Abstract
Bradyrhizobium strains isolated in Europe from Genisteae and serradella legumes form a distinct lineage, designated clade II, on nodulation gene trees. Clade II bradyrhizobia appear to prevail also in the soils of Western Australia and South Africa following probably accidental introduction with seeds of their lupine and serradella hosts. Given this potential for dispersal, we investigated Bradyrhizobium isolates originating from a range of native New World lupines, based on phylogenetic analyses of nodulation (nodA, nodZ, noeI) and housekeeping (atpD, dnaK, glnII, recA) genes. The housekeeping gene trees revealed considerable diversity among lupine bradyrhizobia, with most isolates placed in the Bradyrhizobium japonicum lineage, while some European strains were closely related to Bradyrhizobium canariense. The nodA gene tree resolved seven strongly supported groups (clades I to VII) that correlated with strain geographical origins and to some extent with major Lupinus clades. All European strains were placed in clade II, whereas only a minority of New World strains was placed in this clade. This work, as well as our previous studies, suggests that clade II diversified predominately in the Old World, possibly in the Mediterranean. Most New World isolates formed subclade III.2, nested in a large “pantropical” clade III, which appears to be New World in origin, although it also includes strains originating from nonlupine legumes. Trees generated using nodZ and noeI gene sequences accorded well with the nodA tree, but evidence is presented that the noeI gene may not be required for nodulation of lupine and that loss of this gene is occurring.
The papilionoid legume genus Lupinus comprises ca. 275 species of annual and perennial herbs and shrubs with an amphi-Atlantic distribution. The majority of species are distributed in the New World, with ca. 100 species in the western part of North America and ca. 85 species in the Andes. Only 15 species are found in the Old World, mainly surrounding the Mediterranean (15). Lupines, in part due to their highly effective nitrogen-fixing symbiosis with root nodule bacteria, have been grown since antiquity as a green manure and are an important pulse crop. Their adaptation to nutrient-poor, often acid soils and arid climates means that lupines can be grown in areas where cultivation of more demanding crops, such as soybeans, is problematic (12).
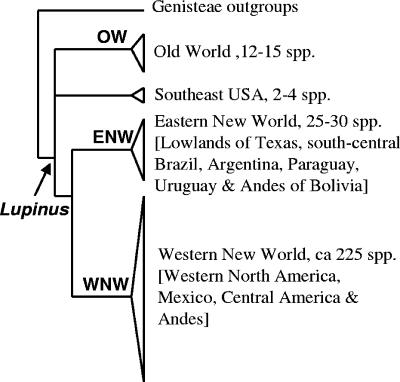
Lupines constitute an isolated lineage within the tribe Genisteae sensu stricto (2). The remaining Genisteae form two assemblages, one comprising the genera Anarthrophyllum, Argyrolobium, Dichilus, and Melolobium, which diversified predominantly in the Southern Hemisphere, and the other (called Genistinae) comprising Cytisus, Chamaecytisus, Genista, Retama, Spartium, Teline, Ulex, and several other small genera that have their centers of diversity in the Mediterranean basin. The geographic origin of the genus Lupinus remains unclear. However, phylogenetic analyses reveal four robustly supported clades that are congruent with lupine geography and chromosome number (1, 2, 3, 15). Notably, all the Old World species are placed in a single clade, here labeled OW, while the New World species comprise three strongly supported lineages; a large western New World (WNW) group distributed in western North America, Mexico, and the Andes; a small group centered in the southeast United States; and a predominately lowland eastern New World (ENW) group distributed mainly in the south-central United States and eastern South America (Fig. 1). The two large WNW and ENW lineages have largely allopatric distributions, but species from both clades are sympatric in limited areas, notably, in the south-central Andes in Bolivia. The Andean species (81 out of 85), which are nested in a strongly supported subclade within the WNW clade, provide one of the most spectacular examples of recent explosive plant species diversification driven by the recent uplift of the Andes (15).
FIG. 1.
Schematic Lupinus phylogeny showing major clades, which are robustly supported in individual and combined parsimony and Bayesian analyses of the nuclear DNA sequence loci internal transcribed spacer and LEGCYC1A that include up to 140 accession numbers (15). No rhizobia were available from lupine species in the small clade labeled Southeast USA for this study.
Cross-inoculation studies have shown that lupines share a common rhizobial pool with other legumes in the tribe Genisteae, including the genera Cytisus, Genista, Retama, and Teline (18, 19, 37, 38, 47). Additionally, lupines are effectively nodulated by rhizobia isolated from serradella (Ornithopus, a genus that belongs to the more distantly related tribe Loteae) and are ineffectively nodulated by rhizobia isolated from the genera Lotus, Anthyllis, and Phaseolus (8, 9, 25).
Lupines are nodulated by fast-growing rhizobia (that are poorly characterized), as well as by slow-growing strains of the genus Bradyrhizobium (8, 25). Phylogenetic studies based on nonsymbiotic genes revealed significant heterogeneity among lupine bradyrhizobia, which group with several additional distinct lineages, including Bradyrhizobium japonicum and Bradyrhizobium canariense. Fewer lupine bradyrhizobia grouped with Bradyrhizobium elkanii in housekeeping gene studies (5, 17, 23, 28, 41, 42, 48).
In contrast to the housekeeping gene phylogenies, most lupine isolates form a single cluster, referred to as clade II, in nodA nodulation gene trees. Notably, nodA clade II comprises bradyrhizobia isolated from other Genisteae species and from serradella species, which corroborates cross-inoculation data (19, 28, 42). Similar grouping was observed in phylogenies of nodC and nifH genes (17, 48, 49), giving rise to the new biovar genistearum for Bradyrhizobium strains nodulating Genisteae legumes, which presumably correspond to clade II Bradyrhizobium strains.
So far, most research has focused on Bradyrhizobium isolates from native Old World lupines growing in the Mediterranean (2) or from Old World species introduced into continental Europe, Australia, and South Africa. Considering that the four major lupine lineages occupy largely isolated present-day geographic distributions, the presumption is that they may be nodulated by rhizobia differing from European clade II strains. Our objective was to address this issue by searching for possible biogeographic patterns preserved in nod gene phylogenies. For this purpose, we selected Bradyrhizobium strains isolated mainly from lupine nodules collected from native Andean and lowland South American lupines (15).
MATERIALS AND METHODS
Bacterial strains.
The Bradyrhizobium strains included in the study and their characteristics are described in Table 1. The majority of lupine strains were isolated from dried nodules collected in the Andes and Brazil. We also included Bradyrhizobium type strains representing B. japonicum (USDA 6T), B. elkanii (USDA 76T), and B. yuanmingense (CCBAU 10071T), as well as several strains isolated from a range of legumes growing in Panama, Brazil, Japan, China, Mexico, and the United States. Yeast extract mannitol agar medium (46) was used for the growth and maintenance of the strains. All Bradyrhizobium strains were grown at 28°C. Bradyrhizobium strains were isolated from dried nodules as described by McInroy (26).
TABLE 1.
Bradyrhizobium strains used in this study
| Strain | Lineagea | Legume host | Lupinus cladec | Sampling origin | Source |
|---|---|---|---|---|---|
| BLUH1 | BC | Lupinus angustifolius | OW | Canary Islands, Spain | M. León Barrios |
| BLUT1 | BC | Lupinus albus | OW | Canary Islands, Spain | M. León Barrios |
| C4 | BJ1 | Lupinus mutabilis | WNW | Ecuador | G. Bernal |
| C8 | BJ1 | Lupinus mutabilis | WNW | Ecuador | G. Bernal |
| CH2310 | BJ1 | Lupinus bandelierae | ENW | Bolivia | This study |
| CH2318 | BJ1 | Lupinus bandelierae | ENW | Bolivia | This study |
| CH2352 | BJ1 | Lupinus sp. | WNW | Peru | This study |
| CH2355 | BJ1 | Lupinus misticola | WNW | Peru | This study |
| CH2391 | BJ1 | Lupinus sp. | WNW | Peru | This study |
| CH2437 | B. sp.b | Lupinus tominensis | WNW | Bolivia | This study |
| CH2438 | BJ1 | Lupinus breviscapus | WNW | Bolivia | This study |
| CH2443 | BJ1 | Lupinus pycnostachys | WNW | Bolivia | This study |
| CH2490 | BJ2 | Lupinus paranensis | ENW | Brazil | This study |
| CH2493 | BE | Lupinus paraguariensis | ENW | Brazil | This study |
| CH2498 | BJ2 | Lupinus rubriflorus | ENW | Brazil | This study |
| CH2506 | BJ1 | Lupinus uleanus | ENW | Brazil | This study |
| CH2509 | BJ2 | Lupinus albescens | ENW | Brazil | This study |
| CH2510 | BJ1 | Lupinus bracteolaris | ENW | Brazil | This study |
| FTA6 | B. sp | Lupinus nootkatensis | WNW | Alaska | M. Parker |
| FTA7 | BJ1 | Lupinus nootkatensis | WNW | Alaska | M. Parker |
| ICEA | BC | Lupinus nootkatensis | WNW | Iceland | M. Jóhannsson |
| ICEB | BC | Lupinus nootkatensis | WNW | Iceland | M. Jóhannsson |
| ICED | BC | Lupinus nootkatensis | WNW | Iceland | M. Jóhannsson |
| Lcamp1 | BJ1 | Lupinus campestris | WNW | Mexico | This study |
| Lcamp6 | B. sp. | Lupinus campestris | WNW | Mexico | This study |
| Lcamp8 | B. sp. | Lupinus campestris | WNW | Mexico | This study |
| Lpol9 | BJ1 | Lupinus polyphyllus | WNW | Poland | This study |
| MCLA07 | BC | Lupinus albus | OW | Salamanca, Spain | E. Velazquez |
| PL41 | BJ1 | Lupinus sp. | WNW | Peru | P. Lezama |
| RLA08 | BJ1 | Lupinus albus | OW | Leon, Spain | E. Velazquez |
| Zaluz80 | BC | Lupinus polyphyllus | WNW | Poland | This study |
| ORSAT6c | BJ1 | Ornithopus sativus | Mexico | This study | |
| ORSAT8c | BJ1 | Ornithopus sativus | Mexico | This study | |
| Cytisus11 | BJ1 | Cytisus sp. | Poland | W. Małek | |
| Os9 | BJ2 | Cytisus scoparius | Japan | W. Małek | |
| AEKY10 | BE | Amphicarpaea edgeworthii | Japan | M. Parker | |
| C10-2 | B. sp. | Inga oerstediana | Mexico | J. Grossman | |
| C8P-1 | BJ2 | Inga pavoniana | Mexico | J. Grossman | |
| CCBAU10071T | BY | Lespedeza cuneata | China | W.-X. Chen | |
| CPAC-M9 | BE | Mucuna aterrima | Brazil | M. Scotti Muzzi | |
| Da3-1 | B. sp. | Desmodium axillare | Panama | M. Parker | |
| F3b | BE | Lespedeza cuneata | Japan | T. Ezawa | |
| Jwc91-2 | BE | Amphicarpaea bracteata | United States | M. Parker | |
| KFR22 | B. sp. | Faidherbia albida | Kenya | D. Odee | |
| Mm1-3 | B. sp. | Machaerium milleflorum | Panama | M. Parker | |
| Ppau 3-41 | B. sp. | Phaseolus pauciflorus | Mexico | M. Parker | |
| Rp2-1 | B. sp. | Rhynchosia pyramidalis | Panama | M. Parker | |
| Q4A | BJ2 | Lespedeza sp. | Japan | T. Ezawa | |
| USDA3002 | BJ2 | Acacia decurrens | Brazil | P. Van Berkum | |
| USDA3259 | BE | Phaseolus lanatus | Illinois | P. Van Berkum | |
| USDA6T | BJ1 | Glycine max | Japan | P. Van Berkum | |
| USDA76 | BE | Glycine max | California | P. Van Berkum |
Molecular techniques.
For PCR sequencing experiments, total genomic DNA was isolated using the sodium dodecyl sulfate-proteinase K lysis procedure described by Moulin et al. (28) or using the QIAGEN QIAamp DNA minikit by following the manufacturer's recommendations. The primers used in this study are listed in Table 2. Primers were designed using OligoAnalyzer 3.0 software (http://www.idtdna.com/analyzer/Applications/OligoAnalyzer/Default.aspx). All PCR amplifications were made by following the procedures described previously (42). In brief, PCR samples were denatured at 95°C for 2 min, followed by 35 cycles of 95°C for 45 s, 58°C (atpD, dnaK, glnII, and recA) for 30 s, and 72°C for 1.5 min (2.5 min for nodA) and a final elongation step of 7 min at 72°C, as recommended for the FastStart high-fidelity PCR system by the manufacturer (Roche Diagnostics GmbH, Germany). The annealing temperature for amplification of nodA, nodZ, and noeI was 53°C, or 51°C for some strains in noeI amplification. PCR products were purified using the QIAquick gel extraction kit (QIAGEN, Germany) and sequenced using the BigDye Terminator v1.1 cycle sequencing kit on an ABI3100 automated capillary DNA sequencer (Applied Biosystems).
TABLE 2.
Oligonucleotides used for PCR amplificationa
| Primer | Sequence (5′-3′) | Target gene (primer position) | Reference or source |
|---|---|---|---|
| TSnodA2b | GATTCCVWGBCCYTCVAGATC | nodA (345-325b) | 42 |
| TSnodD1-1c | CAGATCNAGDCCBTTGAARCGCAT | nodD1 (24-1b) | 42 |
| TSnodB2N | CTGTGRTTHGCRAYCTYRTGYCC | nodB (239-217b) | 42 |
| TSnodZ3 | GGTTTCGGYGAYTGYCTBTGGTC | nodZ (40-62b) | 28 |
| TSnodZ4 | AATRTCTTCGCCRTTRCCRTGCC | nodZ (552-530b) | 28 |
| TSnoeI1 | CTGGATATCGGYTGCAATGATGG | noeI (64-86c) | 28 |
| TSnoeI2 | TCGGCYCCCTGMACRTCCATCCA | noeI (452-430c) | 28 |
| TSrecAf | CAACTGCMYTGCGTATCGTCGAAGG | recA (8-32c) | 42 |
| TSrecAr | CGGATCTGGTTGATGAAGATCACCATG | recA (620-594c) | 42 |
| TSatpDf | TCTGGTCCGYGGCCAGGAAG | atpD (189-208c) | 42 |
| TSatpDr | CGACACTTCCGARCCSGCCTG | atpD (804-784c) | 42 |
| TSglnIIf | AAGCTCGAGTACATCTGGCTCGACGG | glnII (13-38c) | 42 |
| TSglnIIr | SGAGCCGTTCCAGTCGGTGTCG | glnII (681-660c) | 42 |
| TSdnaK2 | GTACATGGCCTCGCCGAGCTTCA | dnaK (1794-1772c) | 40 |
| TSdnaK4 | GGCAAGGAGCCGCAYAAGG | dnaK (1057-1075c) | This study |
All primers were used for amplification and sequencing of PCR products.
Position of the primer in the corresponding sequence of lupine Bradyrhizobium sp. strain WM9.
Position of the primer in the corresponding sequence of B. japonicum USDA 110.
Phylogenetic analyses.
The multiple nucleotide sequence alignments were generated using ClustalX (44) and optimized manually with GeneDoc (30). All phylogenetic analyses were performed using PAUP version 4.0b10 (43). Due to the large number of sequences, parsimony and maximum likelihood (ML) analyses were performed using the heuristic search option of PAUP.
For the nodA, nodZ, noeI, atpD, dnaK, glnII, and recA data sets, ML and neighbor-joining (NJ) analyses were performed using PAUP. The best-fit model for each gene was assessed using MODELTEST3.6 (34). The best-substitution models (by Akaike information criterion in MODELTEST) found for each marker were, for atpD, dnaK, glnII, recA, and noeI, GTR+I+G (number of substitutions [NST] = 6; general time reversible with invariant sites and a gamma rate distribution) and, for nodZ, TVM+I+G (transversional model with invariant sites and a gamma rate distribution). The nodA phylogeny was constructed using the same ML model as described by Moulin et al. (28), i.e., base frequencies, transition/transversion ratios, and substitution rates were estimated at each position from the data using ML analyses (GTR+G+I model); third-codon positions were excluded from the analyses as they accumulated too much saturation (28). Starting from a random tree, five independent heuristic searches were performed to reach the best ML tree. The Bradyrhizobium-Methylobacterium-Burkholderia branch extracted from the best ML tree found (score: −ln L = 8,372.71 at 362,916 rearrangements, where L is likelihood) is shown (see Fig. 4). For the atpD, recA, dnaK, glnII, and nodZ gene trees, bootstrap analyses were performed using heuristic searches under distance and ML models (using PAUP), with 1,000 and 100 replicates, respectively. Due to the large datasets, bootstrap values for nodA phylogeny were assessed using only NJ analyses.
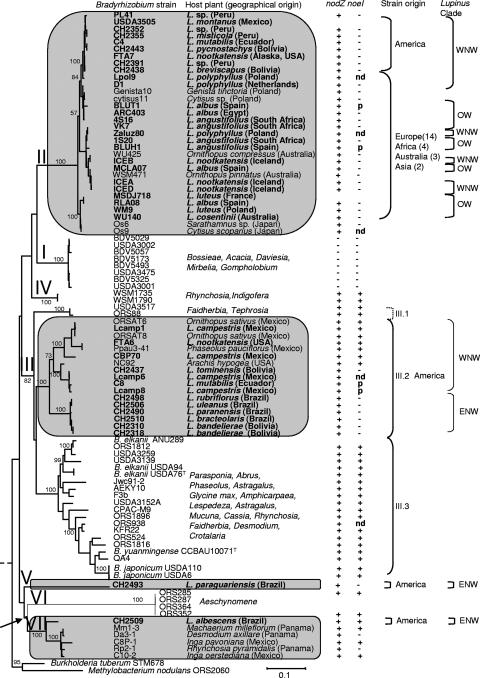
FIG. 4.
ML phylogenetic tree of Bradyrhizobium nodA genes. The tree was constructed by a ML approach described in Materials and Methods. Lupinus strains are indicated in boldface; clades to which they belong are in shaded blocks. The scale bar indicates the number of substitutions per site. Bootstrap values >70% (percentage of 1,000 replicates calculated under distance criteria) are given at the branching nodes. Clades I to IV correspond to the nodA phylogenies described elsewhere (28, 42). Host plant species are indicated on the right side of the tree together with geographical origins in nodA clades containing Lupinus strains. Succeeding columns show results of nodZ and noeI PCR amplifications on test strains, geographical origin of lupine strains, nodA subclades defined by bootstrap values >80%, and finally Lupinus clades corresponding to the phylogeny presented in Fig. 1 including WNW, ENW, and OW. Accession numbers of strains included in this tree are as follows: Bradyrhizobium japonicum USDA 110, NC_004463; B. elkanii USDA 94, U04609; Bradyrhizobium sp. strain ARC403, AJ430731; BDV5029, AJ890291; BDV5057, AJ890311; BDV5173, AJ891168; BDV5325, AJ890290; BDV5493, AJ890312; CBP70, AJ430730; D1, AJ430727; Genista10, AJ430726; NC92, U33192; ORS88, AJ430716; ORS285, AF284858; ORS287, AJ437607; ORS352, AJ438775; ORS364, AJ437613; ORS524, AJ430717; ORS938, AJ430715; ORS1812, AJ430723; ORS1816, AJ430722; ORS1896, AJ430718; 1S20, AJ890296; 4S16, AJ890289; USDA 3001, AJ430713; USDA 3139, AJ430712; USDA 3152A, AJ430711; USDA 3475, AJ430710; USDA 3505, AJ430709; USDA 3517, AJ430708; WSM471, AJ890307; WSM1735, AJ890293; WSM1790, AJ890286; WU140, AJ890313; WU425, AJ890300; VK7, AJ890295; Methylobacterium nodulans ORS2060, AF266748; Burkholderia phymatum STM678, AJ302321. Abbreviations: +, amplified by PCR; −, no PCR product; nd, not determined; p, pseudogene.
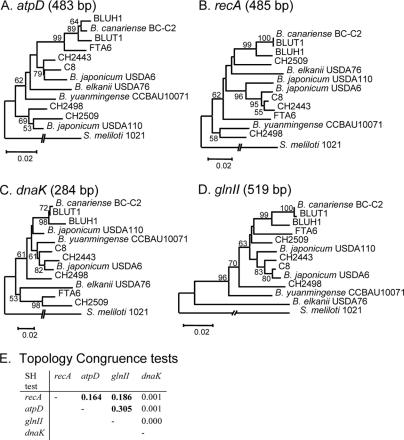
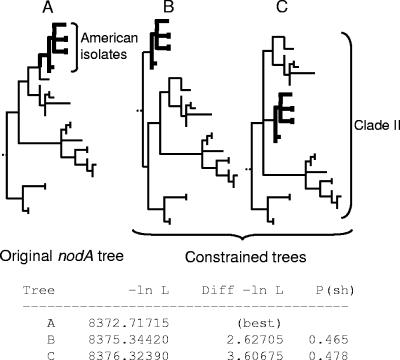
A Shimodaira-Hasegawa (S-H) test of congruence of tree topologies (trees constructed with an ML model [NST = 2], gamma distribution of sites, and ML estimates of base frequencies) was performed using PAUP on a restricted data set of 12 Bradyrhizobium strains for which all markers were available (i.e., BLUH1, BC-C2, BLUT1, FTA6, USDA76, C8, CH2443, USDA6, CCAU10071, CH2509, USDA110, and CH2498), with Sinorhizobium meliloti strain 1021 as a root. The results of the tests are shown in Fig. 2; see also Fig. S2 in the supplemental data. S-H tests were also performed on nodA-constrained trees, using the best ML models to build the original tree shown in Fig. 4 and to test several hypotheses on strain origin (see Fig. 5).
FIG. 2.
Comparison of the atpD (A), recA (B), dnaK (C), and glnII (D) gene fragment phylogenies (size of the alignment used for each marker is in parentheses). Trees shown were constructed by NJ, from a Kimura 2P distance matrix. Node robustness was evaluated by using 1,000 bootstrap replications. (E) Summary of S-H tests of congruence of tree topologies among ML phylogenies of the four markers (trees not shown). ML settings for each marker were of two types: a gamma distribution of site substitution and an estimating-base-frequency substitution. Boldface values indicate P values greater than 5% between two tree topologies, meaning the topologies are not statistically different by the S-H test.
FIG. 5.
S-H tests on several constrained topologies of clade II of the nodA phylogeny. The American isolate subclade II branch is in boldface, and its place in clade II was constrained in two alternative topologies. S-H tests were performed with the original tree. Tree A, best ML tree obtained; trees B and C, two alternative constrained topologies in which American isolates are at the base of clade II (tree B) or mixed with clade II European and African isolates (tree C). ML tree and S-H test scores are summarized below trees. P values greater than 0.05 indicate tree congruence.
Nucleotide sequence accession numbers.
The accession numbers of sequences generated in this study are listed in Table 3.
TABLE 3.
PCR amplification and accession numbers of sequenced PCR productsa
+, amplification of PCR products that were not sequenced; −, lack of amplification. Amplification conditions for all genes are described in Materials and Methods.
noeI pseudogene.
ND, not determined.
RESULTS AND DISCUSSION
Strains of the diverse Bradyrhizobium lineages nodulate Lupinus spp.
In this study, we characterized a collection of 52 Bradyrhizobium strains, of which 32 were isolated from lupine nodules sampled from three (OW, ENW, and WNW) of the four major Lupinus lineages, with 23 isolates originating from native New World lupines (Fig. 1). Prior to this study, it was known that Bradyrhizobium strains isolated in Europe from Genisteae and serradella legumes form a distinct group (clade II) in nod gene trees (19, 28). Furthermore we showed that bradyrhizobia of this clade have become established in acid soils of Western Australia and South Africa following their probably accidental introduction with lupine and serradella seeds (42). Similar transfer with seeds of introduced legumes could explain the presence of clade II nodA sequences in New Zealand strains isolated from invasive Cytisus scoparius and Ulex europeus plants (50, 51). Considering the dynamic dispersion of clade II strains we wondered whether strains from a much wider range of lupine species, including native New World lupines which are geographically distant from the core Genistineae area, could also fall predominately within clade II.
To determine the taxonomic status of the Bradyrhizobium lupine strains, we first evaluated the congruence of four phylogenetic markers (483 bp of atpD, 483 bp of recA, 284 bp of dnaK, and 519 bp of glnII) used in Bradyrhizobium classification studies (10, 40, 42, 49, 51) for a subset of 13 strains (Fig. 2A to D). We did not include the 16S rRNA gene marker in our phylogenetic analyses as this gene is too conserved to resolve genospecies in Bradyrhizobium (49, 52). The congruence of the tree topologies was evaluated using S-H tests, and the results are shown in Fig. 2E. Congruence (P > 0.05) was found between recA and atpD trees (P = 0.164), recA and glnII trees (P = 0.186), and atpD and glnII trees (P = 0.305), but there was no congruence between the dnaK tree and any other tree. We thus decided to evaluate the taxonomic status of the strains by sequencing 485 bp of the atpD gene in all 52 strains included in the study (except for strain Lcamp6, for which no amplification product could be obtained) and to sequence fragments of recA (559 bp) and glnII (594 bp) genes in several randomly selected strains to confirm the atpD phylogeny clustering (see Fig. S1 of the supplemental material).
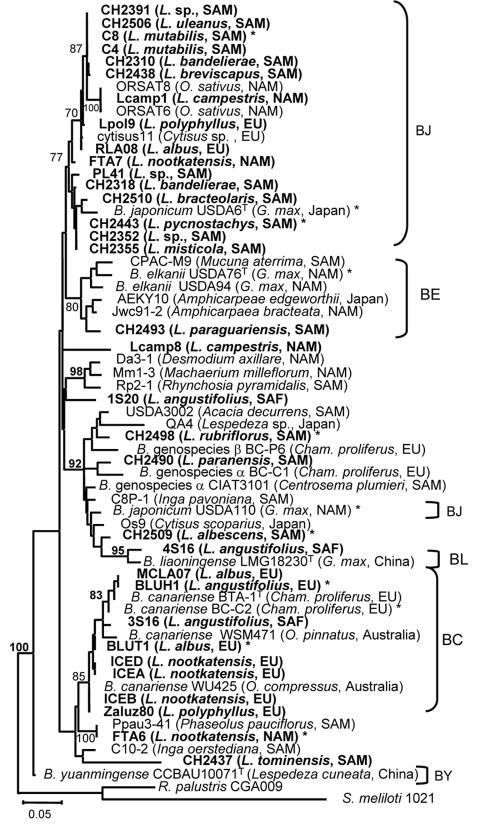
The atpD ML tree of all strains (Fig. 3) reveals several clades, usually with limited internal resolution, placed within the major Bradyrhizobium branch. Some of these clades correspond to the previously delineated species B. japonicum, B. canariense B. liaoningense, and B. yuanmingense, as well as to the genotypes α and β (Fig. 3; see Fig. S1 in the supplemental material). B. japonicum is polyphyletic in the atpD phylogeny, with sequences placed in two separate groups, one containing strain USDA 6T, which forms a group with various Lupinus strains, and the other containing B. japonicum USDA 110, which groups together with the type strains representing the genospecies α and β and with the B. liaoningense type strain LMG18230. Similar grouping of B. japonicum strains has recently been observed by Vinuesa et al. (49) in their atpD tree. The B. yuanmingense strain CCBAU 10071T forms a single clade at the base of the atpD tree (Fig. 3).
FIG. 3.
ML trees based on partial atpD sequences. The tree shown was constructed unrooted. Lupine strains are indicated in boldface. Host plants and geographical origins of the strains are in parentheses. The scale bar indicates the number of substitutions per site. Bootstrap values indicated are percentages of 1,000 replicates (calculated under distance criteria); they are in boldface when a bootstrap value >70% was found under ML criteria (100 replicates). Abbreviations: BC, B. canariense; BJ, B. japonicum; BL, B. liaoningense; BY, B. yuanmingense; EU, Europe; NAM, North America; SAM, South America; SAF, South Africa. Asterisks indicate strains used for S-H tests (strains for which all markers were sequenced). Reference strains and their accession numbers were as follows: USDA 110, NP_767080; 1S20, AJ891286; BTA-1, AY386739; WSM471, AJ891292; 3S16, AJ891287; WU425, AJ891293; BC-C2, AY386736; BC-P6, AY653751; LMG18230T, AY386752; 4S16, AJ891288; BC-C1, AY386735; CIAT3101, AY653762; Rhodopseudomonas palustris CGA009, NC_005296).
These results corroborate earlier reports describing great diversity among lupine Bradyrhizobium strains, which were found to group in several distinct branches in housekeeping gene trees (5, 23, 28, 42, 48, 49). The majority of lupine strains in all trees grouped in the B. japonicum cluster or with B. canariense. In the atpD tree, 16 lupine strains plus 2 strains from serradella and 1 from Cytisus form a moderately supported clade (77% bootstrap) containing B. japonicum strain USDA 6T. Seven lupine strains, all originating from Europe and Iceland, form a well-supported cluster (85% bootstrap) with B. canariense strains. Notably, only one lupine strain groups consistently with B. elkanii, which implies that B. elkanii is seemingly rare among lupine isolates. Six lupine strains and one strain from Cytisus isolated in Japan occupy variable or weakly supported positions in the trees, albeit distinct from B. japonicum, B. canariense, or B. elkanii. These strains may therefore be regarded as unresolved. Despite the differences found between trees, all gene marker phylogenies presented here clearly show that lupine strains constitute a heterogeneous phylogenetic group within the genus Bradyrhizobium.
The evolution of the nodA nodulation gene in the genus Bradyrhizobium.
Previous studies indicated that the nodA genes in strains of Bradyrhizobium are longer than nodA genes from the genera Mesorhizobium, Rhizobium, and Sinorhizobium, due to the presence of codons for an additional 13 amino acid residues that form the N-terminal part of the deduced NodA protein (28, 42). Such a segment is also present in nodA sequences of Methylobacterium nodulans ORS2060 and Burkholderia tuberum STM678, which group with Bradyrhizobium sequences. This segment is present in the majority of the Bradyrhizobium strains studied here. However, in 14 American strains, the aligned nodA sequences reveal that the predicted NodA proteins could be shorter, as their sequences carry a second ATG (ATG2) codon that matches the start codon of the fast-growing rhizobia. In three strains isolated from lupine and serradella, Lcamp1, ORSAT6, and ORSAT8, the first putative codon is GTG, while the second is ATG. Moreover, six other strains, including strain CH2509, originating from a native Brazilian lupine, may produce a NodA protein identical to those of the fast-growing species. These strains carry only one ATG codon, which overlaps with the start codon for NodA proteins of the fast-growing strains (see Fig. S5 in the supplemental material). This finding strengthens our earlier interpretation that the nodA gene may be undergoing an evolutionary progression towards a shorter nodA sequence, which is typical for the majority of rhizobia (28). Strain CH2509, which belongs to new clade VII, as well as photosynthetic bradyrhizobia that here have been grouped in clade VI (Fig. 4), illustrates the loss of this N-terminal segment. On the other hand, the finding that some strains of clade III carry the second ATG codon, which in sequence alignment overlaps with start codons of other rhizobia (and potentially could be an alternative start codon), suggests that in some strains evolution toward a shorter nodA gene is ongoing.
Biogeographic structure shown by the nodA gene phylogenetic tree.
This study supports previous reports showing the dissimilarity between the nodulation and housekeeping gene trees (Fig. 3 and 4), which invoke the hypothesis of multiple lateral transfers of symbiotic loci among Bradyrhizobium lineages (17, 28, 41, 42, 48, 49). Previous studies showed that, despite their great diversity, the nodA gene sequences in the genus Bradyrhizobium formed a monophyletic group that splits into four major branches, referred to as clades I, II, III, and IV. Clades I and IV comprise strains isolated from legumes native to Australia. Clade II includes Genisteae and serradella isolates of mainly European origin, while clade III is a large group comprising nodA sequences of mainly subtropical strains (28, 42). Here, we reveal even greater complexity in the nodA tree, with seven clades being distinguished (Fig. 4). Among these, the new clade VI represents photosynthetic strains that were previously classified in clade III.4.
The nodA gene tree included data from rhizobia isolated from 134 legume taxa that represented all genera and species for which a nodA sequence was available (alignment is available as supplemental material). In the ML nodA tree shown in Fig. 4, all Old World lupine strains isolated in Europe, the Canary Islands, and Iceland, as well as strain cytisus11, are placed in clade II, whereas only eight New World lupine strains are placed in clade II, and these are all from lupine species belonging to the WNW clade. The majority of New World lupine strains are found in clade III, which includes strains from both of the large New World Lupinus lineages (ENW and WNW) but no Old World lupine strains. Two new clades that include lupine strains are evident in the nodA tree of Fig. 4: clades V (strain CH2493) and VII (strain CH2509). Clade VII, apart from the Brazilian lupine strain CH2509, comprises strains, Da3-1, C10-2, C8P-1, Mm1-3, and Rp2-1, originating from Central America (Panama) and Mexico, which were isolated from Desmodium axillare (Desmodieae), Inga spp. (Ingeae), Machaerium milleflorum (Dalbergieae), and Rhynchosia pyramidalis (Phaseoleae), respectively (Table 1).
The New World lupine clade III strains form a strongly (100% bootstrap) supported subgroup, designated subclade III.2. Also included in subgroup III.2 are strains ORSAT6 and ORSAT8, from a Mexican soil sample (trapped using Ornithopus sativus), Ppau3-41, isolated in Mexico from Phaseolus pauciflorus (35), and peanut strain NC92, originating from North Carolina (11). Therefore, III.2 cannot be regarded as an exclusively lupine group. The nodA sequences in subclade III.2 show high sequence divergence (pairwise similarity ranges from 83% to 100%), with the lupine strains divided between two subgroups of closely related sequences (Fig. 4). It is notable that strains from the two independent ENW and WNW Lupinus lineages fall into separate subgroups. This could be interpreted as an example of codivergence of the New World lupine Bradyrhizobium strains with their lupine hosts. These New World lupine lineages both include elements from North and South America, suggesting that the two independent dispersals between North and South America postulated for lupines may have been accompanied by similar dispersal of their symbiotic Bradyrhizobium partners (15).
Although four strains of subclade III.2 originate from nonlupines, all strains originate from the Americas, and therefore it seems justified to name clade III.2 the “Pan-American” subgroup. Notably, a geographical structure in symbiotic gene trees was reported for nodA and nifH genes in Sinorhizobium isolates (14) and later for nifD (but not for the 16S rRNA locus) among Bradyrhizobium strains isolated in Australia, Central America, and eastern Asia (31, 35). Likewise, we previously showed that clade I isolates in the nodA tree have an Australian affinity, comprising strains isolated either in Australia or from native Australian legumes growing elsewhere (42). Clade I strains were also described among isolates collected in Papua New Guinea from the nodules of the nonlegume Parasponia andersonii and from legumes indigenous to New South Wales (22), which further strengthened our belief in the Australasian origin of this group. In our study, the clade VII strains originated exclusively from Central and South America, although only strain CH2509 was isolated from lupine. Thus, the four nodA clades I, II, III.2, and VII each reflect the geographical origins of their respective strains (Fig. 4).
The present study with much wider sampling confirms the earlier findings that all “European” strains isolated from Genisteae (and serradella) group exclusively in nodA gene clade II, implying the predominance of clade II strains among Genisteae bradyrhizobia in European soils (19, 28, 41, 42). Conversely, only 8 of the 23 lupine isolates from New World lupines are placed in clade II, while the remainder are either in clade III or in two new clades, V and VII (Fig. 4). Notably, the American clade II strains form a strongly supported (100% bootstrap) subclade of sequences with very low divergence, despite the fact that they originate from an extensive area spanning Alaska to Bolivia. This area coincides with the distribution of the WNW Lupinus clade and may imply that nodA gene sequences of all these strains diverged from a common ancestor more recently than those in the European strains and then spread rapidly across the Americas. Although the S-H test cannot discriminate between alternative placements of the American clade II strains (Fig. 5), clade II strain predominance in European soils and higher sequence diversity argue for a European rather than American origin. This would imply possible long-distance dispersal of the clade II strains, presumably across the Atlantic Ocean to North and South America, or, alternatively, their transfer from Europe following colonization of the Americas in the late 15th century. A similar explanation was earlier proposed for the spread of common bean (Phaseolus vulgaris) rhizobia outside the Americas (32) and recently for clade II bradyrhizobia in Australia, South Africa, and New Zealand (42, 50).
Clade II is characterized by low sequence divergence and a lack of resolution compared to clade III, which implies that the divergence in clade II was relatively recent compared to that in clade III. We hypothesize that clade II bradyrhizobia diversified initially in the Mediterranean basin, possibly in parallel with the divergence of their legume hosts belonging to the tribes Genisteae and Loteae, which radiated in this region (4). It is likely that radiation of these tribes was triggered by climatic changes leading to formation of the Mediterranean climate during the Pliocene epoch. It appears that lower temperatures and increased aridity after the end of the Miocene eliminated mesophytic boreotropical flora, giving rise to the expansion of cold- and drought-tolerant genera, such as those in the Genisteae and Loteae tribes (21, 33, 45). Thus, the predominance of clade II strains in Europe's soils might be perceived as a consequence of climatic changes that eliminated tropical elements from the European legume flora, while formation of the Sahara Desert prevented migration of subtropical legumes (and their rhizobia) from Africa. In contrast, the closure of the Isthmus of Panama enabled a wide-scale migration of tropical legumes and their rhizobia from South America to the north (21, 27, 45), which could explain the presence of clade III strains among the American lupine isolates.
Amplification of nodulation genes conferring modifications of the Nod factor reducing end.
The structure of the nodulation (Nod) factor is an important determinant of rhizobial host specificity and can be related to the activity of genes involved in modification of the Nod factor reducing end, such as nodZ, noeI, noeE, and nolL (6, 13, 16, 36). Previous studies indicated that the nodZ gene, which is involved in fucosylation of the Nod factor reducing end (36, 39), is present in all strains belonging to nodA clades II, III, and IV but is probably missing in strains from clade I. Moreover, it was shown that the nodZ gene trees are congruent with the nodA gene phylogenies (28, 42). The present study further supports these findings. We were unable to amplify the nodZ gene from new strain USDA 3002 (isolated from Acacia decurrens in Brazil), which was placed in clade I in the nodA gene tree, whereas nodZ was detected in all strains belonging to nodA clades II and III, as well as in the strains of the new clades V and VII (Fig. 4). The nodZ gene tree is similar to the nodA tree, forming separate branches that group in a similar pattern and are composed of the same strains as the nodA clades (Fig. 4; see Fig. S3A in the supplemental material).
Methylation of carbon C-2 of the fucose molecule bound to the Nod factor reducing end is conferred by the noeI gene (16). This modification is particularly common among clade III strains, but the noeI gene has so far not been detected in clades I and II (28, 42). The use of additional isolates in the present study partially contradicts this finding. The noeI gene was missing in the majority of the clade II strains, although faint PCR bands were visible in some strains. For these strains we repeated the PCR amplification using a lower annealing temperature (51°C instead of 53°C). In strains BLUH1 and BLUT1 we sequenced the PCR products, which proved to be two noeI sequences carrying one nucleotide deletion that disrupted the coding sequence continuity. The noeI gene was detected in 17 of the 24 clade III strains (Fig. 4). However, a notable exception was the lack of the PCR products in a group of strains within subclade III.2 that were isolated from the ENW Lupinus clade (strains CH2310, 2318, 2490, 2498, 2506, and 2510). In two other lupine strains belonging to III.2 (C8 and Lcamp8), the amplified noeI fragments harbor an identical nonsense mutation and, additionally, strain C8 harbors a deletion, suggesting that these are nonfunctional copies (see Fig. S4 in the supplemental material). The noeI gene was also amplified and sequenced in the clade VII lupine strain CH2509 but not the clade V lupine strain CH2493 (Fig. 4; see Fig. S3B in the supplemental material).
Unlike nodA and nodZ genes, for which we have not detected pseudogenes, mutated and potentially inactive noeI gene copies suggest the loss of this Nod factor modification gene. In all these strains, the mutated sequences preserved extensive similarity to functional noeI gene sequences, implying that these mutations must have occurred recently. Interestingly, mutated noeI sequences were reported in lupine clade II strain WM9 and on the symbiosis plasmid of Rhizobium etli strain CFN42 (41). Similarly, noeE pseudogenes were detected in soybean modulating Bradyrhizobium strains USDA 110 and USDA 94, both of which are known to produce Nod factors that bear 2-O-methylated, but not 3-O-sulfated, fucose (20, 39). The presence of noeI pseudogenes indicates that the loss of these two genes reflects a more general evolutionary trend related to adaptation to the legume host requirements. We assume that the American clade III.2 strains diverged together with legumes other than lupines. The loss of noeI could therefore be a relatively recent adaptation to their new hosts that has occurred independently in various strains (28, 41).
Our conclusion that the nodA nodulation gene trees show strong geographical structure implies that divergence in geographical isolation, in parallel with host plant diversification, plays a significant role in the evolution of symbiotic loci (24). Comparison of nod gene trees with those of housekeeping genes shows that the latter retain much less geographic structure. Thus, geographic structure appears to be preserved in phylogenies of rapidly evolving accessory genes, such as nodulation genes, rather than in conserved housekeeping genes. In the latter, geographic structure was presumably erased by a series of dispersal and recombination events (49). Given the difficulty of defining bacterial species according to ecological or biological concepts (7, 29), it could be concluded that the biogeography of bacteria relates to genes rather than species, which appears to be corroborated by our results.
Supplementary Material
Acknowledgments
We thank Janet Sprent for facilitating our work on the New World lupine rhizobia and Alison McInnes for useful comments on the manuscript. We also thank Matthew Parker, Mario Ruiz Lopez, Tatsuo Ezawa, Peter van Berkum, Encarna Velazquez, Milagros León Barrios, Julie Grossman, Maria Rita Scotti Muzzi, Gustavo Bernal, David Odee, Magnus Jóhannsson, Maria Teresa Schifino Wittmann, Wen-Xin Chen, and Wanda Małek for providing Bradyrhizobium strains.
This work was financed by grant 2P04C 063 26 from the Polish Ministry of Education and Science. Colin Hughes is supported by the Royal Society.
Footnotes
Published ahead of print on 30 March 2007.
Supplemental material for this article may be found at http://aem.asm.org/.
REFERENCES
- 1.Ainouche, A. K., and R. J. Bayer. 1999. Phylogenetic relationships in Lupinus (Fabaceae: Papilionoideae) based on internal transcribed spacer sequences (ITS) of nuclear ribosomal DNA. Am. J. Bot. 86:590-607. [PubMed] [Google Scholar]
- 2.Ainouche, A. K., R. J. Bayer, and M. T. Misset. 2004. Molecular phylogeny, diversification and character evolution in Lupinus (Fabaceae) with special attention to Mediterranean and African lupines. Plant Syst. Evol. 246:211-222. [Google Scholar]
- 3.Ainouche, A. K., R. J. Bayer, P. Cubas, and M. T. Misset. 2003. Phylogenetic relationships within tribe Genisteae (Papilionaceae) with special reference to the genus Ulex, p. 239-252. In B. B. Klitgaard and A. Bruneau (ed.), Advances in legume systematics, part 10. Royal Botanic Gardens, Kew, United Kingdom.
- 4.Allan, G. J., E. A. Zimmer, W. L. Wagner, et al. 2003. Molecular phylogenetic analyses of tribe Loteae (Leguminosae): implications for classification and biogeography, p. 371-393. In B. B. Klitgaard and A. Bruneau (ed.), Advances in legume systematics, part 10. Royal Botanic Gardens, Kew, United Kingdom.
- 5.Barrera, L. L., M. E. Trujillo, M. Goodfellow, et al. 1997. Biodiversity of bradyrhizobia nodulating Lupinus spp. Int. J. Syst. Bacteriol. 47:1086-1091. [DOI] [PubMed] [Google Scholar]
- 6.Berck, S., X. Perret, D. Quesada-Vincens, et al. 1999. NolL of Rhizobium sp. NGR234 is required for O-acetyltransferase activity. J. Bacteriol. 181:957-964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Doolittle, W. F., and R. T. Papke. 2006. Genomics and the bacterial species problem. Genome Biol. 7:116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Eckhardt, M. M., I. R. Baldwin, and E. B. Fred. 1931. Studies on the root-nodule bacteria of Lupinus. J. Bacteriol. 21:273-285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Fred, E. B., I. L. Baldwin, and E. McCoy. 1932. Root nodule bacteria and leguminous plants. In Studies in science, vol. 5. University of Wisconsin, Madison.
- 10.Gaunt, M. W., S. L. Turner, L. Rigottier-Gois, et al. 2001. Phylogenies of atpD and recA support the small subunit rRNA-based classification of rhizobia. Int. J. Syst. Evol. Microbiol. 51:2037-2048. [DOI] [PubMed] [Google Scholar]
- 11.Gillette, W. K., and G. H. Elkan. 1996. Bradyrhizobium (Arachis) sp. strain NC92 contains two nodD genes involved in the repression of nodA and a nolA gene required for the efficient nodulation of host plants. J. Bacteriol. 178:2757-2766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gladstones, J. S. 1998. Distribution, origin, taxonomy, history and importance, p 1-40. In J. S. Gladstones, C. Atkins, and J. Hamblin (ed.), Lupins as crop plants: biology, production and utilization. CAB International, Cambridge, United Kingdom.
- 13.Hanin, M., S. Jabbouri, D. Quesada-Vincens, et al. 1997. Sulphation of Rhizobium sp. NGR234 Nod factors is dependent on noeE, a new host-specificity gene. Mol. Microbiol. 24:1119-1129. [DOI] [PubMed] [Google Scholar]
- 14.Haukka, K., K. Lindstrom, and J. P. Young. 1998. Three phylogenetic groups of nodA and nifH genes in Sinorhizobium and Mesorhizobium isolates from leguminous trees growing in Africa and Latin America. Appl. Environ. Microbiol. 64:419-426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hughes, C., and R. Eastwood. 2006. Island radiation on a continental scale: exceptional rates of plant diversification after uplift of the Andes. Proc. Natl. Acad. Sci. USA 103:10334-10339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Jabbouri, S., B. Relic, M. Hanin, et al. 1998. nolO and noeI (HsnIII) of Rhizobium sp. NGR234 are involved in 3-O-carbamoylation and 2-O-methylation of Nod factors. J. Biol. Chem. 273:12047-12055. [DOI] [PubMed] [Google Scholar]
- 17.Jarabo-Lorenzo, A., R. Perez-Galdona, J. Donate-Correa, et al. 2003. Genetic diversity of bradyrhizobial populations from diverse geographic origins that nodulate Lupinus spp. and Ornithopus spp. Syst. Appl. Microbiol. 26:611-623. [DOI] [PubMed] [Google Scholar]
- 18.Kalita, M., and W. Malek. 2004. Phenotypic and genomic characteristics of rhizobia isolated from Genista tinctoria root nodules. Syst. Appl. Microbiol. 27:707-715. [DOI] [PubMed] [Google Scholar]
- 19.Kalita, M., T. Stępkowski, B. Łotocka, and W. Małek. 2006. Phylogeny of nodulation genes and symbiotic properties of Genista tinctoria bradyrhizobia. Arch. Microbiol. 186:87-97. [DOI] [PubMed] [Google Scholar]
- 20.Kaneko, T., Y. Nakamura, S. Sato, et al. 2002. Complete genomic sequence of nitrogen-fixing symbiotic bacterium Bradyrhizobium japonicum USDA110. DNA Res. 9:189-197. [DOI] [PubMed] [Google Scholar]
- 21.Kovar-Eder, J., Z. Kvaček, E. Martinetto, and P. Roiron. 2006. Late Miocene to early Pliocene vegetation of southern Europe (7-4Ma) as reflected in the megafossil plant record. Palaeogeog. Palaeoclimat. Palaeoecol. 238:321-339. [Google Scholar]
- 22.Lafay, B., E. Bullier, and J. J. Burdon. 2006. Bradyrhizobia isolated from root nodules of Parasponia (Ulmaceae) do not constitute a separate coherent lineage. Int. J. Syst. Evol. Microbiol. 56:1013-1018. [DOI] [PubMed] [Google Scholar]
- 23.Laguerre, G., P. Van Berkum, N. Amarger, and D. Prévost. 1997. Genetic diversity of rhizobial symbionts isolated from legume species within the genera Astragalus, Oxytropis, and Onobrychis. Appl. Environ. Microbiol. 63:4748-4758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Martiny, J. B., B. J. Bohannan, J. H. Brown, et al. 2006. Microbial biogeography: putting microorganisms on the map. Nat. Rev. Microbiol. 4:102-112. [DOI] [PubMed] [Google Scholar]
- 25.McInnes, A. 2002. Bradyrhizobium sp. (Lupinus) associated with serradella in Western Australia. Ph.D. thesis. The University of Western Australia, Perth, Australia.
- 26.McInroy, S. G. 1997. Rhizobia for African species of Acacia: characterisation, conservation and selection of inoculants. M.Sc. thesis, p. 142. University of Dundee, Dundee, United Kingdom.
- 27.Morley, R. J. 2003. Interplate dispersal paths for megathermal angiosperms. Perspect. Plant Ecol. Evol. Syst. 6:5-20. [Google Scholar]
- 28.Moulin, L., G. Béna, C. Boivin-Masson, and T. Stępkowski. 2004. Phylogenetic analyses of symbiotic nodulation genes support vertical and lateral gene co-transfer within the Bradyrhizobium genus. Mol. Phylogenet. Evol. 30:720-732. [DOI] [PubMed] [Google Scholar]
- 29.Nesbo, C. L., M. Dlutek, and W. F. Doolittle. 2006. Recombination in Thermotoga: implications for species concepts and biogeography. Genetics 172:759-769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Nicholas, K. B., H. B. Nicholas, and D. W. Deerfield. 1997. GeneDoc: analysis and visualization of genetic variation. EMBNEW News 4:14. [Google Scholar]
- 31.Parker, M. A., B. Lafay, J. J. Burdon, and P. Van Berkum. 2002. Conflicting phylogeographic patterns in rRNA and nifD indicate regionally restricted gene transfer in Bradyrhizobium. Microbiology 148:2557-2565. [DOI] [PubMed] [Google Scholar]
- 32.Pérez-Ramírez, N. O., M. A. Rogel, E. Wang, et al. 1998. Seeds of Phaseolus vulgaris bean carry Rhizobium etli. FEMS Microbiol. Ecol. 4:289-296. [Google Scholar]
- 33.Petit, R. J., A. Hampe, and R. Cheddadi. 2005. Climate changes and tree phylogeography in the Mediterranean. Taxon 54:877-885. [Google Scholar]
- 34.Posada, D., and K. A. Crandall. 1998. MODELTEST: testing the model of DNA substitution. Bioinformatics 14:817-818. [DOI] [PubMed] [Google Scholar]
- 35.Qian, J., S. W. Kwon, and M. A. Parker. 2003. rRNA and nifD phylogeny of Bradyrhizobium from sites across the Pacific Basin. FEMS Microbiol. Lett. 219:159-165. [DOI] [PubMed] [Google Scholar]
- 36.Quesada-Vincens, D., R. Fellay, T. Nasim, et al. 1997. Rhizobium sp. strain NGR234 NodZ protein is a fucosyltransferase. J. Bacteriol. 179:5087-5093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Rodríguez Echeverría, S., M. A. Pérez Fernández, S. Vlaar, and T. M. Finan. 2003. Analysis of the legume-rhizobia symbiosis in shrubs from central western Spain. J. Appl. Microbiol. 95:1367-1374. [DOI] [PubMed] [Google Scholar]
- 38.Sajnaga, E., W. Małek, B. Łotocka, et al. 2001. The root-nodule symbiosis between Sarothamnus scoparius L. and its microsymbionts. Antonie Leeuwenhoek Int. J. Gen. Mol. Microbiol. 79:385-391. [DOI] [PubMed] [Google Scholar]
- 39.Sanjuan, J., R. W. Carlson, H. P. Spaink, U. R. Bhat, W. M. Barbour, J. Glushka, and G. Stacey. 1992. A 2-O-methylfucose moiety is present in the lipo-oligosaccharide nodulation signal of Bradyrhizobium japonicum. Proc. Natl. Acad. Sci. USA 89:8789-8793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Stępkowski, T., M. Czaplińska, K. Miedzinska, and L. Moulin. 2003. The variable part of the dnaK gene as an alternative marker for phylogenetic studies of rhizobia and related alpha proteobacteria. Syst. Appl. Microbiol. 26:483-494. [DOI] [PubMed] [Google Scholar]
- 41.Stępkowski, T., A. Świderska, K. Miedzinska, et al. 2003. Low sequence similarity and gene content of symbiotic clusters of Bradyrhizobium sp. WM9 (Lupinus) indicate early divergence of “lupin” lineage in the genus Bradyrhizobium. Antonie Leeuwenhoek Int. J. Gen. Mol. Microbiol. 84:115-124. [DOI] [PubMed] [Google Scholar]
- 42.Stępkowski, T., L. Moulin, A. Krzyżańska, et al. 2005. European origin of Bradyrhizobium populations infecting lupins and serradella in soils of Western Australia and South Africa. Appl. Environ. Microbiol. 71:7041-7052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Swofford, D. L. 1998. PAUP. Phylogenetic analysis using parsimony (and other methods), version 4. Sinauer Associates, Sunderland, MA.
- 44.Thompson, J. D., T. J. Gibson, F. Plewniak, et al. 1997. The ClustalX Windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res. 25:4876-4882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Tiffney, B. H., and S. R. Manchester. 2001. The use of geological and paleontological evidence in evaluating plant phylogeographic hypotheses in the Northern Hemisphere tertiary. Int. J. Plant Sci. 162(Suppl.):S3-S17 [Google Scholar]
- 46.Vincent, J. M. 1970. A manual for the practical study of root-nodule bacteria. Blackwell Scientific Publications, Ltd., Oxford, United Kingdom.
- 47.Vinuesa, P., J. L. Rademaker, F. J. de Bruijn, and D. Werner. 1998. Genotypic characterization of Bradyrhizobium strains nodulating endemic woody legumes of the Canary Islands by PCR-restriction fragment length polymorphism analysis of genes encoding 16S rRNA (16S rDNA) and 16S-23S rDNA intergenic spacers, repetitive extragenic palindromic PCR genomic fingerprinting, and partial 16S rDNA sequencing. Appl. Environ. Microbiol. 64:2096-2104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Vinuesa, P., M. Leon-Barrios, C. Silva, et al. 2005. Bradyrhizobium canariense sp. nov., an acid-tolerant endosymbiont that nodulates genistoid legumes (Papilionoideae: Genisteae) from the Canary Islands, along with Bradyrhizobium japonicum bv. genistearum, Bradyrhizobium genospecies α and Bradyrhizobium genospecies β. Int. J. Syst. Evol. Microbiol. 55:569-575. [DOI] [PubMed] [Google Scholar]
- 49.Vinuesa, P., C. Silva, D. Werner, and E. Martinez-Romero. 2005. Population genetics and phylogenetic inference in bacterial molecular systematics: the roles of migration and recombination in Bradyrhizobium species cohesion and delineation. Mol. Phylogenet. Evol. 34:29-54. [DOI] [PubMed] [Google Scholar]
- 50.Weir, B. S. 2006. Systematics, specificity, and ecology of New Zealand rhizobia. Ph.D. thesis. The University of Auckland, Auckland, New Zealand.
- 51.Weir, B. S., S. L. Turner, W. B. Silvester, et al. 2004. Unexpectedly diverse Mesorhizobium strains and Rhizobium leguminosarum nodulate native legume genera of New Zealand, while introduced legume weeds are nodulated by Bradyrhizobium species. Appl. Environ. Microbiol. 70:5980-5987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Willems, A., R. Coopman, and M. Gillis. 2001. Comparison of sequence analysis of 16S-23S rDNA spacer regions, AFLP analysis and DNA-DNA hybridizations in Bradyrhizobium. Syst. Appl. Microbiol. 51:623-632. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.