Abstract
The Aspergillus nidulans velvet (veA) gene encodes a global regulator of gene expression controlling sexual development as well as secondary metabolism. We have identified the veA homologue AcveA from Acremonium chrysogenum, the major producer of the β-lactam antibiotic cephalosporin C. Two different disruption strains as well as the corresponding complements were generated as a prelude to detailed functional analysis. Northern hybridization and quantitative real-time PCR clearly indicate that the nucleus-localized AcVEA polypeptide controls the transcriptional expression of six cephalosporin C biosynthesis genes. The most drastic reduction in expression is seen for cefEF, encoding the deacetoxycephalosporine/deacetylcephalosporine synthetase. After 120 h of growth, the cefEF transcript level is below 15% in both disruption strains compared to the wild type. These transcriptional expression data are consistent with results from a comparative and time-dependent high-performance liquid chromatography analysis of cephalosporin C production. Compared to the recipient, both disruption strains have a cephalosporin C titer that is reduced by 80%. In addition to its role in cephalosporin C biosynthesis, AcveA is involved in the developmentally dependent hyphal fragmentation. In both disruption strains, hyphal fragmentation is already observed after 48 h of growth, whereas in the recipient strain, arthrospores are not even detected before 96 h of growth. Finally, the two mutant strains show hyperbranching of hyphal tips on osmotically nonstabilized media. Our findings will be significant for biotechnical processes that require a defined stage of cellular differentiation for optimal production of secondary metabolites.
The filamentous fungus Acremonium chrysogenum is the exclusive producer of the pharmaceutically relevant β-lactam antibiotic cephalosporin C, which is structurally related to penicillins. The latter is synthesized by two other filamentous fungi, namely Penicillium chrysogenum or Aspergillus nidulans (11). While P. chysogenum is used mainly for the industrial production of penicillin, work with A. nidulans has focused on the understanding of the complex regulation of β-lactam biosynthesis in filamentous fungi. The first two steps of the biosynthesis of penicillin and cephalosporin C are identical in all fungal producers and involve the activity of N-(5-amino-5-carboxypentanoyl)-l-cysteinyl-d-valine synthase (ACV) synthetase (ACVS) and isopenicillin N-synthetase (IPNS). Interestingly, the genes for the two enzymes pcbAB and pcbC are not only highly homologous in the three fungal species but also have a very similar gene organization. They are located adjacently on the chromosome in a divergent orientation and share a common promoter sequence (5, 32, 40). This clustered gene organization has been found for many fungal genes encoding enzymes for specific metabolic pathways. In addition to β-lactam antibiotics, this includes secondary metabolites such as aflatoxin, gibberellins, or lovastatin (27, 48). Mainly molecular genetic investigations have shown that the coordinated regulation of gene clusters is controlled by narrow or broad domain regulators (20).
In A. chrysogenum, global regulators such as CPCR1, CRE1, and PACC control expression of the pcbAB and pcbC genes (16, 39, 41), whereas in A. nidulans, another set of factors has been discovered to be involved in the coregulation of the homologous genes (5, 20). One such global regulator is velvet (veA), a developmental gene first isolated and described in A. nidulans as a regulator of fungal morphogenesis (18, 29). While wild-type strains conidiate in the presence of light and are aconidial in the dark, mutation of the veA gene eliminates this light dependency, thus allowing conidiation to occur also in the dark (29). In addition, veA deletion strains are unable to develop sexual structures (cleistothecia), even under inducing conditions, and the overexpression of the veA gene leads to the formation of cleistothecia also under unfavorable conditions, where the wild type produces few or no sexual structures (21). These findings led to the conclusion that veA negatively regulates asexual development, while positively regulating sexual development. Recent studies have demonstrated the involvement of veA also in the secondary metabolism of different aspergilli. Examples include versicolorin synthesis in Aspergillus parasiticus, cyclopiazonic acid, aflatrem, and aflatoxin synthesis in Aspergillus flavus (7, 10) or sterigmatocystin and β-lactam biosynthesis in Aspergillus nidulans (19). In the latter, veA regulates transcriptional expression of the two penicillin biosynthesis genes. After deleting the veA gene, an increase in the ipnA (pcbC) gene expression was observed, while acvA (pcbAB) gene expression was no longer detectable (19). This led to an overall decrease in penicillin production in the deletion strain.
Until now, it has been unknown whether velvet, a highly conserved gene in the aspergilli, is also involved in the control of cephalosporin C biosynthesis. Here, we present the molecular characterization and functional analysis of the veA homologue from A. chrysogenum. This is the first report showing that a velvet homologue controls biosynthesis of the β-lactam antibiotic cephalosporin C. In addition, we demonstrate that the time-dependent onset of hyphal fragmentation and hyphal branching in A. chrysogenum requires the presence of a full-size A. chrysogenum VEA (AcVEA) polypeptide. Our findings will have a significant impact on the implementation of further strain improvement programs.
MATERIALS AND METHODS
Strains, DNA extraction, culture conditions, and transformation of A. chrysogenum.
Competent Escherichia coli cells of strain K-12 XL1-Blue (Stratagene) were used for routine DNA manipulations (6). Construction, maintenance, and isolation of recombinant plasmids were performed using standard techniques (37). The cephalosporin C producer strain A3/2 (36) from A. chrysogenum was used as a recipient, and all transformants (Table 1) were obtained by a procedure described previously (24, 47). The transgenic strains were selected on media containing either 10 U/ml of hygromycin B or 25 μg/ml nourseothricin. Cultures were grown in liquid complex culture medium (CCM) (28) or production medium (38) with 55 g/liter dextrin instead of saccharose at 27°C and 180 rpm in Erlenmeyer flasks and in the dark. Fungal DNA extraction was done as described previously (14). Cultures for microscopic studies were started with a 5% inoculum from a 2.5-day preculture as described previously (14).
TABLE 1.
A. chrysogenum strains used in this study
| Strain | Characteristic(s) | Reference |
|---|---|---|
| A3/2 | Producer strain (recipient) | 36 |
| ΔveA | ΔveA::hygB | This work |
| ΔveA::veA | ΔveA::hygB; veA(p)::nat | This work |
| KiveA | veA::hygB | This work |
| KiveA::veA | veA::hygB; veA(p)::nat | This work |
| ΔveA::veA_egfp | ΔveA::hygB; gpd(p)::veA::egfp::trpC(t) | This work |
| ΔveA::veAt_egfp | ΔveA::hygB; gpd(p)::veAt::egfp::trpC(t) | This work |
Isolation and sequencing of genomic DNA.
For the isolation of a veA gene fragment of A. chrysogenum, heterologous primers 8/10 (see Table 2) were designed on the basis of the N. crassa genome sequence (available at http://www-genome.wi.mit.edu/annotation/fungi/neurospora/) and used for PCR amplification with genomic DNA from A3/2 as template. The resulting amplicon was cloned into vector pDrive (QIAGEN, Hilden, Germany) and used for subsequent DNA sequencing by a custom sequencing service (MWG Biotech, Ebersberg, Germany). The amplicon obtained by this PCR approach was further employed to screen an A. chrysogenum cosmid library (custom-made cosmid library using vector P1920; MWG Biotech, Ebersberg, Germany). The screening resulted in the isolation of cosmid E11 containing the entire AcveA open reading frame (ORF), together with flanking regions, and FASTA (31) was used for comparisons of nucleotide and amino acid sequences. Alignments were made using the CLUSTAL W program (44) provided by the European Bioinformatics Institute and formatted using the multiple sequence alignment editor GeneDoc (http://www.psc.edu/biomed/genedoc).
TABLE 2.
Oligonucleotides used in this study
| Oligonucleotide | Specific sequence (5′ to 3′)a | Specificity (position)b |
|---|---|---|
| 8 | CAATCCTGGAAACTTTTTGGCGCT | AcveA (2021-2044) |
| 10 | ACTTTTCAGTACAACGCCAACTTTTTCCTC | AcveA (1681-1710) |
| 30 | GAGCTCCTTGTCCTGTAGCGCTTGCC | AcveA (91-110) |
| 31 | GCGGCCGCACGATATGGCTTCGGATG | AcveA (1364-1381) |
| 32 | GCGGCCGCCATCTACTCTATTCCTTTGC | hph from pZHK2 |
| 33 | TCTAGATGCCAACACATTTGCGTGCC | AcveA (3001-3020) |
| 34 | ACTAGTGGTATGGCGAGGTTACGCGT | AcveA (5069-5088) |
| 35 | TCTAGAAACGACGGCCAGTGCCAAGC | trpC(p) from pZHK2 |
| 42 | AGATCAAGACTTCGCCCTTC | AcveA (1991-2010) |
| 49 | GGCCGGCTACATGGTGAATA | AcveA (2981-3000) |
| 52 | GATCCTTGCTAGCGCCGGAC | AcveA (26-45) |
| 57 | TCTCGTTTCACCTCCACCCA | AcveA (1301-1320) |
| 59 | CAGGTATCAAAACGCAACAG | AcveA (3061-3080) |
| Ac_IPNS_U1 | ACCAGTCCGACGTGCAGAAT | pcbC gene |
| Ac_IPNS_L1 | TCGGTGATATGGGCCATGTAG | pcbC gene |
| Ac_cefEF_U1 | CCGTAACCACCAAGGGTATCT | cefEF genes |
| Ac_cefEF_L1 | CTCCTCGCTTCCGTTCTTGA | cefEF genes |
| Ac_ACVS_U1 | CGTTGCGCACCGTAACC | pcbAB genes |
| Ac_ACVS_L1 | CACTGCCTGGATAAGTCCATGA | pcbAB genes |
| Ac_cefG_U1 | AAGAGCAAACCTGCGATGGA | cefG gene |
| Ac_cefG_L1 | TCTGTGCCGTTGATTTCCTTCT | cefG gene |
| Ac_cefD1_U1 | GAACGACTTGATGAGCGGAATA | cefD1 gene |
| Ac_cefD1_L1 | TCCCCGACATAGACAAATATTGTTG | cefD1 gene |
| Ac_cefD2_U1 | CTAGCCCCAGGCCCATTCT | cefD2 gene |
| Ac_cefD2_L1 | CGATGCAGATGGACGACTTGT | cefD2 gene |
| Ac_act_U3 | GCGACGTCGATGTCCGTAA | Actin gene |
| Ac_act_L3 | AGAAGGAGCAAGAGCAGTGATCTC | Actin gene |
| 66 | CCATGGCCACCCCTTCGCTCAT | AcveA (1390-1405) |
| 74 | CCATGGCGAGCGCGGGCGGCGG | AcveA (2219-2235) |
| 75 | CCATGGTGAATACGCTGAACTT | AcveA (2971-2986) |
Recognition sequences for endonucleases that were used for the construction of knockout cassettes are underlined. For further details, see the text.
Nucleotide positions for AcveA gene are from accession no. AM410093 in the EMBL data library.
Generation of an insertional mutant.
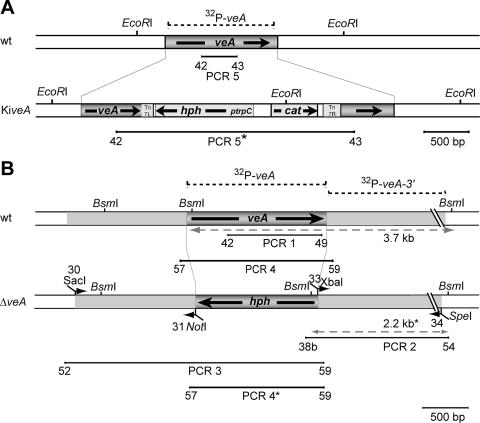
The screening of an A. chrysogenum cosmid library using the AcveA gene as a probe led to the isolation of cosmid E11, carrying the entire AcveA open reading frame together with flanking sequences. For in vitro transposition, we used a novel transposition vector, pGPS2.1-hph, that was constructed as follows. The SwaI-restricted plasmid pGPS2.1 (New England Biolabs) was ligated with an HpaI fragment from vector pZHK2 (23) carrying the hph gene. The recombinant vector contains the hph gene under the control of the trpC promoter together with the chloramphenicol acetyltransferase resistance (CmR) gene. Both genes are flanked by the right- and left-hand borders from transposon Tn7 (New England Biolabs), designated Tn7R and Tn7L, respectively. pGPS2.1-hph was used in the genome priming system for in vitro transposition of the hph gene into cosmid E11, according to the manufacturer's recommendations. The recombinant transposon inserts randomly into the target DNA (in vitro), and selection of clones with the mutagenized DNA was performed on chloramphenicol-containing media. The resulting recombinant cosmid, E11-hph, carries an insertion at nucleotide position 850 of the AcveA ORF and received the designation AcveAt. A. chrysogenum insertional mutants, designated as knock-in (KiveA), carrying E11-hph can be selected on hygromycin B-containing media.
Deletion of the AcveA gene.
To replace the AcveA open reading frame with the hygromycin phosphotransferase gene (hph), deletion vector pVEAKO was constructed as follows. Approximately 1.3 kb and 2.0 kb of the upstream and downstream regions of the AcveA gene together with primer pairs 30/31 and 33/34 (see Table 2), respectively, were amplified by PCR using genomic DNA from A3/2 as template. Both amplicons are terminated by SacI/NotI (upstream region) or XbaI/SpeI (downstream region) restriction sites. Together with the hph cassette derived from plasmid pZHK2 and terminated by NotI/XbaI restriction sites, these fragments were inserted into vector pBluescript KS+. The resulting vector, pVEAKO, contains a 4.8-kb insert with the hph gene flanked by genomic sequences from A. chrysogenum. This insert is terminated by SacI and SpeI restriction sites, as shown in Fig. 1B. Transformation of recipient strain A3/2 was done with the 4.8-kb SacI/SpeI fragment by standard protocols (46). DNA extraction and Southern blot analysis were performed as described previously (14) using standard procedures (37). Screening for deletion mutants was performed by a PCR-based approach using primer pair 57/59 (see Table 2), leading to amplicons of 1,780 bp in the wild-type gene and 1,594 bp for the knockout cassette. Transformants lacking the 1,780-bp PCR product were further tested by PCR using primer pairs 42/49 and 52/59 (see Table 2) in order to amplify fragments representing the AcveA ORF or upstream region of the AcveA gene.
FIG. 1.
Disruption of the AcveA gene in A. chrysogenum. The AcveA ORF is represented by the dark gray bar. (A) Generation of an insertional mutant. Schematic representation of the AcveA open reading frame before and after insertion of the transposon. (B) Generation of a ΔveA strain: genomic organization of the A. chrysogenum veA locus from wild-type (wt) and knockout strains. The disruption cassette was constructed by amplifying the AcveA flanking regions by PCR. The restriction sites necessary for further cloning were inserted through the primers. The 5′ and 3′ flanking regions present on the disruption cassette are depicted by the light gray regions. Fragments expected in Southern blot analysis with the 3′ AcveA-flanking probe against BsmI-restricted DNA from the wild-type or the knockout strain are indicated by light-gray dashed double-headed arrows. The locations of the oligonucleotide primers used for PCR amplifications and the sizes of the expected amplicons are indicated by the numbers at the respective lines. The DNA fragment used for Southern blot analysis is represented by the dashed line over the map of the wild-type locus.
Complementation of AcveA disruption strains.
For complementation of the AcveA gene in both disruption strains, cosmid E11 was modified by insertion of the nourseothricin resistance cassette. For this purpose, we used pG-Nat1 (22), which carries a modified Transprimer element, derived from pGPS2.1 (New England Biolabs) that includes the nat1 gene for selection of transformants. Screening of recombinant cosmids was done by Southern blot analysis using the AcveA ORF as probe. A single clone (E11-nat1), with a transposon insertion outside of the AcveA ORF was used for further experiments, and transformation into ΔveA and KiveA strains generated the ΔveA::veA and KiveA::veA complements, respectively.
Construction of plasmids for fluorescence microscopy.
pEHN1-nat is a fungal expression vector carrying the enhanced green fluorescent protein (egfp) gene under the control of the Aspergillus nidulans gpd promoter and trpC terminator; it also contains the nat1 gene for selection of transgenic strains (22). Using the unique NcoI restriction site, this vector was used to generate fusions of the egfp gene with the entire or truncated AcveA ORF. For amplification of the AcveA derivatives, primer pairs 66/75 and 66/74 were used (see Table 2). The length of the ORF in AcveAt corresponds to the truncated open reading frame that is contained on cosmid E11-hph (see above).
Microscopy and image analysis.
Hyphal morphology at different cultivation times (48 to 120 h) was analyzed using a Zeiss Axiophot microscope (Carl Zeiss, Jena, Germany). Images were captured with an Axiovision digital imaging system and processed with Adobe Photoshop 6.0 software. Fluorescence microscopy was carried out with an AxioImager microscope (Zeiss, Jena, Germany) using an XBO 75 xenon lamp for fluorescence excitation. Fluorescence was studied using Chroma filter sets 41017 (exciter, HQ470/40; emitter, HQ525/50; beam splitter, Q560Ip) (Chroma Technology Corp., Rockingham, VT) for EGFP detection. Images were captured with a Photometrix Cool SnapHQ camera (Roper Scientific) and MetaMorph (version 6.3.1; Universal Imaging). Recorded images were edited with MetaMorph and Adobe Photoshop 6.0 software.
RNA isolation.
RNA extraction from mycelia, grown in production media as mentioned above, was done using the TRIzol solution (Invitrogen) and RNeasy kit (QIAGEN, Hilden, Germany) according to the manufacturer's protocol. Total RNA from mycelia grown in CCM was isolated as previously described (15), and the integrity of RNAs was verified by agarose gel and Northern blot analysis according to standard procedures (37).
qRT-PCR.
Quantification of mRNA levels derived from the pcbAB, pcbC, cefD1, cefD2, cefEF, and cefG genes was performed in two independent experiments. Total RNA was reversed transcribed using the high-capacity cDNA Archive kit (Applied Biosystems) according to the manufacturer's protocol. cDNA was analyzed in quantitative real-time PCR (qRT-PCR) assays in triplicates on an ABI 7900HT Fast real-time PCR machine (Applied Biosystems) using Power SYBR green master mix (Applied Biosystems) according to the manufacturer's recommendation. As a reference, the mean cycle threshold values from an amplicon derived from the actin mRNA were used for normalization. All primers used are given in Table 2.
Titer analysis of cephalosporin C.
Culture broth from 100-ml liquid shake flask cultures, grown in production medium, was used for high-performance liquid chromatography (HPLC). Quantification of cephalosporin C was performed as previously described (39).
Nucleotide sequence accession number.
The sequence of the AcveA gene has been deposited in the EMBL data library under accession no. AM410093.
RESULTS
Isolation and characterization of the veA homologue gene from Acremonium chrysogenum.
In a pilot study, we have compared five protein-coding genes (coding for diphthamine biosynthesis methyltransferase, eIF-2α, eIF5, ATP synthase subunit beta, and DNA-directed RNA polymerase III second largest chain) from A. chrysogenum with the corresponding sequences from filamentous fungi that were used in genome sequencing projects. We found that A. chrysogenum shares a sequence similarity of 80% with N. crassa at the amino acid sequence level (J. Kamerewerd and U. Kück, unpublished data). Therefore, heterologous primers 8/10 (see Table 2) based on conserved domains of the N. crassa velvet sequence were used to obtain a 360-bp amplicon using A. chrysogenum genomic DNA as template. This product was further used to screen an A. chrysogenum cosmid library leading to the isolation of cosmid E11. Using a primer-walking strategy, the complete nucleotide sequence of the A. chrysogenum homologue of NcveA, including the 1.3-kb upstream and 2.0-kb downstream region, was determined. Annotation of the sequence identified an ORF of 514 amino acids that is interrupted by a single intron. The predicted gene was named AcveA, and an alignment of the derived amino acid sequence with those from velvet proteins of other ascomycetes shows high conservation in the N-terminal regions (data not shown). Overall, the amino acid sequence has 54%, 37%, 33% and 28% identity with FvVEA from Fusarium verticillioides, NcVEA from Neurospora crassa, the hypothetical MgVEA of Magnaporthe grisea, and the VEA from Aspergillus nidulans. We also compared the sequence with a putative classical bipartite nuclear localization signal (NLS) motif, NLSI, which recently was predicted for the A. nidulans VEA polypeptide (43). Comparing this N-terminal sequence with the A. chrysogenum AcVEA sequence, we found considerable conservation between both sequences, although two highly conserved Lys residues are missing in the A. chrysogenum sequence, as well as in the velvet sequences from Fusarium verticillioides and Magnaporthe grisea. Using the computer program PSORTII (http://psort.nibb.ac.jp), another nuclear localization signal, PIGSKRK (NLSII), was predicted close to the C-terminal end of the putative polypeptide. This motif is less conserved in the compared sequences from other filamentous fungi. As demonstrated below, the function of the predicted NLSII is further supported by localization studies of AcVEA using fluorescent microscopy. To determine the gene copy number of AcveA in the A. chrysogenum genome, Southern blot analyses were performed with the complete AcveA gene as a probe. Using BsmI-, EcoRI-, or PstI- restricted genomic DNA from A. chrysogenum as a template, single hybridizing restriction fragments were obtained, indicating that AcveA is a single-copy gene (data not shown).
Generation of strains with a deleted or disrupted AcveA gene.
To study the function of velvet in A. chrysogenum, two different strains carrying a disrupted copy of AcveA were constructed. In addition, the two strains were complemented with a wild-type copy of AcveA, and the resulting strains were used for further analysis.
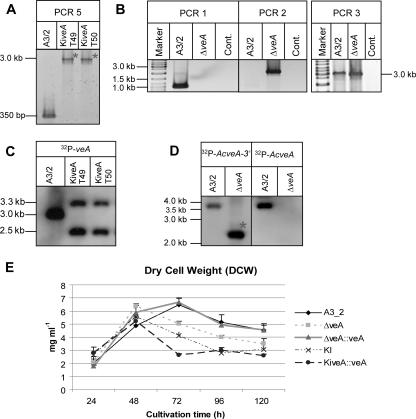
In the first attempt, we used recombinant cosmid E11-hph for generation of a fungal insertional mutant (KiveA). As described in Materials and Methods, E11-hph carries an AcveA ORF that is disrupted at nucleotide position 850 by the modified transposon Tn7 (Fig. 1A). Transformation of the wild-type strain of A. chrysogenum gave transformants that were characterized by a PCR-based strategy. Using DNA from all transformants as a template, primer pair 42/43 (see Table 2) generates two different PCR products (PCR 5 and 5*). While the 350-bp fragment from PCR 5 is derived from the wild-type AcveA gene, the 3.0-kb fragment of PCR 5* is obtained from the disrupted AcveA gene. As expected, most fungal transformants produced the PCR 5 as well as the PCR 5* product. However, we identified 2 transformants out of 90 that showed only the larger 3.0-kb fragment of PCR 5* (Fig. 2A). Both are therefore likely to contain a disrupted AcveA gene that was received by homologous recombination with cosmid E11-hph.
FIG. 2.
Characterization of AcveA disruption strains. Shown are the results of PCR analysis of the recipient strain A3/2 and the disruption strains KiveA (A) and ΔveA (B) using genomic DNA as template. The 2.2-kb amplicon generated by PCR 2 can only be obtained after insertion of the veA knockout cassette by homologous recombination into genomic DNA. Cont., control. (C and D) Southern analysis of strains A3/2, KiveA (C), and ΔveA (D). (C) The genomic DNA of the recipient and two insertional mutants (KiveA T49 and KiveA T50) was digested with EcoRI and hybridized with the 32P-AcveA probe. The hybridization bands of 3.3 kb and 2.5 kb in the transformants indicate the occurrence of the homologous recombination event depicted in Fig. 1A. (D) Genomic DNA was digested with BsmI. Expected band sizes after hybridization with the 32P-AcveA-3′ probe are as follows: A3/2, 3.7 kb; ΔveA, 2.2 kb (marked with an asterisk). As expected, hybridization with the 32P-AcveA probe gained a single band of 3.7 kb for the A3/2 strain. (E) Growth curves of the Α3/2, ΔveA, ΔveA::veA, KiveA (KI), and KiveA::veA strains determined as mg DCW per ml liquid CCM. The mean values of three independent experiments with respective standard deviations are shown.
To confirm further the disruption event, both transformants were analyzed by Southern hybridization. When probed with the AcveA gene, EcoRI-restricted genomic DNA from both strains showed two fragments of 2.5 kb and 3.3 kb after hybridization. In contrast, the wild-type strain gave only a single band of 3.0 kb (Fig. 2C). Both insertional mutants displayed identical phenotypes on all media tested (data not shown), and thus a single clone, designated KiveA, was used for further analysis.
In the second attempt, a knockout strain was constructed by a strategy previously described for A. chrysogenum (14). Plasmid pVEAKO contains a genomic sequence of A. chrysogenum of about 5.0 kb, in which the AcveA ORF was substituted for by the 1.4-kb hph resistance gene. The SacI/SpeI fragment from pVEAKO (Fig. 1B) was used to generate hygromycin-resistant A. chrysogenum transformants. Using a PCR-based screening procedure, fungal clones were identified in which the AcveA locus was substituted for by the hph gene via homologous recombination. As seen in Fig. 1B, the use of primer pair 57/59 (see Table 2) leads to an amplicon of 1.7 kb (PCR 4) in the wild-type gene, while the 1.5-kb band of PCR 4* originates from the knockout cassette. Transformants lacking PCR 4 were further tested with primer pairs 42/49 (PCR 1) and 38b/54 (PCR 2) for verification of the AcveA deletion by homologous integration (Fig. 2B). PCR 1 is only feasible in the recipient strain A3/2, whereas genomic DNA from a ΔveA strain lacks any binding sites for primers 42/49. Further verification of the homologous integration event came from PCR 2. While primer 38b binds to the knockout cassette, primer 54 binds specifically to a sequence outside of the homologous targeting region. The 2.3-kb fragment of PCR 2 is indicative for the disruption event that occurred through homologous recombination in the ΔveA strain. As expected, A3/2 lacks any amplification product.
To analyze the deletion by Southern blotting, genomic DNA was digested with BsmI and hybridized with probes corresponding either to the 3′ region of the veA gene or the veA ORF. The 3′ probe detected a 3.7-kb fragment in the wild-type strain, which was absent in the knockout strain, in which a 2.2-kb band resulted from an additional BsmI site of the hph cassette. Rehybridization of the membrane with the veA ORF probe resulted in the expected 3.7-kb band for the recipient strain and complete absence of a signal in ΔveA, thereby confirming the construction of a knockout strain (Fig. 2D).
In summary, using A3/2 as a recipient, we constructed two different strains as a prelude to functional analysis of the homologue of the Aspergillus velvet gene in A. chrysogenum. KiveA carries a truncated version of AcveA that codes for a predicted polypeptide of 264 amino acids, while ΔveA lacks any codon of the AcveA ORF. Our strain construction was completed by generating two strains with the mutants described above as recipients. Using cosmid E11-nat1, both strains were complemented with the full-size AcveA wild-type gene that is contained in the transforming DNA. These strains were further characterized and carry ectopically integrated copies of the AcveA gene (data not shown). As summarized in Table 1, five different strains served as a basis for the detailed functional analysis. It is important to note that the dry cell weight (DCW) of all strains was measured at different time points (24 to 120 h) (Fig. 2E). No significant differences in DCW were observed, although the strains are distinguishable by their morphological and physiological features.
Subcellular localization of AcVEA.
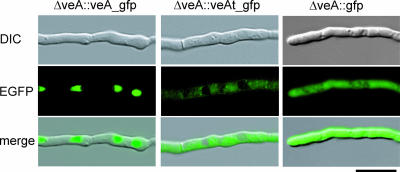
To determine the subcellular localization of the AcVEA protein and its truncated version, we generated egfp-tagged fusion constructs. While plasmid pveA_egfp contains the full-size AcveA gene fused to the egfp reporter gene, pveAt_egfp contains a truncated version of AcveA coding for the 264 N-terminal amino acids of the wild-type gene. Transformation of both plasmids into recipient ΔveA resulted in the ΔveA::veA-egfp and ΔveA::veAt-egfp strains. Microscopic investigation of both strains gave clearly different fluorescence patterns (Fig. 3). The ΔveA::veA-egfp strain shows that the complete AcVEA protein is targeted into the nucleus, producing a fluorescence pattern that coincides with the staining pattern of nucleic acid dyes (data not shown). The ΔveA::veAt-egfp strain provides a fluorescence pattern that is distributed throughout the cytoplasm and seems to be excluded from nuclei and vacuoles of the fungal cell. The fluorescence of the ΔveA::veAt-egfp strain can clearly be distinguished from the bright uniformly distributed fluorescence of strains synthesizing solely the EGFP. These results indicate that the predicted NLSII of AcveA is most probably functional. The truncated mRNA of AcveAt lacking the NLSII sequence is translated into a shortened version of the AcveA polypeptide that is unable to enter the nucleus.
FIG. 3.
Fluorescence microscopy of two strains carrying gfp fusions with the full-length (ΔveA::veA_gfp) or truncated (ΔveA::veAt_gfp) AcveA gene as indicated. Transformants carrying solely the egfp gene (ΔveA::gfp) served as a control. DIC, differential interference contrast.
Transcriptional expression of AcveA.
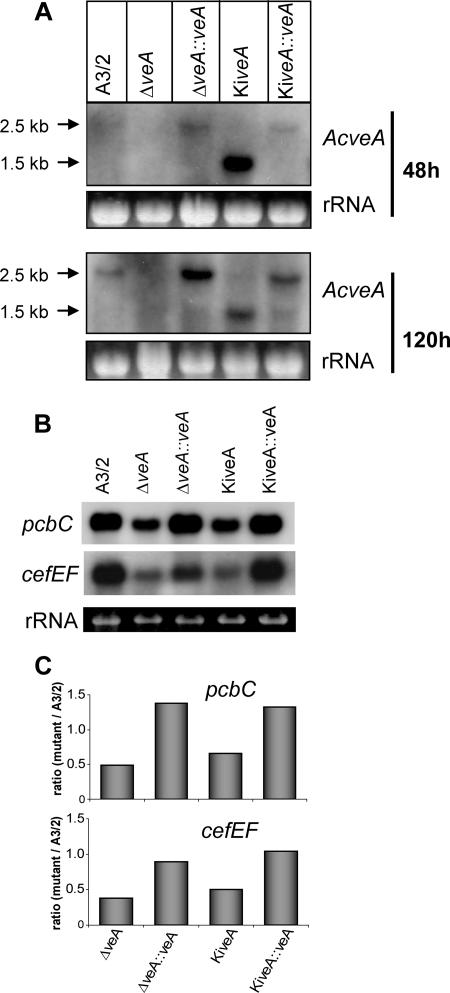
Next, we investigated the transcriptional expression of AcveA in the five different strains described above. For the hybridization analysis, RNA from all strains grown for 48, 72, 96, and 120 h on rich medium was used in a comparative investigation. As an example, Fig. 4A shows a hybridization pattern which was obtained with RNA from the 48- and 120-h-grown cultures. Northern blot analysis shows that the wild-type veA transcript is ∼2.5 kb in length, while the insertional mutant KiveA produces a transcript of only ∼1.5 kb. The generation of a stable 1.5-kb transcript can be explained by the presence of eukaryotic polyadenylation signals that are preceded by stop codons in all reading frames in the bacterial transposon (26). This leads to the premature polyadenylation of the mRNA, and thus most probably to a truncated AcveA polypeptide (AcVEAT). Interestingly, the 1.5-kb transcript seems to accumulate already after 48 h, while for the larger transcript of 2.5 kb, the highest transcript level was detectable after 120 h of growth (Fig. 4A).
FIG. 4.
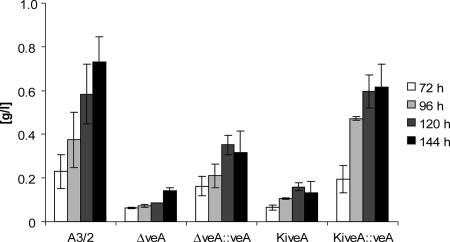
Transcript analysis of AcveA, pcbC, and cefEF expression in five strains as indicated. Total RNA was isolated after cultivation on CCM. (A) Northern hybridization of RNA isolated at two different time points with the AcveA gene as a probe. (B) Northern analysis with RNA of cultures grown for 120 h. Probes were used as indicated. (C) Quantification of the autoradiogram shown in panel B by densitometry using Scion Image software (Scion Corporation, Frederick, MD) and normalized with rRNA. Gene products: cefEF, deacetoxycephalosporine synthase; pcbC, IPN synthase.
Altered expression of cephalosporin biosynthesis genes in strains with a disturbed AcveA gene.
For Aspergillus nidulans, velvet is known to be a global regulator of genes involved in secondary metabolism of this fungus. This includes the three penicillin N biosynthesis genes, two of which are homologous to the pcbAB and pcbC from A. chrysogenum. Both encoded enzymes are involved in the first two steps of cephalosporin C biosynthesis in A. chrysogenum. We investigated the transcriptional expression of pcbAB and pcbC together with the cefD1, cefD2, cefEF, and cefG genes that are specific for cephalosporin C biosynthesis. RNA derived from all five strains was isolated at different time points of growth in batch culture with production media. In Fig. 4B, Northern blot analysis for the pcbC and cefEF genes at 120 h is shown, and for standardization, the amount of loaded RNA is shown. pcbC and cefEF show the highest transcriptional expression of all cephalosporin C biosynthesis genes and were therefore chosen for a quantitative analysis. As can be seen in Fig. 4B and C, the level of transcripts of both genes is highly abundant in A3/2, but clearly reduced in both disruption strains. The wild-type level was partially restored when the full-length AcveA gene was reinserted into the knockout strains. Both complements show an increase of the pcbC and cefEF transcript level. A more quantitative analysis of the transcripts was done by qRT-PCR using actin mRNA as internal standard. As can be seen in Fig. 5, the transcript levels of all genes were clearly decreased in both disruption strains. Quantification by qRT-PCR of the pcbC and cefEF transcripts thereby confirms data obtained by Northern blot analysis (Fig. 4C). The most drastic reduction is seen in the case of the cefEF transcript. While pcbAB, pcbC, cefD1, cefD2, and cefG transcript levels are reduced by about 20 to 50%, the cefEF transcript level decreases to less than 13% in both disruption strains. In both complements, the wild-type phenotype is almost fully restored, and these data were further confirmed by cephalosporin C titer quantification.
FIG. 5.
Quantitative real-time PCR analysis of the pcbAB, pcbC, cefD1, cefD2, cefEF, and cefG transcripts in the indicated strains. Data are given as ratio of mutant to A3/2 with the corresponding standard deviations. Results were tested for the significance of differential expression at P = 0.001 using REST (33). Gene products: pcbAB, ACV synthethase; cefG, deacetylcephalosporine acetyltransferase. All other products are given in the legend to Fig. 4.
Disruption of the AcveA gene results in reduction of cephalosporin C.
In the next set of experiments, cephalosporin C was quantified by HPLC. For this analysis, culture broths of those mycelia were used, which were employed for the RNA extractions as described above. For this comparative analysis, five different strains were investigated in parallel and HPLC values for all strains were taken at four different time points (Fig. 6). The amounts produced by the ΔveA and KiveA strains are similar and account for only approximately 20% of the amount produced by A3/2. Compared to the disruption strains, the ΔveA::veA and KiveA::veA complemented strains show an increase in cephalosporin C titer reaching amounts similar to that of the wild type. These results correlate well with the overall decreased expression of the cephalosporin C biosynthesis genes in disruption strains observed by Northern blot and qRT-PCR analyses.
FIG. 6.
HPLC quantification of cephalosporin C. All strains were grown in production media and harvested after the indicated time. The supernatant of the cultures was analyzed by HPLC as described in Materials and Methods.
Disturbance of AcveA leads to an altered hyphal morphology.
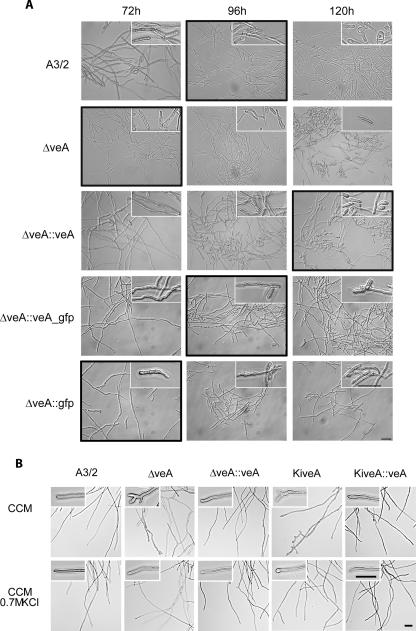
We have previously shown that ACFKH1 and CPCR1, two global regulators of secondary metabolism in A. chrysogenum, have a strong impact on fungal arthrospore morphology (14). The fragmentation of long hyphae is an active developmental process in cellular differentiation of A. chrysogenum and results in the formation of spherical cells called arthrospores (9, 30, 35). It was therefore logical to investigate also the impact of AcveA on fungal morphology, since AcveA homologues in aspergilli have a significant effect on fungal development. For our microscopic investigations, fungal morphology was monitored after 48, 72, 96, and 120 h of growth in liquid medium, using the five different strains described above. Arthrospore formation in the A3/2 strain starts at 96 h and is correlated with an increase in the yield of cephalosporin C biosynthesis (1, 30). As shown in Fig. 7A, microscopic examinations revealed, however, that the observed hyphal fragmentation in the disruption strain is different from arthrospore formation. While A3/2 arthrospores can be described as round-ended short hyphal fragments with one or two compartments, the hyphal fragments of the ΔveA strain are on average larger and often have one tapered end. Measurements of the length of the hyphal fragments smaller than 20 μm showed that in the recipient strain about 57% of the cells are larger than 10 μm and the remaining cells are smaller than 10 μm. In contrast, the ΔveA strain showed a distribution of 84% of the hyphal fragments being larger than 10 μm and only 16% being smaller. This hyphal fragmentation can already be seen after 72 h in ΔveA. Most importantly, the A3/2 phenotype is restored when ΔveA is complemented with AcveA (ΔveA::veA) and arthrospores are not formed before 96 h. After reintroduction of the AcveA gene, arthrospores have an even more spherical appearance than in the recipient strain, with 53.7% of the fragments being larger than 10 μm and the rest (46.3%) being smaller. These data were confirmed when the ΔveA strain was complemented with the veA_gfp fusion gene or with the gfp gene. While the fusion protein obviously restored the A3/2 phenotype, GFP itself is not able to prevent hyphal fragmentation at 72 h.
FIG. 7.
Microscopic investigations of A. chrysogenum strains as indicated. (A) For arthrospore formation, all strains were grown at 27°C and 180 rpm in liquid CCM. At the assigned time points, mycelial morphology was analyzed by differential interference contrast microscopy, and representative microscopic fields are depicted. Insets show an enlargement of characteristic mycelial structures, and black frames indicate the beginning of arthrospore formation. (B) Growth of the recipient strain and transformants in complete medium with (CCM plus 0.7 M KCl) and without (CCM) the addition of the osmotic stabilizer KCl. Insets show an enlargement of characteristic mycelial tips. The scale bar represents 20 μm in all panels.
The two strains with a disturbed veA (ΔveA and KiveA) gene did not show any obvious phenotype when grown on solid CCM (data not shown). In static liquid culture, the mycelial growth of recipient A3/2 typically consists of slender hyphal filaments that almost exclusively display lateral branching. In contrast, both disruption strains show altered hyphal tip morphology with frequent dichotomous branching or hyperbranching (Fig. 7B). However, this phenotype could be restored by the addition of osmotic stabilizers, indicating that the change in morphology could be due to an alteration in cell wall composition.
DISCUSSION
The homologue of the Aspergillus velvet gene from A. chrysogenum shows a high degree of sequence similarity and carries a putative nuclear localization signal at the C-terminal end.
During her pioneering work with Aspergillus nidulans, E. Käfer (18) generated the velvet mutant, showing delayed and reduced sexual reproduction together with enhanced conidiation in the dark. Molecular investigations later identified the velvet gene encoding a 537-amino-acid-long polypeptide that is conserved in all filamentous ascomycetes investigated so far (21, 25). It was further shown that in the original velvet mutant, a VEA polypeptide shorter by 36 amino acids than the wild-type protein is most probably produced. This truncation is located at the N terminus and affects a functional NLS that could be specifically recognized by the general and nuclear importin α (43). Yeast two-hybrid analysis has shown that the A. nidulans NLS interacts with importin α and deletion of the VEA-NLS motif weakens this interaction, indicating that additional VEA motifs interact with importin α (34). An appropriate candidate sequence might be NLSII, predicted by us to be between amino acid positions 454 and 460. This prediction is consistent with our observation that only the full-length AcVEA polypeptide, and not the truncated version, shows nuclear localization by fluorescent microscopy. Two nuclear localization signals have frequently been reported for proteins from higher as well as lower eukaryotes (2, 34). The transcriptional activator Gcn4p from yeast, for example, has been shown to carry two nuclear localization signals, NLS1 and NLS2. While the nuclear localization function of NLS1 is relatively unspecific, NLS2 mediates specific translocation by a heterodimeric importin α/β complex (34). Another case was described for the Aspergillus oryzae HapB subunit of the CCAAT-binding complex. This polypeptide carries two C-terminal NLS motifs. Only when both were mutated was a complete loss of the nuclear localization ability observed (12).
Expression of the late cluster gene cefEF is severely affected by disruption of AcveA.
The involvement of veA in the regulation of secondary metabolism has been shown for different species of aspergilli, where veA is necessary for the synthesis of different metabolites (7, 10, 19). In A. nidulans, the first two genes of penicillin biosynthesis were analyzed and veA was shown to repress the transcription of the isopenicillin synthase gene ipnA and to be necessary for the expression of acvA. Since the expression of acvA is the rate-limiting step in penicillin biosynthesis, this led to a reduction in penicillin production (19). However, recently, the opposite was observed using two different veA disruption strains (42). These authors used reporter genes to demonstrate that VeA acts as a repressor mainly on pcbAB (acvA) expression. In addition, penicillin V synthesis was increased under veA-inducing conditions.
While in A. nidulans and P. chrysogenum the penicillin biosynthesis genes are found in a single cluster, in A. chrysogenum the genes involved in cephalosporin biosynthesis are organized in at least two clusters located on different chromosomes. The pcbAB, pcbC, cefD1, and cefD2 genes are linked in the so-called “early” cephalosporin cluster, while the “late” cluster contains the cefEF and cefG genes. These genes are involved in the last two steps, which are specific for cephalosporin C biosynthesis (4, 40). Knowledge about the molecular regulation of β-lactam biosynthesis in A. chrysogenum is, however, still very limited. Here, we analyzed the effect of AcveA on the cephalosporin C titer and transcriptional expression of the “early” genes as well as the “late” genes, cefEF and cefG. The quantification of Northern hybridizations as well as qRT-PCR experiments showed that disruption of AcveA represses transcription of all cephalosporin C biosynthesis genes. Similarly, all downstream-acting enzymes are downregulated in the disruption strains. The strongest reduction was observed for cefEF transcription, which decreased to less than 15% in both disruption strains. We also determined the cephalosporin C content by HPLC and saw that it correlates well with the reduction in transcription levels. The amount of penicillin N produced, however, was less drastically altered (data not shown), thus indicating that the effect of AcveA is more pronounced on the late cluster of cephalosporin biosynthesis genes. The strong reduction of cefEF gene expression together with a reduction to less than 50% for the cefG gene can account for the strong decline in cephalosporin C biosynthesis, since deacetylcephalosporin C acetyltransferase, encoded by cefG, is a very labile enzyme requiring continuous resynthesis throughout the fermentation process (8). Moreover, cefG is very poorly expressed in A. chrysogenum compared to other genes of the cephalosporin pathway (pcbAB, pcbC, and cefEF) (45), and the overexpression of this gene was shown to result in increased cephalosporin C production (13), indicating that it is a limiting factor.
AcveA seems to be involved in the control of hyphal fragmentation and cell wall biosynthesis.
In filamentous fungi, secondary metabolism and morphological alterations are often linked phenomena, suggesting that their onset has a common regulation. The molecular determinants connecting both processes, however, remain largely undefined. It was therefore the aim of this study to see whether global regulators that have been reported to connect secondary metabolism and morphogenesis in A. nidulans (19) play a similar role in another β-lactam producer. In A. chrysogenum, the formation of uni- or bicellular swollen hyphal fragments called arthrospores seems to be correlated with high-yield cephalosporin C production in the corresponding production strains (14). This is consistent with observations that the phase of hyphal differentiation coincides with the maximum rate of β-lactam biosynthesis (30). We recently have shown that the RFX1 transcription factor CPCR1, but not the interacting forkhead transcription factor AcFKH1, controls arthrospore formation in A. chrysogenum (14). The fragmentation of hyphae observed in this contribution is different from typical round-ended short hyphal arthrospores. All hyphal fragments observed in the AcveA disruption strains are longer than typical arthrospores, indicating that cell wall integrity in these strains is disturbed. The fungal cell wall is critical for cell viability and pathogenicity, serving as a protective shell and providing cell morphology. The main macromolecular components in the cell walls of higher fungi (Ascomycetes, Basidiomycetes, and Deuteromycetes) are mannoproteins, 1,3-β-glucan, and chitin, although wall composition frequently varies markedly between species of fungi. In yeast, it is known that the major mannoproteins are present both at the surface and in deeper layers of the cell wall, shielding the cell wall glucan. Removal of most of the mannoproteins results in an increased porosity of the cell wall (49). Deletion of the veA gene in A. nidulans resulted in reduced transcript levels of the mannoprotein-encoding gene mnpA, without altering cell wall integrity (17). In Fusarium verticillioides, however, the veA homologue Fvve1 was shown to be necessary for normal cell wall composition and integrity (25). Disruption of the AcveA gene led to changes in hyphal tip morphology and hyphal fragmentation, indicating that similar to FvVE1 it might have an influence on cell wall composition. The hyperbranching and dichotomous branching observed in the disruption strains were restored by the addition of osmotic stabilizers, a further indication of an altered cell wall composition. A disturbance in cell wall composition, and thus rigidity, could also be the cause of the premature fragmentation observed under liquid shaking culture conditions. This could indicate that the ΔveA and KiveA strains do not produce regular arthrospores. This effect is more prominent in the ΔveA strain, where the hyphal fragments often displayed an altered morphology and the culture was also depleted of the typical spherical arthrospores that are present in large amounts in the ΔveA::veA strain.
Are velvet homologues differently regulated in different fungi?
The cellular function of velvet in A. nidulans is so far poorly understood. It was recently demonstrated that velvet most probably acts downstream of a phytochrome that represses sexual development under red-light conditions (3). It was also suggested that the phytochrome acts in the cytoplasm on red-light photoperception since it is excluded from the nuclei. Fungal phytochromes show a close relationship to bacterial phytochromes carrying an additional response regulator domain. These modules function in bacterial two-component signal transduction systems, and it can therefore be assumed that AcVEA, as a component of a signal transduction pathway, shuffles between the cytoplasm and the nucleus and thus controls directly or indirectly differential gene expression. This assumption is supported by the recent findings of a light-dependent subcellular mobility of the A. nidulans VEA polypeptide (43). In the dark, VEA is located mainly in the nuclei, while in the presence of light, VEA associates with filamentous bodies in the cytoplasm. As preliminary data, we found a similar light-dependent subcellular localization in A. chrysogenum cells grown in liquid CCM (J. Dreyer and U. Kück, unpublished observation). Stinnett and coworkers (43) discuss how phytochromes, similar to data obtained with velvet, are associated with electrodense particles and possibly are part of a single cytoplasmic complex. Whether this complex controls identical cellular functions in different filamentous fungi remains to be determined. However, Li et al. (25) have pointed out that the mode of control by velvet homologues in different fungi may be different, since velvet deletion phenotypes observed in Fusarium and Aspergillus species suggest different regulation of cellular processes. This would also explain the different regulation of early β-lactam biosynthesis genes in A. nidulans and A. chrysogenum, as demonstrated by us and others (19, 42).
Acknowledgments
We thank Ingeborg Godehard for her excellent technical assistance, Eva Szczypka and Gabriele Frenßen-Schenkel for the artwork, and B. Hoff and M. Nowrousian for fruitful discussions. Help by M. Nowrousian with the real-time experiments is acknowledged.
This work was funded by Sandoz GmbH (Kundl, Austria). J.D. was partially supported by a grant of the European Graduate College (EGC) 795, funded by the Deutsche Forschungsgemeinschaft (DFG) (Bonn-Bad Godesberg, Germany).
Footnotes
Published ahead of print on 30 March 2007.
REFERENCES
- 1.Bartoshevich, Y. E., P. L. Zaslavskaya, M. J. Novak, and O. D. Yudina. 1990. Acremonium chrysogenum differentiation and biosynthesis of cephalosporin. J. Basic Microbiol. 30:313-320. [DOI] [PubMed] [Google Scholar]
- 2.Beals, C. R., N. A. Clipstone, S. N. Ho, and G. R. Crabtree. 1997. Nuclear localization of NF-ATc by a calcineurin-dependent, cyclosporin-sensitive intramolecular interaction. Genes Dev. 11:824-834. [DOI] [PubMed] [Google Scholar]
- 3.Blumenstein, A., K. Vienken, R. Tasler, J. Purschwitz, D. Veith, N. Frankenberg-Dinkel, and R. Fischer. 2005. The Aspergillus nidulans phytochrome FphA represses sexual development in red light. Curr. Biol. 15:1833-1838. [DOI] [PubMed] [Google Scholar]
- 4.Brakhage, A. A., Q. Al-Abdallah, A. Tuncher, and P. Spröte. 2005. Evolution of beta-lactam biosynthesis genes and recruitment of trans-acting factors. Phytochemistry 66:1200-1210. [DOI] [PubMed] [Google Scholar]
- 5.Brakhage, A. A., P. Spröte, Q. Al-Abdallah, A. Gehrke, H. Plattner, and A. Tuncher. 2004. Regulation of penicillin biosynthesis in filamentous fungi. Adv. Biochem. Eng. Biotechnol. 88:45-90. [DOI] [PubMed] [Google Scholar]
- 6.Bullock, W. O., J. M. Fernandez, and J. M. Short. 1987. XL1-Blue: a high efficiency plasmid transforming recA Escherichia coli strain with beta-galactosidase selection. BioTechniques 5:376-378. [Google Scholar]
- 7.Calvo, A. M., J. Bok, W. Brooks, and N. P. Keller. 2004. veA is required for toxin and sclerotial production in Aspergillus parasiticus. Appl. Environ. Microbiol. 70:4733-4739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Demain, A. L. 1986. Regulation of secondary metabolism in fungi. Pure Appl. Chem. 58:219-226. [Google Scholar]
- 9.Drew, S. W., D. J. Winstanley, and A. L. Demain. 1976. Effect of norleucine on mycelial fragmentation in Cephalosporium acremonium. Appl. Environ. Microbiol. 31:143-145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Duran, R. M., J. W. Cary, and A. M. Calvo. 2007. Production of cyclopiazonic acid, aflatrem, and aflatoxin by Aspergillus flavus is regulated by veA, a gene necessary for sclerotial formation. Appl. Microbiol. Biotechnol. 73:1158-1168. [DOI] [PubMed] [Google Scholar]
- 11.Elander, R. P. 2003. Industrial production of beta-lactam antibiotics. Appl. Microbiol. Biotechnol. 61:385-392. [DOI] [PubMed] [Google Scholar]
- 12.Goda, H., T. Nagase, S. Tanoue, J. Sugiyama, S. Steidl, A. Tuncher, T. Kobayashi, N. Tsukagoshi, A. A. Brakhage, and M. Kato. 2005. Nuclear translocation of the heterotrimeric CCAAT binding factor of Aspergillus oryzae is dependent on two redundant localising signals in a single subunit. Arch. Microbiol. 184:93-100. [DOI] [PubMed] [Google Scholar]
- 13.Gutiérrez, S., J. Valasco, A. T. Marcos, F. J. Fernández, F. Fierro, J. L. Barredo, B. Díez, and J. F. Martin. 1997. Expression of the cefG gene is limiting for cephalosporin biosynthesis in Acremonium chrysogenum. Appl. Microbiol. Biotechnol. 48:606-614. [DOI] [PubMed] [Google Scholar]
- 14.Hoff, B., E. K. Schmitt, and U. Kück. 2005. CPCR1, but not its interacting transcription factor AcFKH1, controls fungal arthrospore formation in Acremonium chrysogenum. Mol. Microbiol. 56:1220-1233. [DOI] [PubMed] [Google Scholar]
- 15.Jekosch, K., and U. Kück. 2000. Glucose dependent transcriptional expression of the cre1 gene in Acremonium chrysogenum strains showing different levels of cephalosporin C production. Curr. Genet. 37:388-395. [DOI] [PubMed] [Google Scholar]
- 16.Jekosch, K., and U. Kück. 2000. Loss of glucose repression in an Acremonium chrysogenum beta-lactam producer strain and its restoration by multiple copies of the cre1 gene. Appl. Microbiol. Biotechnol. 54:556-563. [DOI] [PubMed] [Google Scholar]
- 17.Jeong, H. Y., H. Kim, D. M. Han, K. Y. Jahng, and K. S. Chae. 2003. Expression of the mnpA gene that encodes the mannoprotein of Aspergillus nidulans is dependent on fadA and flbA as well as veA. Fungal Genet. Biol. 38:228-236. [DOI] [PubMed] [Google Scholar]
- 18.Käfer, E. 1965. Origins of translocations in Aspergillus nidulans. Genetics 52:217-232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kato, N., W. Brooks, and A. M. Calvo. 2003. The expression of sterigmatocystin and penicillin genes in Aspergillus nidulans is controlled by veA, a gene required for sexual development. Eukaryot. Cell 2:1178-1186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Keller, N. P., G. Turner, and J. W. Bennett. 2005. Fungal secondary metabolism—from biochemistry to genomics. Nat. Rev. Microbiol. 3:937-947. [DOI] [PubMed] [Google Scholar]
- 21.Kim, H., K. Han, K. Kim, D. Han, K. Jahng, and K. Chae. 2002. The veA gene activates sexual development in Aspergillus nidulans. Fungal Genet. Biol. 37:72-80. [DOI] [PubMed] [Google Scholar]
- 22.Kück, U., and B. Hoff. 2006. Application of the nourseothricin acetyltransferase gene (nat1) as a dominant marker for the transformation of filamentous fungi. Fungal Genet. Newsl. 53:9-11. [Google Scholar]
- 23.Kück, U., and S. Pöggeler. 2004. pZHK2, a bifunctional transformation vector, suitable for two step gene targeting. Fungal Genet. Newslett. 51:4-6. [Google Scholar]
- 24.Kück, U., M. Walz, G. Mohr, and M. Mracek. 1989. The 5′-sequence of isopenicillin N-synthetase gene (pcbC) from Cephalosporium acremonium directs the expression of the prokaryotic hygromycin B phosphotransferase gene (hph) in Aspergillus niger. Appl. Microbiol. Biotechnol. 31:358-365. [Google Scholar]
- 25.Li, S., K. Myung, D. Guse, B. Donkin, R. H. Proctor, W. S. Grayburn, and A. M. Calvo. 2006. FvVE1 regulates filamentous growth, the ratio of microconidia to macroconidia and cell wall formation in Fusarium verticillioides. Mol. Microbiol. 62:1418-1432. [DOI] [PubMed] [Google Scholar]
- 26.Lo, C., K. Adachi, J. R. Shuster, J. E. Hamer, and L. Hamer. 2003. The bacterial transposon Tn7 causes premature polyadenylation of mRNA in eukaryotic organisms: TAGKO mutagenesis in filamentous fungi. Nucleic Acids Res. 31:4822-4827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Martín, J. F., and P. Liras. 1989. Organization and expression of genes involved in the biosynthesis of antibiotics and other secondary metabolites. Annu. Rev. Microbiol. 43:173-206. [DOI] [PubMed] [Google Scholar]
- 28.Minuth, W., P. Tudzynski, and K. Esser. 1982. Extrachromosomal genetics of Cephalosporium acremonium. Curr. Genet. 5:227-231. [DOI] [PubMed] [Google Scholar]
- 29.Mooney, J. L., and L. N. Yager. 1990. Light is required for conidiation in Aspergillus nidulans. Genes Dev. 4:1473-1482. [DOI] [PubMed] [Google Scholar]
- 30.Nash, C. H., and F. M. Huber. 1971. Antibiotic synthesis and morphological differentiation of Cephalosporium acremonium. Appl. Microbiol. 22:6-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Pearson, W. R. 1990. Rapid and sensitive sequence comparison with FASTP and FASTA. Methods Enzymol. 183:63-98. [DOI] [PubMed] [Google Scholar]
- 32.Peñalva, M. A., R. T. Rowlands, and G. Turner. 1998. The optimization of penicillin biosynthesis in fungi. Trends Biotechnol. 16:483-489. [DOI] [PubMed] [Google Scholar]
- 33.Pfaffl, M. W., G. W. Horgan, and L. Dempfle. 2002. Relative expression software tool (REST) for group-wise comparison and statistical analysis of relative expression results in real-time PCR. Nucleic Acids Res. 30:e36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Pries, R., K. Bomeke, O. Draht, M. Kunzler, and G. H. Braus. 2004. Nuclear import of yeast Gcn4p requires karyopherins Srp1p and Kap95p. Mol. Genet. Genomics 271:257-266. [DOI] [PubMed] [Google Scholar]
- 35.Queener, S. W., and L. F. Ellis. 1975. Differentiation of mutants of Cephalosporium acremonium in complex medium: the formation of unicellular arthrospores and their germination. Can. J. Microbiol. 21:1981-1996. [DOI] [PubMed] [Google Scholar]
- 36.Radzio, R., and U. Kück. 1997. Efficient synthesis of the blood-coagulation inhibitor hirudin in the filamentous fungus Acremonium chrysogenum. Appl. Microbiol. Biotechnol. 48:58-65. [DOI] [PubMed] [Google Scholar]
- 37.Sambrook, J., and D. W. Russell. 2001. Molecular cloning. A laboratory manual. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- 38.Scheidegger, A., M. T. Kuenzi, and J. Nüesch. 1984. Partial purification and catalytic properties of a bifunctional enzyme in the biosynthetic pathway of beta-lactams in Cephalosporium acremonium. J. Antibiot. (Tokyo) 37:522-531. [DOI] [PubMed] [Google Scholar]
- 39.Schmitt, E. K., A. Bunse, D. Janus, B. Hoff, E. Friedlin, H. Kürnsteiner, and U. Kück. 2004. Winged helix transcription factor CPCR1 is involved in regulation of β-lactam biosynthesis in the fungus Acremonium chrysogenum. Eukaryot. Cell 3:121-134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Schmitt, E. K., B. Hoff, and U. Kück. 2004. Regulation of cephalosporin biosynthesis. Adv. Biochem. Eng. Biotechnol. 88:1-43. [DOI] [PubMed] [Google Scholar]
- 41.Schmitt, E. K., R. Kempken, and U. Kück. 2001. Functional analysis of promoter sequences of cephalosporin C biosynthesis genes from Acremonium chrysogenum: specific DNA-protein interactions and characterization of the transcription factor PACC. Mol. Genet. Genomics 265:508-518. [DOI] [PubMed] [Google Scholar]
- 42.Spröte, P., and A. Brakhage. The light dependent regulator velvet A of Aspergillus nidulans acts as a repressor of the penicillin biosynthesis. Arch. Microbiol., in press. [DOI] [PubMed]
- 43.Stinnett, S. M., E. A. Espeso, L. Cobeno, L. Araujo-Bazan, and A. M. Calvo. 2007. Aspergillus nidulans VeA subcellular localization is dependent on the importin alpha carrier and on light. Mol. Microbiol. 63:242-255. [DOI] [PubMed] [Google Scholar]
- 44.Thompson, J. D., D. G. Higgins, and T. J. Gibson. 1994. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 22:4673-4680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Velasco, J., S. Gutiérrez, F. J. Fernandez, A. T. Marcos, C. Arenos, and J. F. Martín. 1994. Exogenous methionine increases levels of mRNAs transcribed from pcbAB, pcbC, and cefEF genes, encoding enzymes of the cephalosporin biosynthetic pathway, in Acremonium chrysogenum. J. Bacteriol. 176:985-991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Walz, M., and U. Kück. 1993. Targeted integration into the Acremonium chrysogenum genome: disruption of the pcbC gene. Curr. Genet. 24:421-427. [DOI] [PubMed] [Google Scholar]
- 47.Walz, M., and U. Kück. 1995. Transformation of Sordaria macrospora to hygromycin B resistance: characterization of transformants by electrophoretic karyotyping and tetrad analysis. Curr. Genet. 29:88-95. [DOI] [PubMed] [Google Scholar]
- 48.Woloshuk, C. P., and R. Prieto. 1998. Genetic organization and function of the aflatoxin B1 biosynthetic genes. FEMS Microbiol. Lett. 160:169-176. [DOI] [PubMed] [Google Scholar]
- 49.Zlotnik, H., M. P. Fernandez, B. Bowers, and E. Cabib. 1984. Saccharomyces cerevisiae mannoproteins form an external cell wall layer that determines wall porosity. J. Bacteriol. 159:1018-1026. [DOI] [PMC free article] [PubMed] [Google Scholar]