotal ANME-1 archaea (one phylotype) as well as the crenarchaeotal groups MBGC (two phylotypes) and marine group I archaea (six phylotypes) (Fig. 4). Members of the ANME-2 group were detected with FISH in sediments at the base of the tubeworm roots (54) but were not found in the clone library from the surface sediments.
Identification of dominant microbial groups and quantification of in situ cell numbers.
In situ cell abundances were determined with FISH or CARD-FISH to investigate which groups within the methanotrophic guilds dominate the different habitats of the HMMV. To study spatial variability, three sediment cores as replicates of the Beggiatoa site and the tubeworm field and two cores of the center sediments were analyzed in replicate.
Free-living methylotrophs and Bacteroidetes in center sediments.
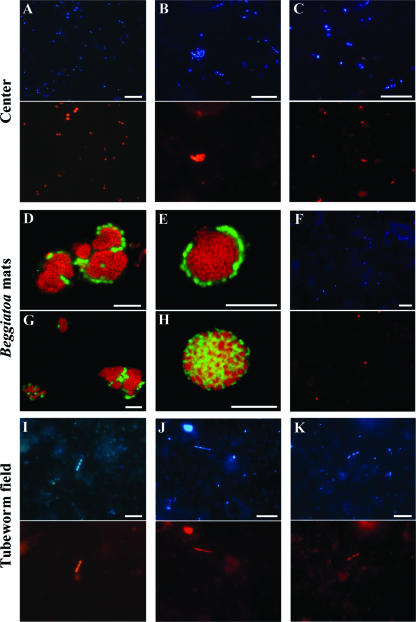
Total cell numbers were up to 3.6 × 109 ± 2.1 × 109 cells cm−3 in the uppermost sediment layer (<1-cm sediment depth) and decreased to less than 1 × 108 cells cm−3 at 6- to 10-cm sediment depths (Table 2). Detection rates of FISH with monolabeled oligonucleotide probes were high, with up to 98% DAPI-stained cells. Thus, no CARD-FISH was needed. Bacteria of cluster HMMV-MetI (probe MetI-444) were most abundant, with 48% ± 2% of total cells, and clearly dominated the microbial community in center sediments (Table 2). This percentage corresponds to an absolute number of 1.7 × 109 ± 0.9 × 109 cells cm−3 sediment. HMMV-MetI cells were mainly diplococci with a diameter of 0.7 μm. A second morphotype of MetI-444 was constituted of large cocci with a diameter of about 2 μm. They occurred in clusters of three or more cells (Fig. 5A). The DAPI staining of the clustered cells was irregular, indicating the presence of intracellular membrane systems which are characteristic for aerobic methanotrophs (22). HMMV-MetI abundance decreased with sediment depth to 26% ± 11% of the total cells at 9- to 10-cm sediment depths. HMMV-MetII (probe MetII-844) could also be detected in situ but in a lower abundance, with a maximum of 1.1 × 108 ± 0.8 × 108 cells cm−3 (3% ± 2%) at 0- to 1-cm sediment depths (Table 2). In general, HMMV-MetII cells occurred as cocci with an average diameter of 1 to 2 μm (Fig. 5B). The third abundant clone group in center sediments, HMMV-MPH, accounted for 5% ± 4% at the sediment surface and abundance decreased with depth. Target cells were morphologically uniform and occurred as small (<1-μm) oval cells (Fig. 5C). Hence, free-living aerobic methanotrophs accounted in total for 56% ± 8% at 0- to 1-cm sediment depths. In addition to aerobic methanotrophs, up to 33% of the cells could be assigned to the Cytophaga/Flavobacterium cluster of the Bacteroidetes by use of probe CF319a. Deltaproteobacterial groups were detected only in low percentages (<0.5%).
TABLE 2.
In situ quantificationa
| Depth (cm) | % Cellsb
|
Mean ± SDc
|
||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Bacteria (EUB338 I-III) | Archaea (Arch915) |
Deltaproteobacteria by probe:
|
Cytophaga/Flavobacterium (Cf319a) | % HMMV Met I (Met I-444) | % HMMV Met II (Met II-844) | % HMMV MPH (MPH-732) | Aggregate counts (106/ml) | Cell counts (109 cells/ml)
|
Cells in aggregates (% of total cells) | |||||
| DSS658 | DSV698 | DSR651 | DSB985 | Single | Total | |||||||||
| Center sediment | ||||||||||||||
| 0-1 | 91 | <1 | 0 | <0.5 | <1 | <0.5 | 23 | 48 ± 2 | 3 ± 2 | 5 ± 4 | 0.0 ± 0.0 | 3.6 ± 2.1 | 3.6 ± 2.1 | 0 ± 0 |
| 1-2 | 97 | <1 | 0 | 0 | 0 | 0 | 33 | 44 ± 5 | 5 ± 0 | 3 ± 4 | 0.0 ± 0.0 | 1.1 ± 0.1 | 1.1 ± 0.1 | 0 ± 0 |
| 2-3 | 89 | 6 | 0 | 0 | <0.5 | <0.5 | 22 | 43 ± 11 | 4 ± 2 | 3 ± 4 | 0.0 ± 0.0 | 0.3 ± 0.3 | 0.3 ± 0.3 | 0 ± 0 |
| 3-4 | 90 | 9 | 0 | <0.5 | 0 | 0 | 14 | 43 ± 2 | 2 ± 1 | 3 ± 4 | 0.0 ± 0.0 | 0.1 ± 0.1 | 0.1 ± 0.1 | 0 ± 0 |
| 4-5 | 83 | 8 | 0 | 0 | <0.5 | 0 | 24 | 33 ± 8 | 6 ± 4 | <1 ± 0 | 0.0 ± 0.0 | 0.1 ± 0.1 | 0.1 ± 0.1 | 0 ± 0 |
| 6-7 | 64 | 17 | ND | ND | ND | ND | 22 | 36 ± 2 | 1 ± 1 | ND | 0.0 ± 0.0 | <0.1 ± <0.1 | <0.1 ± <0.1 | 0 ± 0 |
| 7-8 | 80 | 14 | 0 | 0 | 0 | 0 | 23 | 35 ± 13 | 0 ± 0 | <1 ± 0 | 0.0 ± 0.0 | <0.1 ± 0.0 | <0.1 ± 0.0 | 0 ± 0 |
| 9-10 | 61 | 13 | 0 | <0.5 | ND | ND | 21 | 26 ± 11 | 0 ± 0 | ND | 0.0 ± 0.0 | <0.1 ± 0.0 | <0.1 ± 0.0 | 0 ± 0 |
| Beggiatoa mat | ||||||||||||||
| 0-1 | 51 | <1 | 4 | <0.5 | 3 | <0.5 | 23 | 8 ± 1 | 1 ± 1 | <1 ± 0 | 8.7 ± 6.3 | 3.7 ± 1.1 | 10.2 ± 5.0 | 60 ± 16 |
| 1-2 | 40 | 2 | 4 | <0.5 | 3 | <0.5 | 16 | 4 ± 1 | 1 ± 1 | <1 ± 0 | 19.7 ± 0.6 | 4.8 ± 1.4 | 19.4 ± 1.6 | 76 ± 5 |
| 2-3 | 38 | 2 | 6 | 0 | <0.5 | 0 | 7 | 4 ± 1 | <1 ± 0 | ND | 10.9 ± 2.4 | 0.5 ± 0.2 | 8.6 ± 1.9 | 94 ± 2 |
| 3-4 | 42 | 4 | 4 | 0 | 0 | 0 | 18 | 1 ± 0 | 0 ± 0 | 0 ± 0 | 1.0 ± 0.4 | 0.2 ± 0.1 | 1.0 ± 0.4 | 83 ± 8 |
| 4-5 | 35 | 4 | 16 | <0.5 | 0 | 0 | 10 | <1 ± 0 | 0 ± 0 | ND | 0.8 ± 0.3 | 0.2 ± 0.2 | 0.7 ± 0.1 | 77 ± 26 |
| 6-7 | 32 | 6 | 7 | 0 | ND | ND | ND | ND | ND | 0 ± 0 | 0.7 ± 1.1 | <0.1 ± <0.1 | 0.6 ± 0.8 | 57 ± 50 |
| 7-8 | 33 | 7 | 12 | 0 | 0 | 0 | 10 | 0 ± 0 | 0 ± 0 | ND | 0.2 ± 0.3 | 0.1 ± 0.0 | 0.2 ± 0.2 | 30 ± 52 |
| 8-9 | 30 | 22 | 17 | 0 | ND | ND | 10 | ND | ND | 0 ± 0 | 0.1 ± 0.1 | 0.1 ± 0.0 | 0.2 ± 0.1 | 39 ± 34 |
| 9-10 | 27 | 25 | 5 | 0 | 0 | 0 | 6 | 0 ± 0 | 0 ± 0 | ND | 0.3 ± 0.1 | 0.1 ± 0.0 | 0.2 ± 0.1 | 51 ± 45 |
| Tubeworm field | ||||||||||||||
| 0-1 | 27 | <0.5 | 3 | <0.5 | <0.5 | 0 | 4 | 0 ± 0 | 0 ± 0 | ND | 0.1 ± 0.1 | 0.8 ± 0.6 | 0.8 ± 0.6 | 3 ± 5 |
| 1-2 | 22 | 2 | 4 | <0.5 | <0.5 | 0 | 10 | ND | ND | 0 ± 0 | 0.1 ± 0.1 | 0.9 ± 0.3 | 0.9 ± 0.3 | 4 ± 6 |
| 2-3 | 28 | 3 | 4 | 0 | <0.5 | 0 | 4 | 0 ± 0 | 0 ± 0 | ND | 0.3 ± 0.6 | 1.4 ± 0.5 | 1.6 ± 0.7 | 11 ± 19 |
| 3-4 | 18 | <0.5 | 2 | <0.5 | 2 | <0.5 | <1 | ND | ND | 0 ± 0 | 0.1 ± 0.1 | 3.2 ± 3.1 | 3.3 ± 3.0 | 3 ± 5 |
| 6-7 | 12 | 1 | 6 | <0.5 | <0.5 | <0.5 | 2 | 0 ± 0 | 0 ± 0 | ND | 0.2 ± 0.2 | 1.8 ± 1.2 | 2.0 ± 1.1 | 16 ± 19 |
| 7-8 | 16 | <1 | 5 | <1 | <0.5 | <0.5 | 2 | 0 ± 0 | 0 ± 0 | ND | 0.2 ± 0.2 | 0.7 ± 0.7 | 0.8 ± 0.8 | 20 ± 24 |
| 8-9 | 12 | <1 | 4 | 1 | <0.5 | 0 | <1 | ND | ND | 0 ± 0 | 0.4 ± 0.5 | 0.6 ± 0.6 | 0.8 ± 0.9 | 10 ± 17 |
| 9-10 | 22 | 1 | 7 | 2 | <0.5 | 0 | <0.5 | 0 ± 0 | 0 ± 0 | ND | 0.0 ± 0.0 | 0.6 ± 0.5 | 0.6 ± 0.5 | 0 ± 0 |
| 12-13 | 18 | 1 | 12 | <0.5 | <0.5 | 0 | <0.5 | ND | ND | 0 ± 0 | 0.2 ± 0.3 | 0.6 ± 0.6 | 0.8 ± 0.5 | 26 ± 41 |
| 13-14 | 19 | <1 | 12 | <0.5 | <1 | 0 | <0.5 | 0 ± 0 | 0 ± 0 | ND | 0.6 ± 0.5 | 0.4 ± 0.6 | 0.9 ± 0.9 | 41 ± 42 |
FISH probe names are in parentheses; for probe specificity, see Table 1. ND, not determined.
Percentages of detected DAPI-stained single cells.
Multiple independent MUC cores were examined for spatial variability: two cores of center sediments and three cores each of sediments covered with Beggiatoa mats and sediments from the tubeworm field.
FIG. 5.
Epifluorescence micrographs of different microbial populations at HMMV visualized by FISH (panels A to C, center; panels D to H, Beggiatoa site; panels I to K, tubeworms field). DAPI staining (upper picture) and FISH with probes specific for aerobic methanotrophic/methylotrophic bacteria MetI-444 (A), MetII-844 (B), and MPH-732 (C). (D, E, and G) Color overlays of shell-type consortia. Probe ANME3-1249, specific for ANME-3 archaea, is shown in red, and probe DBB660, specific for Desulfobulbus spp., is shown in green. (H) Color overlay of mixed-type consortia. Probe ANME3-1249, specific for ANME-3 archaea, is shown in red, and probe EUB338 I-III, specific for Bacteria, is shown in green. (F) DAPI staining and CARD-FISH with probe ANME3-1249 showing single ANME-3 cells in green. DAPI staining and FISH with probe ANME1-350, specific for ANME-1 archaea (I); probe CF319a, specific for Cytophaga/Flavobacterium (J); and probe DSS658, specific for the Desulfosarcina/Desulfococcus branch (K), are shown. Scale bars, 10 μm.
Detection rates of archaea in center sediments ranged from <1% to 17% of total cells, with a maximum in 6- to 7-cm sediment depths. The crenarchaeotal group HMMV-Cren was the only clone group in our archaeal clone library from center sediments, but its members were detected in situ only in low abundance (<1%). Target cells were on average 5-μm-long slender rods and occurred as single cells or in chains of up to 10 cells. ANME-1, ANME-2, and ANME-3 archaea could not be detected in situ, and the major fraction of Archaea at the center site remained unidentified. Further probes need to be developed and/or CARD-FISH must be applied to achieve higher detection rates.
ANME-3 aggregates in sediments below Beggiatoa mats.
In our previous study, we demonstrated the dominance of ANME-3/Desulfobulbus aggregates (ANME-3/DBB) in sediments below Beggiatoa mats (54). In this study, we give a detailed description of their morphological characteristics and abundance along vertical profiles at three Beggiatoa sites. Using two probes (ANME3-1249 and ANME3-397) specific for ANME-3 archaea, we identified the coccoid cells in the consortia as ANME-3 cells (0.7 μm diameter). All ANME-3 cells were autofluorescent under UV excitation, indicating the presence of coenzyme F420, which is a characteristic feature of methanogens (78). Hybridization signals of the monolabeled probe ANME3-1249 were dim; therefore, we developed the helper probes (17) ANME-3-1249H3 and ANME-3-1249H5 (Table 1) and added them to the hybridization reaction or used HRP-labeled probes for CARD-FISH (60). Both procedures resulted in bright signals. The associated bacteria were identified as rod-shaped SRB (1.1 by 0.5 μm) affiliated with Desulfobulbus species (DBB) by using probe 660 (11), which has one mismatch to the clone sequences retrieved from HMMV, and a newly designed probe (DBB305; zero mismatches).
The ANME-3/DBB aggregates mainly occurred as shell-type spheres (35). Sometimes acridine orange staining showed the presence of an exopolysaccharide matrix enclosing several aggregates. Often several spheres were aligned to an overall cylindrical morphology and the DBB layer formed only an incomplete outer shell consisting of a few cells (Fig. 5D, E, and G). The size spectrum of the diameters of ANME-3/DBB consortia ranged from 2 to 50 μm, with an average diameter of 5 ± 3 μm for spherical aggregates and a width of 5 ± 3 μm and a height of 10 ± 5 μm for cylindrical aggregates (several spherical aggregates together).
High ANME-3/DBB aggregate numbers were restricted to the sediment layers directly below the Beggiatoa mat (1- to 3-cm depths) with up to 19.7 × 106 ± 0.6 × 106 cm−3 sediment (Table 2). Aggregate numbers decreased by two orders of magnitude below a 7-cm sediment depth. The average spherical aggregate comprised approximately 550 ANME-3 cells, whereas cylindrical aggregates were built up on average with 1,250 ANME-3 cells. The observed ratio of spheres to cylinders (73:27) was taken into account, resulting in an average of 740 ANME-3 cells per HMMV aggregate. Calculated cell numbers in ANME-3 aggregates accounted for 75 to 94% of total cells, comprising the major part of the microbial biomass in a 0- to 5-cm sediment depth interval at the Beggiatoa site. The distribution of ANME-3/DBB aggregates was almost identical in three sediment cores sampled from different areas of the Beggiatoa species-covered zone of the HMMV (Table 2).
Very few consortia were detected with a yet-unidentified bacterial partner. These ANME-3/bacterial aggregates showed a mixed-type morphology and appeared to grow in direct association with each other (Fig. 6H). Almost 25% of HMMV aggregates occurred without any obvious bacterial partners. These probably monospecific ANME-3 aggregates were more tightly packed and were enclosed by a thick exopolysaccharide layer. In the horizon with highest ANME-3/DBB abundance (1- to 2-cm depths) we also found a remarkably high number of single ANME-3 cells, accounting for 25% of total single cells (1.2 × 109 cells cm−3) (Fig. 5F and data not shown). Single DBB cells were only rarely detected (≪0.5% of total single cells).
FIG. 6.
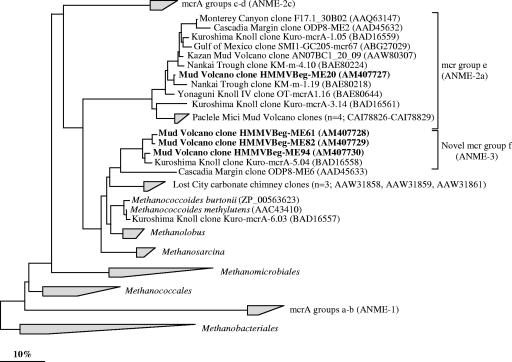
Phylogenetic tree showing the affiliations of HMMV genes sequences coding for the alpha subunit of methyl-coenzyme M-reductase (mcrA) to selected sequences of the domain Archaea. The tree was generated from deduced amino acid sequences (>247 amino acids) by neighbor-joining analysis with a 30% amino acid frequency filter. Clone sequences from sediments below Beggiatoa mats are in boldface type. mcrA sequences from sorted ANME-3-aggregates (not shown in the tree) were identical to those of the HMMV clones in group f. The bar indicates 10% estimated phylogenetic divergence. ANME groups (shown in brackets) were assigned to mcrA groups based on the assumption that mcrA and 16S rRNA genes show congruent phylogenies.
Abundance of other bacterial groups below Beggiatoa mats.
Members of Bacteroidetes dominated the bacterial community, constituting up to 23% of total single cells in the upper sediment layers. Gammaproteobacteria of the aerobic methanotrophic clusters HMMV-MetI and HMMV-MetII were also detected in 0- to 4-cm sediment depths, with a maximum of 9% ± 2% of total single cells (3.9 × 108 ± 2.5 × 108 cells cm−3) in 0- to 1-cm sediment depths. Methanotrophic bacteria were not detectable in deeper sediments layers. Members of the Desulfosarcina/Desulfococcus branch dominated in these sediment depths (4 to 10 cm), constituting up to 17% of the total single cells.
Cooccurrence of different ANME groups in sediments of the tubeworm field.
In sediments populated with siboglinid tubeworms, ANME-3/DBB aggregates were detected throughout three sediment cores with a low abundance, having a maximum of 6 × 105 ± 5 × 105 aggregates cm−3 (Table 2). In our previous study, we found the highest AOM rate between the base of the worm tubes and hydrate layers (60- to 90-cm depths) coinciding with a subsurface peak of ANME-2/DSS aggregates (5.5 × 106 aggregates cm−3 [54]). However, in the uppermost 14 cm of the tubeworm sites, ANME-2/DSS aggregates were not detected. Few ANME-1 cells (Fig. 5I) were present in a single sediment horizon (9- to 10-cm depth; <0.5% of total single cells).
Although bacterial diversity was highest at the tubeworm site, FISH detection rates of single cells at this site were much lower than the detection rates at the other sites, with a maximum rate of 32% of total single cells. Cells detected by CF319a were most abundant, at up to 10% (Table 2; Fig. 5J). Gammaproteobacteria of HMMV-specific sequence clades HMMV-MetI, HMMV-MetII, and HMMV-MPH were not detectable.
Sorting of ANME-3/DBB aggregates.
ANME-3/DBB aggregates were sorted by flow cytometry in order to further characterize the DBB cells associated with ANME-3 archaea, because only a single DBB-related sequence (clone HMMVBeg-DBB4) was retrieved from the Beggiatoa site clone library. The purity of the sorted fraction was high, with >99% of the cells belonging to ANME-3/DBB aggregates. The archaeal clone library of the sorted cells contained three types of highly similar ANME-3 sequences, which differed only in a single base and were >99% similar to sequences originally retrieved from the Beggiatoa site (data not shown). In the bacterial clone library of the sorted aggregates, we found DBB sequences (clones HMMV-sortedANME-3agg-Bac104 and HMMV-sortedANME-3agg-Bac125) (Fig. 2) with 99.6% similarity to clone sequence HMMVBeg-DBB4. These sequences form a separate new clade affiliated with Desulfobulbus. This clade includes only few other sequences from a Mediterranean mud volcano (GenBank accession no. AY592579), the Guaymas Basin (76), and two other aquatic sediments (GenBank accession no. AY193761 and AY123207/AY123218). The closest cultivated relative of the HMMV DBB sequences is Desulfobulbus mediterraneus (94%). By use of a new, specific FISH probe (DBBA665) (Table 1; Fig. 3), members of this phylogenetic clade were confirmed to be the bacterial partners of ANME-3 archaea.
Diversity of mcrA genes.
To further describe the diversity of methane-oxidizing archaea in sediments below Beggiatoa mats, clone libraries for the mcrA gene were constructed. The mcrA gene codes for the alpha subunit of methyl-coenzyme M-reductase, which is the key enzyme in methanogenesis and supposed to be involved in AOM (21, 36, 69). Comparative sequence analysis of deduced amino acid sequences (>247 amino acids) of 170 clones showed the presence of two lineages. Most HMMV sequences (165 of 170 clone sequences) were closely related (90 to 93%) to a clone sequence from methane-rich Kuroshima Knoll sediments (GenBank accession no. BAD16558), forming a novel clade of mcrA sequences within the order Methanosarcinales (Fig. 6). We call this clade “group f” and extend the existing naming scheme (groups a, b, c, d, and e) suggested by Hallam et al. (21). Four HMMV phylotypes within the clade could be distinguished (93 to 98% sequence identity). Phylogenies of mcrA and 16S rRNA genes are congruent: mcrA group f is closely related to cultivated genera, namely, Methanococcoides and Methanolobus (79 to 83% sequence identity). Furthermore, we obtained mcrA sequences from sorted ANME-3 aggregates which were identical with group f HMMV sequences. Thus, we conclude that these sequences originated from ANME-3 archaea.
A small number of HMMV sequences (5 of 170 sequences) grouped with sequences of mcrA group e (Fig. 6), which most likely represents ANME-2a archaea.
DISCUSSION
Diversity of methanotrophic guilds.
Our detailed molecular characterization of the three main habitats of HMMV shows that habitat variations (Fig. 1) are clearly reflected by differences in the bacterial and archaeal diversity and in situ abundance. Horizontally, the HMMV habitats cover hundreds of meters of seafloor and differ with respect to environmental factors, such as fluid flow and energy availability (see the introduction). Vertically, the niches for methanotrophic microorganisms can be described as microhabitats, according to their extension of only a few millimeters or centimeters, which are controlled by the distribution of electron acceptors such as oxygen and sulfate. The habitats at HMMV seem to be rather stable on time scales of years, since the analysis of replicate cores distributed over tens and hundreds of meters resulted in highly similar abundances and distributions of aerobic methanotrophs (groups HMMV-MetI, HMMV-MetII, and HMMV-MPH) at the volcano center and anaerobic methanotrophs (ANME-3) at the Beggiatoa site. This indicates stable selection mechanisms for the methanotrophic community.
The methanotrophic guilds of HMMV are composed of aerobic and anaerobic microorganisms which are responsible for the remarkably large microbial biomass present in the mud volcano surface sediments. Different aerobic and anaerobic methanotrophic groups accounted for up to 94% of the microbial biomass (Beggiatoa site, 2- to 3-cm sediment depths). In the volcano center and at the Beggiatoa site, carbon sources other than methane are not available. Only one phylogenetic group strongly dominated, suggesting that the limited carbon substrate spectrum appears to limit the diversity of the microbial community. Both habitats are characterized by high methane concentrations over the entire vertical profile (54) and a limited oxygen penetration depth of a few millimeters (9). There are only four studies published (5, 35, 53, 56) providing quantitative in situ data on different anaerobic methanotrophic communities and SRB at methane-seep sites. Other studies on environmental methanotrophs used methods such as lipid biomarker analysis (e.g., (4) or slot blot hybridizations (77). In these studies, the dominance of one methanotrophic group over several cooccurring groups was reported for different habitats, namely, the Hydrate Ridge methane seeps (5, 35), which are strongly dominated by ANME-2a and ANME-2c groups, and the Black Sea microbial mats (4, 35, 46, 77) and the Gulf of Mexico sediments (40, 56), which are both dominated by the ANME-1 group.
The high dominance of aerobic methanotrophic bacteria at the HMMV center and anaerobic methanotrophic archaea at the Beggiatoa site is reflected in a low microbial diversity (16 and 26 phylotypes, respectively) at these sites. The archaeal diversity was extremely low compared to most other methane-seep sites investigated so far (24, 28, 35, 40, 48, 49, 57, 74, 76). This might be explained by the limited vertical extension of the mud volcano habitat of only a few centimeters below seafloor and/or other constraints in populating muds emitted from subsurface depths.
The microbial diversity increased considerably in zones outside of highest fluid flow (51 microbial phylotypes) which were populated by siboglinid tubeworms. Here, aerobic and anaerobic methane oxidizers were less abundant in the top 10 cm of sediment and the total microbial biomass in the uppermost three centimeters was only about 9% of that at the Beggiatoa site (the tubeworm field biomass in 0- to 10-cm depth was ∼30% of that at the Beggiatoa site). The tubeworm site is characterized by the dense colonization of the sediments with tubeworms above thin layers of gas hydrates. Electron acceptors such as oxygen and sulfate are transported by bioirrigation deep into the sediments (9, 54). The presence of the tubeworms likely enhanced availability of organic carbon other than methane, e.g., worm exudates and sediments particles trapped by the worm tubes, and increased the diversity of heterotrophic groups of bacteria and crenarchaeotal groups.
Habitat and diversity of aerobic methanotrophs.
The HMMV is the first cold seep for which high in situ abundance of aerobic methanotrophs could be demonstrated. Ten phylotypes of aerobic methanotrophs dominate the highly gassy sediments of the center of the volcano and reach high abundances in a very narrow oxic sediment horizon at the seafloor. The specific biogeochemistry of the freshly expelled center sediments may promote the growth of aerobic methanotrophs. The sediments of the volcano center are permanently vented by ascending methane-rich fluids but also mixed with deep-subsurface sediments during events of mud eruption. The organic content of the center sediments is very low, and methane is most likely the main carbon source for microbial life. A high upward flow of sulfate-free subsurface fluids limits the penetration of sulfate and oxygen from the seawater to the uppermost centimeter of the sediment (9) and thereby excludes ANME archaea from these sediments. Sulfide levels at the center were below the detection limit (9), confirming that AOM was not a significant process. High concentrations of δ13C-depleted (−89‰) bacterial fatty acids (e.g., C16:1ω8c) assigned to type 1 methanotrophs and high aerobic methane oxidation rates (0.9 mol m−2 yr−1 [54]) support the 16S rRNA-based results of this study.
Habitat and diversity of anaerobic methanotrophs.
The sediments below Beggiatoa mats at the HMMV represent the first habitat known to host a microbial community dominated by ANME-3 biomass. The maximum abundance of ANME-3 cells occurred in the upper sediment layers below Beggiatoa mats, with up to 94% ± 2% of total cells, and correlated with high concentrations of archaeal lipids (dominance of pentamethylicosene with four double bonds [PMI:4] and PMI:5) from which δ13C (−109‰) was highly depleted (54). However, the ANME-3 habitat was limited to a rather narrow zone below oxygen penetration, in the zone of sulfate diffusion between 1- and 3-cm sediment depth (9). In this layer, AOM rates were high, at 4.5 mol m−2 yr−1 (54). Whereas ANME-1 and ANME-2 archaea are associated with SRB of the Desulfosarcina-Desulfococcus branch (83), ANME-3 archaea are the first anaerobic methanotrophs known to associate with Desulfobulbus species. Only a few environmental sequences from this bacterial group have been obtained. Only two out of the six known ANME-3 habitats also hosted DBB sequences (Mediterranean mud volcano [GenBank accession no. AY592579] and Eel River Basin [57]). Curiously, although ANME-3/DBB aggregates were responsible for up to 94% ± 2% of the microbial biomass below Beggiatoa mats at HMMV, DBB-related clones were not found among 132 analyzed clones in our general 16S rRNA gene library of Bacteria. Only a single clone (HMMVBeg-DBB4) (Fig. 3) out of 102 clones from a second library constructed by using a DBB-specific PCR (primer DBB305/GM4) (data not shown) was identified as a DBB relative. Organisms of this group might have mismatches to one of the general PCR primers and therefore may be underrepresented in the clone libraries.
In contrast to the sulfate-reducing partner of ANME-2 (DSS) (5, 35, 57, 58), DBB cells seem to be only loosely associated with the ANME-3 archaea, and the ratio of DBB cells to ANME-3 cells is ≪1. Interestingly, we also found a substantial number of single ANME-3 cells below Beggiatoa mats at the HMMV. In addition, there was a high percentage of ANME-3 aggregates without a sulfate-reducing partner. These observations suggest that a physical association with DBB cells may not be obligatory for ANME-3 archaea. Alternatively, treatment of samples might have released DBB and/or ANME-3 single cells to the environment. Furthermore, some ANME-3 cells occurred with yet unidentified bacteria, forming mixed-type aggregates, suggesting that ANME-3 cells do not necessarily depend on DBB as a sulfate-reducing partner and might be able to live syntrophically with other partners as well.
A novel type of mcrA in ANME-3 archaea.
Methanotrophic archaea have been shown to harbor genes encoding the enzyme methyl-coenzyme M-reductase, which is involved in the terminal step in methane formation from CO2 in methanogenic archaea (for a review, see reference 69). Previous analyses have revealed considerably diversity within mcrA genes among ANME archaea, and five subgroups have been defined (1, 12, 20, 21, 30, 40, 55): groups a and b and groups c and d have been assigned to ANME-1 and ANME-2 archaea, respectively. A phylogenetic relationship for group e is not clear yet; it may be assigned to ANME-2a archaea (1), while groups c and d probably represent ANME-2c archaea (A. Meyerdierks, personal communication).
Here, we describe a novel cluster of mcrA genes, group f. As shown for all known orders of methanotrophs (42, 73), mcrA group f and ANME-3 16S rRNA gene sequences show congruent tree topologies, having Methanococcoides spp. and Methanolobus spp. as their closest relatives. Furthermore, the microbial biomass at the Beggiatoa site is strongly dominated by ANME-3 and identical sequences of mcrA gene group f were retrieved from sorted ANME-3/DBB aggregates. Thus, we conclude that the mcrA gene group f sequences originate from ANME-3 archaea.
Acknowledgments
We thank the captain, crew, and team of the ROV VICTOR 6000, as well as the shipboard scientific community of the R/V L'Atalante and R/V Polarstern, for their help at sea. We are especially indebted to Michael Klages, the chief scientist of cruise ARKXIX-3b, for his support of the fieldwork. We thank Anita Ritz for help with probe optimization and Thomas Holler for providing enrichment cultures for flow cytometry. Viola Beier and Birgit Rattunde are acknowledged for excellent technical assistance.
This is publication no. GEOTECH-263 of the GEOTECHNOLOGIEN program funded by the German Ministry of Education and Research (BMBF) and the German Research Foundation (DFG), project MUMM (03G0554A and 03G0608A). Further support was provided by the Max Planck Society, Germany.
Footnotes
Published ahead of print on 16 March 2007.
REFERENCES
- 1.Alain, K., T. Holler, F. Musat, M. Elvert, T. Treude, and M. Krüger. 2006. Microbiological investigation of methane- and hydrocarbon-discharging mud volcanoes in the Carpathian Mountains, Romania. Environ. Microbiol. 8:574-590. [DOI] [PubMed] [Google Scholar]
- 2.Baer, M., J. Ravel, J. Chun, R. Hill, and H. Williams. 2000. A proposal for the reclassification of Bdellovibrio stolpii and Bdellovibrio starrii into a new genus, Bacteriovorax gen. nov. as Bacteriovorax stolpii comb. nov. and Bacteriovorax starrii comb. nov., respectively. Int. J. Syst. Evol Microbiol. 50:219-224. [DOI] [PubMed] [Google Scholar]
- 3.Biddle, J. F., J. S. Lipp, M. A. Lever, K. G. Lloyd, K. B. Sorensen, R. Anderson, H. F. Fredricks, M. Elvert, T. J. Kelly, D. P. Schrag, M. L. Sogin, J. E. Brenchley, A. Teske, C. H. House, and K.-U. Hinrichs. 2006. Heterotrophic Archaea dominate sedimentary subsurface ecosystems off Peru. Proc. Natl. Acad. Sci. USA 103:3846-3851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Blumenberg, M., R. Seifert, K. Nauhaus, T. Pape, and W. Michaelis. 2005. In vitro study of lipid biosynthesis in an anaerobically methane-oxidizing microbial mat. Appl. Environ. Microbiol. 71:4345-4351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Boetius, A., K. Ravenschlag, C. Schubert, D. Rickert, F. Widdel, A. Gieseke, R. Amann, B. B. Jørgensen, U. Witte, and O. Pfannkuche. 2000. A marine microbial consortium apparently mediating anaerobic oxidation of methane. Nature 407:623-626. [DOI] [PubMed] [Google Scholar]
- 5a.Brosius, J., T. J. Dull, D. D. Sleeter, and H. F. Noller. 1981. Gene organization and primary structure of a ribosomal RNA operon from Escherichia coli. J. Mol. Biol. 148:107-127. [DOI] [PubMed] [Google Scholar]
- 6.Chandler, D. P., F. J. Brockman, T. J. Bailey, and J. K. Fredrickson. 1998. Phylogenetic diversity of archaea and bacteria in a deep subsurface paleosol. Microb. Ecol. 36:37-50. [DOI] [PubMed] [Google Scholar]
- 7.Daims, H., A. Brühl, R. Amann, and K. H. Schleifer. 1999. The domain-specific probe EUB338 is insufficient for the detection of all Bacteria: development and evaluation of a more comprehensive probe set. Syst. Appl. Microbiol. 22:434-444. [DOI] [PubMed] [Google Scholar]
- 8.Damm, E., and G. Budéus. 2003. Fate of vent-derived methane in seawater above the Haakon Mosby mud volcano (Norwegian Sea). Mar. Chem. 1969:1-11. [Google Scholar]
- 9.De Beer, D., E. Sauter, H. Niemann, U. Witte, M. Schlüter, and A. Boetius. 2006. In situ fluxes and zonation of microbial activity in surface sediments of the Haakon Mosby Mud Volcano. Limnol. Oceanogr. 51:1315-1331. [Google Scholar]
- 10.DeLong, E. F. 1992. Archaea in coastal marine environments. Proc. Natl. Acad. Sci. USA 89:5685-5689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Devereux, R., M. D. Kane, J. Winfrey, and D. A. Stahl. 1992. Genus- and group-specific hybridization probes for determinative and environmental studies of sulfate-reducing bacteria. Syst. Appl. Microbiol. 15:601-609. [Google Scholar]
- 12.Dhillon, A., M. Lever, K. G. Lloyd, D. B. Albert, M. L. Sogin, and A. Teske. 2005. Methanogen diversity evidenced by molecular characterization of methyl coenzyme M reductase A (mcrA) genes in hydrothermal sediments of the Guaymas Basin. Appl. Environ. Microbiol. 71:4592-4601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Distel, D. L., and C. M. Cavanaugh. 1994. Independent phylogenetic origins of methanotrophic and chemoautotrophic bacterial endosymbioses in marine bivalves. J. Bacteriol. 176:1932-1938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Distel, D. L., H. K. Lee, and C. M. Cavanaugh. 1995. Intracellular coexistence of methano- and thioautotrophic bacteria in a hydrothermal vent mussel. Proc. Natl. Acad. Sci. USA 92:9598-9602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Dojka, M. A., P. Hugenholtz, S. K. Haack, and N. R. Pace. 1998. Microbial diversity in a hydrocarbon- and chlorinated-solvent-contaminated aquifer undergoing intrinsic bioremediation. Appl. Environ. Microbiol. 64:3869-3877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Eldholm, O., E. Sundvor, P. R. Vogt, B. O. Hjelstuen, K. Crane, A. K. Nilsen, and T. P. Gladczenko. 1999. SW Barents Sea continental margin heat flow and Haakon Mosby Mud Volcano. Geo-Mar. Lett. 19:29-37. [Google Scholar]
- 17.Fuchs, B. M., F. O. Glöckner, J. Wulf, and R. Amann. 2000. Unlabeled helper oligonucleotides increase the in situ accessibility to 16S rRNA of fluorescently labeled oligonucleotide probes. Appl. Environ. Microbiol. 66:3603-3607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ginsburg, G. D., G. V. Milkov, V. A. Soloviev, A. V. Egorov, G. A. Cherkashev, P. R. Vogt, K. Crane, T. D. Lorenson, and M. D. Khutorskoy. 1999. Gas hydrate accumulation at the Haakon Mosby Mud Volcano. Geo-Mar. Lett. 19:57-67. [Google Scholar]
- 19.Hales, B. A., C. Edwards, D. A. Ritchie, G. Hall, R. W. Pickup, and J. R. Saunders. 1996. Isolation and identification of methanogen-specific DNA from blanket bog peat by PCR amplification and sequence analysis. Appl. Environ. Microbiol. 62:668-675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hallam, S. J., P. R. Girguis, C. M. Preston, P. M. Richardson, and E. F. DeLong. 2003. Identification of methyl coenzyme M reductase A (mcrA) genes associated with methane-oxidizing archaea. Appl. Environ. Microbiol. 69:5483-5491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hallam, S. J., N. Putnam, C. M. Preston, J. C. Detter, D. Rokhsar, P. M. Richardson, and E. F. DeLong. 2004. Reverse methanogenesis: testing the hypothesis with environmental genomics. Science 305:1457-1462. [DOI] [PubMed] [Google Scholar]
- 22.Hanson, R. S., and T. E. Hanson. 1996. Methanotrophic bacteria. Microbiol. Rev. 60:439-471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hinrichs, K.-U., and A. Boetius. 2002. The anaerobic oxidation of methane: new insights in microbial ecology and biogeochemistry, p. 457-477. In G. Wefer, D. Billett, D. Hebbeln, B. B. Jörgensen, M. Schlüter, and T. C. Van Weering (ed.), Ocean margin systems. Springer, Berlin, Germany.
- 24.Hinrichs, K. U., J. M. Hayes, S. P. Sylva, P. G. Brewer, and E. F. DeLong. 1999. Methane-consuming archaebacteria in marine sediments. Nature 398:802-805. [DOI] [PubMed] [Google Scholar]
- 25.Hjelstuen, B. O., O. Eldholm, J. I. Faleide, and P. R. Vogt. 1999. Regional setting of Haakon Mosby Mud Volcano, SW Barents Sea margin. Geo-Mar. Lett. 19:22-28. [Google Scholar]
- 26.Hoehler, T. M., M. J. Alperin, D. B. Albert, and C. S. Martens. 1994. Field and laboratory studies of methane oxidation in an anoxic marine sediment: evidence for a methanogen-sulfate reducer consortium. Glob. Biogeochem. Cycles 8:451-463. [Google Scholar]
- 27.Hong, S.-H., J. Bunge, S.-O. Jeon, and S. S. Epstein. 2006. Predicting microbial species richness. Proc. Natl. Acad. Sci. USA 103:117-122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Inagaki, F., M. M. M. Kuypers, U. Tsunogai, J.-I. Ishibashi, K.-I. Nakamura, T. Treude, S. Ohkubo, M. Nakaseama, K. Gena, H. Chiba, H. Hirayama, T. Nunoura, K. Takai, B. B. Jørgensen, K. Horikoshi, and A. Boetius. 2006. Microbial community in a sediment-hosted CO2 lake of the southern Okinawa Trough hydrothermal system. Proc. Natl. Acad. Sci. USA 103:14164-14169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Inagaki, F., T. Nunoura, S. Nakagawa, A. Teske, M. Lever, A. Lauer, M. Suzuki, K. Takai, M. Delwiche, F. S. Colwell, K. H. Nealson, K. Horikoshi, S. D'Hondt, and B. B. Jørgensen. 2006. Biogeographical distribution and diversity of microbes in methane hydrate-bearing deep marine sediments on the Pacific Ocean Margin. Proc. Natl. Acad. Sci. USA 103:2815-2820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Inagaki, F., U. Tsunogai, M. Suzuki, A. Kosaka, H. Machiyama, K. Takai, T. Nunoura, K. H. Nealson, and K. Horikoshi. 2004. Characterization of C1-metabolizing prokaryotic communities in methane seep habitats at the Kuroshima Knoll, Southern Ryukyu Arc, by analyzing pmoA, mmoX, mxaF, mcrA, and 16S rRNA genes. Appl. Environ. Microbiol. 70:7445-7455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Jurkevitch, E. 2002. The genus Bdellovibrio, p. 12-30. In M. Dworkin, S. Falkow, E. Rosenberg, K.-H. Schleifer, and E. Stackebrandt (ed.), The prokaryotes. Springer, New York, NY.
- 32.Kane, M. D., L. K. Poulsen, and D. A. Stahl. 1993. Monitoring the enrichment and isolation of sulfate-reducing bacteria by using oligonucleotide hybridization probes designed from environmentally derived 16S rRNA sequences. Appl. Environ. Microbiol. 59:682-686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Klages, M., J. Thiede, and J. P. Foucher (ed.). 2004. The expedition ARKTIS XIX/3 of the research vessel Polarstern in 2003, vol. 488. Buchhandlung Kamloth, Bremen, Germany.
- 34.Knittel, K., A. Boetius, A. Lemke, H. Eilers, K. Lochte, O. Pfannkuche, P. Linke, and R. Amann. 2003. Activity, distribution, and diversity of sulfate reducers and other bacteria in sediments above gas hydrate (Cascadia Margin, Oregon). Geomicrobiol. J. 20:269-294. [Google Scholar]
- 35.Knittel, K., T. Lösekann, A. Boetius, R. Kort, and R. Amann. 2005. Diversity and distribution of methanotrophic archaea at cold seeps. Appl. Environ. Microbiol. 71:467-479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Krüger, M., A. Meyerdierks, F. O. Glöckner, R. Amann, F. Widdel, M. Kube, R. Reinhardt, J. Kahnt, R. Böcher, R. K. Thauer, and S. Shima. 2003. A conspicuous nickel protein in microbial mats that oxidize methane anaerobically. Nature 426:878-881. [DOI] [PubMed] [Google Scholar]
- 37.Lane, D. J., B. Pace, G. J. Olsen, D. A. Stahl, M. Sogin, and N. R. Pace. 1985. Rapid determination of 16S ribosomal RNA sequences for phylogenetic analyses. Proc. Natl. Acad. Sci. USA 82:6955-6959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lein, A., P. Vogt, K. Crane, A. Egorov, and M. Ivanov. 1999. Chemical and isotopic evidence for the nature of the fluid in CH4-containing sediments of the Haakon Mosby Mud Volcano. Geo-Mar. Lett. 19:76-83. [Google Scholar]
- 39.Li, L., C. Kato, and K. Horikoshi. 1999. Microbial diversity in sediments collected from the deepest cold-seep area, the Japan trench. Mar. Biotechnol. 1:391-400. [DOI] [PubMed] [Google Scholar]
- 40.Lloyd, K. G., L. Lapham, and A. Teske. 2006. An anaerobic methane-oxidizing community of ANME-1b archaea in hypersaline Gulf of Mexico sediments. Appl. Environ. Microbiol. 72:7218-7230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ludwig, W., O. Strunk, R. Westram, L. Richter, H. Meier, Yadhukumar, A. Buchner, T. Lai, S. Steppi, G. Jobb, W. Förster, I. Brettske, S. Gerber, A. W. Ginhart, O. Gross, S. Grumann, S. Hermann, R. Jost, A. König, T. Liss, R. Lüβmann, M. May, B. Nonhoff, B. Reichel, R. Strehlow, A. Stamatakis, N. Stuckmann, A. Vilbig, M. Lenke, T. Ludwig, A. Bode, and K. H. Schleifer. 2004. ARB: a software environment for sequence data. Nucleic Acids Res. 32:1363-1371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Luton, P. E., J. M. Wayne, R. J. Sharp, and P. W. Riley. 2002. The mcrA gene as an alternative to 16S rRNA in the phylogenetic analysis of methanogen population in landfill. Microbiology 148:3521-3530. [DOI] [PubMed] [Google Scholar]
- 43.Manz, W., M. Eisenbrecher, T. R. Neu, and U. Szewzyk. 1998. Abundance and spatial organization of gram negative sulfate-reducing bacteria in activated sludge investigated by in situ probing with specific 16S rRNA targeted oligonucleotides. FEMS Microbiol. Ecol. 25:43-61. [Google Scholar]
- 44.Massana, R., A. E. Murray, C. M. Preston, and E. F. DeLong. 1997. Vertical distribution and phylogenetic characterization of marine planktonic Archaea in the Santa Barbara Channel. Appl. Environ. Microbiol. 63:50-56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Meyer-Reil, L. A. 1983. Benthic response to sedimentation events during autumn to spring at a shallow water station in the Western Kiel Bight. Mar. Biol. 77:247-256. [Google Scholar]
- 46.Michaelis, W., R. Seifert, K. Nauhaus, T. Treude, V. Thiel, M. Blumenberg, K. Knittel, A. Gieseke, K. Peterknecht, T. Pape, A. Boetius, R. Amann, B. B. Jørgensen, F. Widdel, J. Peckmann, N. V. Pimenov, and M. B. Gulin. 2002. Microbial reefs in the Black Sea fueled by anaerobic oxidation of methane. Science 297:1013-1015. [DOI] [PubMed] [Google Scholar]
- 47.Milkov, A., P. Vogt, G. Cherkashev, G. Ginsburg, N. Chernova, and A. Andriashev. 1999. Sea-floor terrains of Haakon Mosby Mud Volcano as surveyed by deep-tow video and still photography. Geo-Mar. Lett. 19:38-47. [Google Scholar]
- 48.Mills, H. J., R. J. Martinez, S. Story, and P. A. Sobecky. 2005. Characterization of microbial community structure in Gulf of Mexico Gas hydrates: comparative analysis of DNA- and RNA-derived clone libraries. Appl. Environ. Microbiol. 71:3235-3247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Mills, H. J., R. J. Martinez, S. Story, and P. A. Sobecky. 2004. Identification of members of the metabolically active microbial populations associated with Beggiatoa species mat communities from Gulf of Mexico cold-seep sediments. Appl. Environ. Microbiol. 70:5447-5458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Muyzer, G., A. Teske, C. O. Wirsen, and H. W. Jannasch. 1995. Phylogenetic relationships of Thiomicrospira species and their identification in deep-sea hydrothermal vent samples by denaturing gradient gel electrophoresis of 16S rDNA fragments. Arch. Microbiol. 164:165-172. [DOI] [PubMed] [Google Scholar]
- 51.Nauhaus, K., A. Boetius, M. Krüger, and F. Widdel. 2002. In vitro demonstration of anaerobic oxidation of methane coupled to sulphate reduction in sediment from a marine gas hydrate area. Environ. Microbiol. 4:296-305. [DOI] [PubMed] [Google Scholar]
- 52.Nauhaus, K., T. Treude, A. Boetius, and M. Krüger. 2005. Environmental regulation of the anaerobic oxidation of methane: a comparison of ANME-I and ANME-II communities. Environ. Microbiol. 7:98-106. [DOI] [PubMed] [Google Scholar]
- 53.Niemann, H., M. Elvert, M. Hovland, B. Orcutt, A. G. Judd, I. Suck, J. Gutt, S. B. Joye, E. Damm, K. Finster, and A. Boetius. 2005. Methane emission and consumption at a North Sea gas seep (Tommeliten area). Biogeosciences 2:335-351. [Google Scholar]
- 54.Niemann, H., T. Lösekann, D. de Beer, M. Elvert, T. Nadalig, K. Knittel, R. Amann, E. J. Sauter, M. Schlüter, M. Klages, J. P. Foucher, and A. Boetius. 2006. Novel microbial communities of the Haakon Mosby mud volcano and their role as a methane sink. Nature 443:854-858. [DOI] [PubMed] [Google Scholar]
- 55.Nunoura, T., H. Oida, T. Toki, J. Ashi, K. Takai, and K. Horikoshi. 2006. Quantification of mcrA by quantitative fluorescent PCR in sediments from methane seep of the Nankai Trough. FEMS Microbiol. Ecol. 57:149-157. [DOI] [PubMed] [Google Scholar]
- 56.Orcutt, B., A. Boetius, M. Elvert, V. Samarkin, and S. B. Joye. 2005. Molecular biogeochemistry of sulfate reduction, methanogenesis and the anaerobic oxidation of methane at Gulf of Mexico cold seeps. Geochim. Cosmochim. Acta 69:4267-4281. [Google Scholar]
- 57.Orphan, V. J., K.-U. Hinrichs, W. Ussler III, C. K. Paull, L. T. Taylor, S. P. Sylva, J. M. Hayes, and E. F. DeLong. 2001. Comparative analysis of methane-oxidizing archaea and sulfate-reducing bacteria in anoxic marine sediments. Appl. Environ. Microbiol. 67:1922-1934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Orphan, V. J., C. H. House, K.-U. Hinrichs, K. D. McKeegan, and E. F. DeLong. 2001. Methane-consuming archaea revealed by directly coupled isotopic and phylogenetic analysis. Science 293:484-487. [DOI] [PubMed] [Google Scholar]
- 59.Orphan, V. J., C. H. House, K.-U. Hinrichs, K. D. McKeegan, and E. F. DeLong. 2002. Multiple archaeal groups mediate methane oxidation in anoxic cold seep sediments. Proc. Natl. Acad. Sci. USA 99:7663-7668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Pernthaler, A., J. Pernthaler, and R. Amann. 2002. Fluorescence in situ hybridization and catalyzed reporter deposition (CARD) for the identification of marine Bacteria. Appl. Environ. Microbiol. 68:3094-3101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Pimenov, N., A. Savvichev, I. Rusanov, A. Lein, A. Egorov, A. Gebruk, L. Moskalev, and P. Vogt. 1999. Microbial processes of carbon cycle as the base of food chain of Haakon Mosby Mud Volcano. Geo-Mar. Lett. 19:89-96. [Google Scholar]
- 62.Prasolov, E. M., I. V. Tokarev, G. D. Ginsburg, V. A. Soloviev, and G. M. Eltsova. 1999. Helium and other noble gases in gas-hydrate sediments of the Haakon Mosby Mud Volcano. Geo-Mar. Lett. 19:84-88. [Google Scholar]
- 63.Ravenschlag, K., K. Sahm, C. Knoblauch, B. B. Jørgensen, and R. Amann. 2000. Community structure, cellular rRNA content and activity of sulfate-reducing bacteria in marine Arctic sediments. Appl. Environ. Microbiol. 66:3592-3602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Ravenschlag, K., K. Sahm, J. Pernthaler, and R. Amann. 1999. High bacterial diversity in permanently cold marine sediments. Appl. Environ. Microbiol. 65:3982-3989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Reeburgh, W. S. 1996. “Soft spots” in the global methane budget, p. 334-342. In M. E. Lidstrom and F. R. Tabita (ed.), Microbial growth on C1 compounds. Kluwer Academic Publishers, Dordrecht, The Netherlands.
- 66.Schmaljohann, R., and H. J. Flügel. 1987. Methane-oxidizing bacteria in Pogonophora. Sarsia 72:91-98. [Google Scholar]
- 67.Sekar, R., B. M. Fuchs, R. Amann, and J. Pernthaler. 2004. Flow sorting of marine bacterioplankton after fluorescence in situ hybridization. Appl. Environ. Microbiol. 70:6210-6219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Shilov, V. V., N. I. Druzhinina, L. V. Vasilenko, and V. V. Krupskaya. 1999. Stratigraphy of sediments from the Haakon Mosby Mud Volcano area. Geo-Mar. Lett. 19:48-56. [Google Scholar]
- 69.Shima, S., and R. K. Thauer. 2005. Methyl-coenzyme M reductase and the anaerobic oxidation of methane in methanotrophic archaea. Curr. Opin. Microbiol. 8:1-6. [DOI] [PubMed] [Google Scholar]
- 70.Smirnov, R. V. 2000. Two new species of Pogonophora from the arctic mud volcano off northwestern Norway. Sarsia 85:141-150. [Google Scholar]
- 71.Snaidr, J., R. Amann, I. Huber, W. Ludwig, and K. H. Schleifer. 1997. Phylogenetic analysis and in situ identification of bacteria in activated sludge. Appl. Environ. Microbiol. 63:2884-2896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Southward, E. C. 1982. Bacterial symbionts in Pogonophora. J. Mar. Biol. Assoc. UK 46:579-616. [Google Scholar]
- 73.Springer, E., M. S. Sachs, C. R. Woese, and D. R. Boone. 1995. Partial gene sequences for the A subunit of methyl-coenzyme M reductase (mcrI) as a phylogenetic tool for the family Methanosarcinaceae. Int. J. Syst. Bacteriol. 45:554-559. [DOI] [PubMed] [Google Scholar]
- 74.Stadnitskaia, A., G. Muyzer, B. Abbas, M. J. L. Coolen, E. C. Hopmans, M. Baas, T. C. E. van Weering, M. K. Ivanov, E. Poludetkina, and J. S. Sinninghe Damsté. 2005. Biomarker and 16S rDNA evidence for anaerobic oxidation of methane and related carbonate precipitation in deep-sea mud volcanoes of the Sorokin Trough, Black Sea. Mar. Geol. 217:67-96. [Google Scholar]
- 75.Stahl, D. A., and R. I. Amann. 1991. Development and application of nucleic acid probes in bacterial systematics, p. 205-248. In E. Stackebrandt and M. Goodfellow (ed.), Nucleic acid techniques in bacterial systematics. John Wiley & Sons Ltd., Chichester, England.
- 76.Teske, A., K.-U. Hinrichs, V. Edgcomb, A. de Vera Gomez, D. Kysela, S. P. Sylva, M. L. Sogin, and H. W. Jannasch. 2002. Microbial diversity of hydrothermal sediments in the Guaymas Basin: evidence for anaerobic methanotrophic communities. Appl. Environ. Microbiol. 68:1994-2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Treude, T., K. Knittel, M. Blumenberg, R. Seifert, and A. Boetius. 2005. Subsurface microbial methanotrophic mats in the Black Sea. Appl. Environ. Microbiol. 71:6375-6378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Van Bruggen, J. J. A., C. K. Stumm, and G. D. Vogels. 1983. Symbiosis of methanogenic bacteria and sapropelic protozoa. Arch. Microbiol. 136:89-95. [Google Scholar]
- 79.Vetriani, C., H. W. Jannasch, B. J. MacGregor, D. A. Stahl, and A. L. Reysenbach. 1999. Population structure and phylogenetic characterization of marine benthic archaea in deep-sea sediments. Appl. Environ. Microbiol. 65:4375-4384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Vogt, P. R., G. Cherkashev, G. Ginsburg, G. I. Ivanov, A. Milkov, K. Crane, A. Lein, E. Sundvor, N. V. Pimenov, and A. Egorov. 1997. Haakon Mosby Mud Volcano provides unusual example of venting. EOS Trans. Am. Geophys. Union Suppl. 78:556-557. [Google Scholar]
- 81.Vogt, P. R., J. Gardner, and K. Crane. 1999. The Norwegian-Barents-Svalbard (NBS) continental margin: introducing a natural laboratory of mass wasting, hydrates, and ascent of sediment, pore water, and methane. Geo-Mar. Lett. 19:2-21. [Google Scholar]
- 82.Wallner, G., R. Amann, and W. Beisker. 1993. Optimizing fluorescent in situ hybridization with rRNA-targeted oligonucleotide probes for flow cytometric identification of microorganism. Cytometry 14:136-143. [DOI] [PubMed] [Google Scholar]
- 83.Widdel, F., A. Boetius, and R. Rabus. 2006. Anaerobic biodegradation of hydrocarbons including methane, p. 1028-1049. In M. Dworkin, S. Falkow, E. Rosenberg, K.-H. Schleifer, and E. Stackebrandt (ed.), The prokaryotes, vol. 2. Springer-Verlag, New York, NY. [Google Scholar]
- 84.Widdel, F., F. Musat, K. Knittel, and A. Galushko. 2007. Anaerobic degradation of hydrocarbons with sulphate as electron acceptor, p. 265-304. In L. L. Barton and W. A. Hamilton (ed.), Sulphate-reducing bacteria: environmental and engineered systems. Cambridge University Press, Cambridge, United Kingdom.
- 85.Zhou, J., M. A. Brunns, and J. M. Tiedje. 1996. DNA recovery from soils of diverse composition. Appl. Environ. Microbiol. 62:316-322. [DOI] [PMC free article] [PubMed] [Google Scholar]