Abstract
A comparative analysis of the morphology, toxin composition, and ribosomal DNA (rDNA) sequences was performed on a suite of clonal cultures of the potentially toxic dinoflagellate Alexandrium minutum Halim. These were established from resting cysts or vegetative cells isolated from sediment and water samples taken from the south and west coasts of Ireland. Results revealed that strains were indistinguishable, both morphologically and through the sequencing of the D1-D2 domain of the large subunit and the ITS1-5.8S-ITS2 regions of the rDNA. High-performance liquid chromatography fluorescence detection analysis, however, showed that only strains derived from retentive inlets on the southern Irish coast synthesized paralytic shellfish poisoning (PSP) toxins (GTX2 and GTX3), whereas all strains of A. minutum isolated from the west coast were nontoxic. Toxin analysis of net hauls, taken when A. minutum vegetative cells were in the water column, revealed no PSP toxins in samples from Killary Harbor (western coast), whereas GTX2 and GTX3 were detected in samples from Cork Harbor (southern coast). These results confirm the identity of A. minutum as the most probable causative organism for historical occurrences of contamination of shellfish with PSP toxins in Cork Harbor. Finally, random amplification of polymorphic DNA was carried out to determine the degree of polymorphism among strains. The analysis showed that all toxic strains from Cork Harbor clustered together and that a separate cluster grouped all nontoxic strains from the western coast.
Blooms of harmful phytoplankton have become a significant problem worldwide over the past decades due to the negative impacts they cause toward marine ecosystems and shellfish and finfish industries (24). Among the causative organisms leading to human intoxication or mortality of stocks, marine dinoflagellates within the genus Alexandrium have been responsible for paralytic shellfish poisoning (PSP). The compounds involved in these events are potent alkaloid neurotoxins that may cause muscular paralysis, neurological symptoms, and in extreme cases, death (33). The genus Alexandrium has been one of the most studied of the harmful algae because of the threat posed by the potent suite of toxins it synthesizes, the large number of species in the genus (at least 27 have been described [7]), and their wide geographical distribution.
Currently, the identification of Alexandrium species still relies largely on visual observations of the cell morphology, using conventional light microscopy. Some monitoring and research institutions, however, have successfully used molecular diagnostic tools. Despite being more expensive, these tools can be less time consuming and have provided a more specific and sensitive detection of the target organisms (5, 28, 56). For example, real-time PCR and fluorescence in situ hybridization (FISH)-based assays have been designed for laboratory and field applications (21, 30). New fingerprinting methods such as microsatellite, amplified fragment length polymorphism (AFLP), and intersimple sequence repeat (ISSR) markers have also been developed to analyze the genetic variability among isolates (12, 32, 43). In order to determine specific characteristics of locally or globally distributed Alexandrium species, a significant number of key studies are currently being carried out on their taxonomy, toxicity, life cycle, physiology, phylogeny, bloom dynamics, biogeography, and genetic diversity. Characters that have traditionally been used to assess diversity within the genus include morphology (7), isozyme patterns (51), immunological properties (5, 50) and toxin profiles (4, 14). More recent studies of the inter- and intraspecific genetic variability of Alexandrium strains have relied on the application of ribosomal DNA (rDNA)-based sequencing (1, 27, 31, 36, 42, 44, 61). The increasing number of molecular studies has also improved our capability to assess the evolutionary relationships within the genus and to assess the species variability down to the population scale (3, 37, 46, 52). As a result, the species boundaries within the genus have been contested, as taxa, for example, belonging to the Alexandrium tamarense/A. catenella/A. fundyense complex have been shown to cluster by geographical origin rather than by morphotype (52, 53).
Within Ireland, Alexandrium organisms have been assumed to be responsible for the temporary closure of shellfish production sites in Cork Harbor on the south coast, where for a long time, A. tamarense was suspected as the causative organism (9). However, recent investigations suggest that in affected sites the species is nontoxic (27) and have shown that it cooccurs with A. minutum (60a). On a broader geographical scale, Alexandrium has been detected on all Irish coasts, and blooms have not always generated positive results for PSP toxin bioassays carried out on shellfish under the National Biotoxin Monitoring Programme. For example, a bloom of A. minutum on the western Irish coast in August 2001 achieved concentrations of over 8 × 103 cells liter−1. Yet no corresponding toxicity was recorded in shellfish tissue, despite extensive monitoring at the time (Irish Marine Institute, unpublished data). It is therefore possible that the observed lack of toxins in shellfish may have been due to the lack of toxicity of A. minutum.
The present study was initiated by this apparent confusion regarding A. minutum toxicity in Irish waters. Its aim was to generate clonal cultures of A. minutum isolated from diverse regions around the Irish coast and to compare them with respect to their morphology, toxin composition, and genetic diversity.
MATERIALS AND METHODS
Culturing of Alexandrium.
Alexandrium minutum cultures were established from sediment and water samples collected from a number of sites along the coast of Ireland (Fig. 1 and Table 1). Monoclonal cell cultures were obtained by single-cell isolations carried out on board the RV Celtic Voyager, on phytoplankton net haul samples taken from Killary Harbor and from the mouth of the Shannon Estuary. Individual vegetative cells were placed in 96-well plates containing 200 μl f/2 medium minus silicate (23) and maintained for 2 weeks at 15°C under a photon flux density of 75 μmol m−2 s−1 and a 14:10 light/dark cycle before transfer to larger culture vessels. Cultures were also derived from the germination of resting cysts from surface sediment samples taken from Cork Harbor. Sediments were processed by ultrasonication followed by sieving approximately 1 g wet weight of material through 80-μm mesh material and collecting it on a 20-μm mesh sieve. Individual A. minutum cysts were isolated under an inverted light microscope, using a micropipette. Each isolated cyst was rinsed once in autoclaved seawater and was transferred to a multiple well plate containing 200 μl f/2 medium minus silicate. The isolated cysts were incubated at 15°C under a photon flux density of 75 μmol m−2 s−1 and a 14:10 light/dark cycle. After germination, a single cell was transferred to a new well, and after a few divisions, cells were placed in increased volumes of medium until an actively growing culture was established. Cells were grown in natural seawater filter-sterilized through a 0.22-μm membrane and supplemented with the f/2 medium minus silicate. Cultures derived from single vegetative cells or cysts were all maintained in 100-ml conical flasks containing 50 ml of medium under a 14:10 light/dark cycle, with a photon flux density of 150 μmol m−2 s−1 provided by cool-white fluorescent tubes at 15°C in a culture chamber.
FIG. 1.
Map of Ireland showing the geographical origin of collected Alexandrium sp. isolates. (Adapted with permission from reference 39a).
TABLE 1.
Designation and geographical origin of Irish strains of Alexandrium spp. included in this study
| Species | Strain | Toxic | Sample information
|
||
|---|---|---|---|---|---|
| Geographical position | Location | Date | |||
| A. minutum | CK.A02 | Yes | 51°52.95'N 08°15.10'W | Cork Harbor | October 2002 |
| A. minutum | CK.A14 | Yes | 51°52.95'N 08°15.10'W | Cork Harbor | October 2002 |
| A. minutum | CK.A17 | Yes | 51°52.95'N 08°15.10'W | Cork Harbor | October 2002 |
| A. minutum | CK.A20 | Yes | 51°52.95'N 08°15.10'W | Cork Harbor | October 2002 |
| A. minutum | CK.A23 | Yes | 51°52.95'N 08°15.10'W | Cork Harbor | October 2002 |
| A. minutum | CK.D04 | Yes | 51°53.13'N 08°14.55'W | Cork Harbor | October 2002 |
| A. minutum | SHA.A12 | No | 52°26.58'N 09°56.56'W | Shannon Estuary | July 2003 |
| A. minutum | SHA.B11 | No | 52°26.58'N 09°56.56'W | Shannon Estuary | July 2003 |
| A. minutum | SHA.B12 | No | 52°26.58'N 09°56.56'W | Shannon Estuary | July 2003 |
| A. minutum | Kill.A12 | No | 53°33.51'N 10°20.48'W | Killary Harbor | July 2003 |
| A. minutum | Kill.C6 | No | 53°33.51'N 10°20.48'W | Killary Harbor | July 2003 |
| A. minutum | Kill.E4 | No | 53°33.51'N 10°20.48'W | Killary Harbor | July 2003 |
| A. minutum | Kill.G3 | No | 53°33.51'N 10°20.48'W | Killary Harbor | July 2003 |
| A. tamarense | CK.C01 | No | 51°53.00'N 08°14.55'W | Cork Harbor | October 2002 |
Fluorescence microscopy.
Cell morphology and thecal plate pattern were examined under an Olympus CKX-41 inverted microscope fitted with a U-RFL-T epifluorescence attachment and a 100-W mercury lamp. Formalin-fixed vegetative cells were stained with Calcofluor White (18) at a final concentration of 20 μg ml−1 and were examined using a filter arrangement for violet excitation (330- to 385-nm excitation filter, 400-nm dichroic mirror, and 420-nm barrier filter). Identification was based on examination of thecal plate morphology of 300 cells (7). Measurements of cell dimensions were performed at ×400 magnification with a micrometer. Authorities for Alexandrium species are those cited in Balech (7).
Toxin analysis.
PSP toxin analysis was carried out on exponentially growing vegetative cells by high-performance liquid chromatography (HPLC) with postcolumn derivatization using a 5-μm LiChrospher 100 RP-18 (125-by-4-mm inside diameter) column and fluorometric detection. Approximately 5 × 105 cells were centrifuged at 3,000 × g for 10 min, and toxins were extracted with 500 μl of 0.05 M acetic acid by multiple freezing-thawing cycles. Field samples from Cork Harbor and Killary Harbor were also used to determine the presence of PSP toxins. On these occasions (June 13th 2004 and July 28th 2004, respectively), Alexandrium spp. vegetative cells were concentrated by vertical net haul on a 20-μm mesh material phytoplankton net before the centrifugation and acetic acid extraction steps. All the extracts of both cultures and field material were kept frozen at −32°C until analysis. Analytical details were based on those from previous studies (17, 45). The mobile phase for the first isocratic run, allowing the detection of saxitoxin (STX), neosaxitoxin (neoSTX), and decarbamoyl saxitoxin (dcSTX), was composed of 95% 1 mM octanesulfonic acid in a 10 mM ammonium phosphate buffer (pH 7.2) and 5% acetonitrile. The mobile phase for the second isocratic run, allowing the detection of gonyautoxins (GTXs) and their decarbamoyl forms (dcGTXs), was composed of 1.5 mM sodium octanesulfonate in a 10 mM ammonium phosphate buffer (pH 7.0). The postcolumn effluent was oxidized at a reaction temperature of 65°C in a Teflon coil (10-m-by-0.5-mm inside diameter) with a solution of 7 mM periodic acid in 0.5 M phosphate buffer (pH 9.0) and an acidification solution of 0.5 M acetic acid, with flow rates of 0.4 ml min−1 for both solutions. Fluorescence excitation and emission wavelengths were 330 and 390 nm, respectively. PSP toxins were identified by the comparison of their retention times with those of purified standards (National Research Council of Canada, Halifax, NS, Canada). Injection volumes of 10 to 30 μl were used for each crude or diluted extract. The detection limits of toxin standards, with peak heights three times superior to the average background noise, were 0.015 ng (STX), 0.010 ng (dcSTX), 0.070 ng (neoSTX), 0.030 ng (GTX1), 0.007 ng (GTX2), 0.007 ng (GTX3), and 0.030 ng (GTX4) under the selected conditions.
DNA extraction, PCR amplification and sequencing of partial rDNA.
Approximately 15 ml of exponentially growing cultures of A. minutum were concentrated by centrifugation (3,000 × g for 10 min). The pellet was kept frozen at −32°C prior to total genomic DNA extraction. DNA was extracted using a DNeasy plant mini-kit (QIAGEN) and stored at −32°C. The 28S and 5.8S subunits of the rDNA gene together with the flanking internal transcribed spacers 1 and 2 (ITS1 and ITS2) were amplified by PCR. For the large subunit (LSU) rDNA, the D1-D2 domain of the double-stranded DNA was amplified in a 50-μl reaction mixture volume using the forward primer D1R-F and the reverse primer D2C targeting toward conserved positions relative to Prorocentrum micans Ehrenberg LSU rDNA (34) (Table 2). The PCR cocktail contained 1× PCR buffer, 2 mM MgCl2, 200 μM of each deoxynucleoside triphosphate (dNTP), 0.1 μM of each primer, 1 unit of Bioline Taq polymerase, and 1 μl of template DNA. PCRs were performed in a thermocycler as follows: initial cycle of denaturation (94°C for 3 min), 35 cycles of production (denaturation, 94°C for 1 min; annealing, 52°C for 1 min; and extension, 72°C for 3 min), and a final extension step (72°C for 6 min). For the 5.8S and ITS flanking regions, amplifications were done in a 50-μl reaction mixture volume using the forward primer P1 and reverse primer P2 (57). The PCR cocktail contained 1× PCR buffer, 3 mM MgCl2, 250 μM of each dNTP, 0.2 μM of each primer, 1 unit of Taq polymerase, and 1 μl of template DNA. PCRs were performed as follows: initial cycle of denaturation, 95°C for 5 min; 35 cycles of production (denaturation, 95°C for 1 min; annealing, 55°C for 2 min; and extension, 72°C for 2 min); and a final extension step of 72°C for 30 min. The DNA fragments were made visible under UV illumination on 1% agarose-1× TAE (Tris-acetate-EDTA) buffer gel containing ethidium bromide (1 μg ml−1). PCR products were purified using a QIAquick PCR purification kit (QIAGEN). After ice-cold 100% ethanol precipitation and subsequent solvent evaporation, air-dried PCR products were sequenced (MWG Biotech, Ebersberg, Germany).
TABLE 2.
List of primers selected for the PCR amplification of rDNA and random DNA fragments from Irish strains of Alexandrium spp. used in this study
| Primer | Sequence (5′-3′) | Direction | Target region | Length (bp) | Source or reference |
|---|---|---|---|---|---|
| D1R | ACCCGCTGAATTTAAGCATA | Forward | D1-D2 LSU | 20 | 34 |
| D2C | CTTGGTCCGTGTTTCAAGA | Reverse | D1-D2 LSU | 20 | 34 |
| P1 | GTAGGATCCGGTGAACCTTGCAGAAGGA | Forward | ITS1-5.8S-ITS2 | 28 | 57 |
| P2 | ATCGAATTCCTCCGCTTACTTATATGC | Reverse | ITS1-5.8S-ITS2 | 27 | 57 |
| RP1 | CGATCGAGGA | Random | 10 | 41 | |
| RP2 | AAGCAGCAAG | Random | 10 | 41 | |
| RP3 | GTCTGACGGT | Random | 10 | 41 | |
| RP4 | AGCACTTCGG | Random | 10 | 41 | |
| RP5 | CGATAGCTGG | Random | 10 | This study | |
| RP6 | ACGGTAGCCA | Random | 10 | This study | |
| RP7 | GGTCAGTGGG | Random | 10 | This study | |
| RP8 | TTCAGAGACG | Random | 10 | This study | |
| RP9 | CGCCGGTACTTGAGA | Random | 15 | This study | |
| RP10 | CTCCACCTGCGCTTA | Random | 15 | This study |
LSU and ITS rDNA sequence alignment and phylogenetic analysis.
rDNA-derived sequence data were initially evaluated against published sequences in GenBank using a Basic Local Alignment Search Tool (BLAST). Computer-assisted multiple alignment of the sequences was performed with GeneDoc (version 2.6.002; http://www.psc.edu/biomed/genedoc) using reported Alexandrium spp. sequences imported from GenBank (Table 3). The aligned LSU rDNA sequences (D1-D2 domain) were initially subjected to neighbor-joining analysis for a rapid assessment of the relationships between taxa, whereas the ITS sequences were kept aligned and were compared manually. Not enough ITS sequences of A. minutum strains were available in databases to conduct a relevant phylogenetic analysis at the species level. A Modeltest 3.7 program (47) was used to determine the optimal base substitution model for the alignment relative to the LSU rDNA sequences. Maximum likelihood analysis was carried out using PAUP* version 4.0b10 (58), with the parameters derived from the Akaike information criterion in Modeltest. The phylogeny was then reconstructed by performing a heuristic search with the random addition of sequences (10 replicates) and a tri-bisection-reconnection branch-swapping algorithm. Fragilidium subglobosum was used as the out-group. Bootstrap value analysis was conducted using the fast stepwise addition option in PAUP* to evaluate the robust nature of the groupings.
TABLE 3.
Species, strain designations, isolation locales, rDNA regions, and GenBank accession numbers of nucleotide sequence data used in the phylogenetic and sequence analyses
| Species | Strain designation | Isolation localitya | rDNA region | GenBank accession no. |
|---|---|---|---|---|
| A. minutum | CK. A14 | Ireland, south coast | LSU D1-D2 ITS1-5.8S-ITS2 | |
| A. minutum | SHA. B11 | Ireland, west coast | LSU D1-D2 | |
| A. minutum | Kill. E4 | Ireland, west coast | LSU D1-D2 ITS1-5.8S-ITS2 | |
| A. minutum | AL9T | Italy | LSU D1-D2 | AJ535360 |
| A. minutum | 95/4 | France | LSU D1-D2 | AF318264 |
| A. minutum | Anakoha | New-Zealand | LSU D1-D2 | AF033532 |
| A. minutum | 3.9h | United Kingdom | LSU D1-D2 | AY705869 |
| A. minutum | AM89BM | France | LSU D1-D2 | AF318221 |
| A. minutum | AMAD06 | Australia | LSU D1-D2 | U44936 |
| A. lusitanicum | 181NT | Portugal | LSU D1-D2 | AY455828 |
| A. insuetum | AI104 | Japan | LSU D1-D2 | AB088248 |
| A. insuetum | X6 | France | LSU D1-D2 | AF318233 |
| A. tamutum | SZN29 | Italy | LSU D1-D2 | AJ535372 |
| A. tamutum | AT3 | Italy | LSU D1-D2 | AJ535367 |
| A. ostenfeldii | K0324 | Denmark | LSU D1-D2 | AJ535363 |
| A. ostenfeldii | NDb | New-Zealand | LSU D1-D2 | AF033533 |
| A. margalefi | ND | Denmark (?) | LSU D1-D3 | AY154958 |
| A. andersoni | TC02 | United States, east coast | LSU D1-D2 | U44937 |
| A. affine | JHW0210 | Korea | LSU D1-D3 | AY438015 |
| A. concavum | CAWD52 | New-Zealand | LSU D1-D2 | AF032348 |
| A. catenella | ATT98 | France | LSU D1-D2 | AF318220 |
| A. catenella | OF101 | Japan | LSU D1-D2 | U44931 |
| A. catenella | JHW0007-7 | Korea | LSU D1-D3 | AY438016 |
| A. catenella | A3 | United States, west coast | LSU D1-D3 | AF200667 |
| A. excavatum | Ge1V | ND | LSU D1-D2 | L38632 |
| A. tamarense | UW53 | United Kingdom | LSU D1-D2 | AJ303429 |
| A. tamarense | ATBB01 | Australia | LSU D1-D2 | U44933 |
| A. tamarense | pgt183 | United Kingdom | LSU D1-D2 | U44930 |
| A. tamarense | SZN01 | Italy | LSU D1-D2 | AJ535368 |
| A. tamarense | SZN19 | Italy | LSU D1-D2 | AJ535370 |
| A. tamarense | UW4-2 | Scotland | LSU D1-D2 | AJ303448 |
| A. tamarense | Plymouth | United Kingdom | LSU D1-D2 | AF033534 |
| A. tamarense | HAT4 | Japan | LSU D1-D2 | AB088279 |
| A. tamarense | K-0055 | Faroe Islands | LSU D1-D3 | AF200668 |
| A. tamarense | JHW0003-2 | Korea (?) | LSU D1-D3 | AY438021 |
| A. fundyense | AFNFA3.1 | Canada | LSU D1-D2 | U44926 |
| A. cohorticula | ND | Malaysia | LSU D1-D2 | AF174614 |
| A. tamiyavanichi | TAMI2207 | Japan | LSU D1-D2 | AB088267 |
| A. fraterculus | JHW0110-1 | Korea (?) | LSU D1-D3 | AY438017 |
| F. subglobosum | ND | ND | LSU D1-D3 | AF260387 |
| A. lusitanicum | LAC27 | Italy | ITS1-5.8S-ITS2 | ALAJ5050 |
| A. minutum | CNR AMI-A4 | Italy | ITS1-5.8S-ITS2 | AJ318460 |
| A. minutum | Anakoha | New-Zealand | ITS1-5.8S-ITS2 | |
| A. insuetum | S1 | Japan | ITS1-5.8S-ITS2 | AB006996 |
| A. tamutum | SHA. A11 | Ireland, west | ITS1-5.8S-ITS2 | |
| A. ostenfeldii | BY. K04 | Ireland, southwest | ITS1-5.8S-ITS2 | |
| A. margalefi | AM-1 | Italy | ITS1-5.8S-ITS2 | AJ251208 |
?, putative isolation locality.
ND, not determined.
Random amplification of polymorphic DNA analysis.
Total genomic DNA samples obtained from each A. minutum culture were subjected to random amplification of polymorphic DNA (RAPD) analysis using 10 randomly generated primers, including RP1-4, used for Chattonella spp. in another study (41). PCR mixtures were as follows: 1× PCR buffer, 3 mM MgCl2, 200 μM of each dNTP, 1 μM of random primer, 1 unit of Taq polymerase, and 1 μl of template DNA. The amplification conditions were as follows: initial cycle of denaturation, 94°C for 5 min; 40 cycles of production (denaturation, 94°C for 30 sec; annealing, 45°C for 1 min; and extension, 72°C for 30 sec); and a final extension step (72°C for 6 min). The DNA fragments were observed on thin 1.5% agarose-1× TAE buffer gel containing ethidium bromide (1 μg ml−1) at a low migration speed for optimum separation of bands. For each random primer set, all the bands produced in each line of the gel were recorded, and then the absence or presence of each band for each strain was scored as 0 or 1, respectively, to generate a binary matrix. The generated similarity matrix was converted into a dendrogram with the unweighted pair group method with arithmetic averages (UPGMA) cluster analysis provided by the PAUP package.
RESULTS
Morphology.
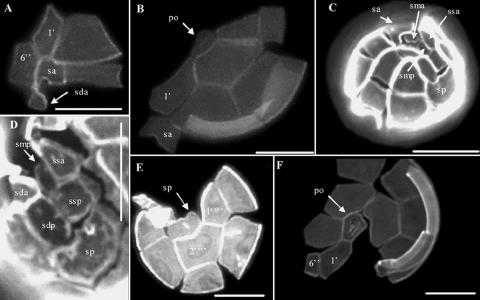
Morphological analysis was carried out on material derived from cultured cells only. Only small solitary pigmented cells were observed, roughly oval in shape and with thick plates. No morphological differences were observed among cultures of A. minutum strains isolated from the south and west coasts of Ireland. Vegetative cells had an average height and width of 24.2 μm (±3.3 μm, standard deviation [SD]) and 22.3 μm (±3.4 μm SD), respectively. Species identification was straightforward when diagnostic plates were visible using Calcofluor White stain (Fig. 2). The plate tabulation was typical for Alexandrium species (7). The apical pore complex was ovoid and quite narrow. The first apical plate (1′, Kofoid notation) was variously shaped but was most often found directly attached to the apical pore; rarely, the 1′ plate was joined to the pore by a narrow connection of variable length. No ventral pore was present on the first apical plate in any of the specimens. The sixth precingular plate (6′) was very narrow, a feature characteristic of A. minutum. In the sulcal area, the anterior lateral plate and the short left posterior lateral plate were, respectively, rhomboidal and short, as described in Balech (7). Finally, the posterior sulcal plate was short and wider than it was long, and no attachment pore was observed.
FIG. 2.
Epifluorescence microscopy of Calcofluor-stained cultured vegetative cells of A. minutum isolated from Irish coastal waters. (A) Ventral view showing the first apical plate (1′) and a narrow sixth precingular plate (6′). (B) Apical view showing a wide sulcal anterior plate (sa) and a narrow connection between the apical pore (po) and the first apical plate (1′). (C and D) Details of the sulcal region with the characteristic rhomboidal left anterior lateral (ssa) and short left posterior lateral plates (ssp). Abbreviations: smp, posterior median plate; sda, right anterior lateral plate; sdp, right posterior lateral plate. (E) Antapical view showing the wide posterior sulcal plate (sp). 1′‴ and 2′‴ are the first and second postcingular plates, respectively. (F) Apical view showing details of a large apical pore (po) with a wide connection to the first apical plate (1′). All scale bars for panels A to F are 10 μm.
PSP toxin analysis.
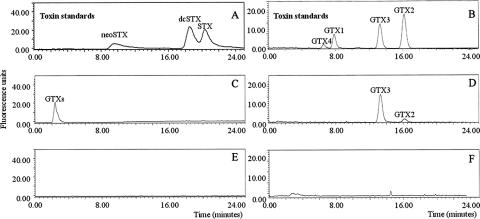
None of the culture extracts of A. minutum strains isolated from the west coast of Ireland contained PSP toxins, in contrast to the toxin-containing strains from the south coast (Fig. 3). In late exponential growth phase, PSP toxins in these strains comprised only GTX2 and GTX3; the latter toxin was always more abundant and constituted more than 65% of the total on a molar basis. Strains from Cork Harbor maintained in f/2 medium minus silicate contained on average 2.5 pg GTX cell−1 (±0.5 pg, SD; n = 5). Traces of dcGTX23 were also detected, but peaks of N-sulfocarbamoyl (C-toxins) or nonsulfated (saxitoxin, neo-saxitoxin, and dc-saxitoxin) toxins were not found during any stage of culture. Cultures of both A. minutum forms were sampled and analyzed for the presence of PSP toxins at various stages of the growth, including the late stationary phase, when nutrients are exhausted. No toxins were detected in the nontoxic A. minutum form. The presence of GTX2 and GTX3 was observed in a concentrated field sample taken from the south coast in Cork Harbor in which the Alexandrium cell density was greater than 2 × 105 cells liter−1, but none was detected in net haul samples obtained from the west coast in Killary Harbor (Alexandrium spp. concentration greater than 2 × 103 cells liter−1).
FIG. 3.
Selected HPLC fluorescence detection chromatograms of PSP toxins obtained from Alexandrium minutum strains isolated from the south and west coasts of Ireland. (A and B) Standards of toxins. (C and D) A. minutum Cork Harbor sample (south coast). (E and F) A. minutum Shannon Estuary sample (west coast). Panels A, C, and E and B, D, and F correspond to the first and second isocratic runs, respectively. Only A. minutum strains from the south coast synthesized PSP toxins (GTX2/GTX3).
LSU rDNA phylogeny, alignment, and comparison of ITS sequences.
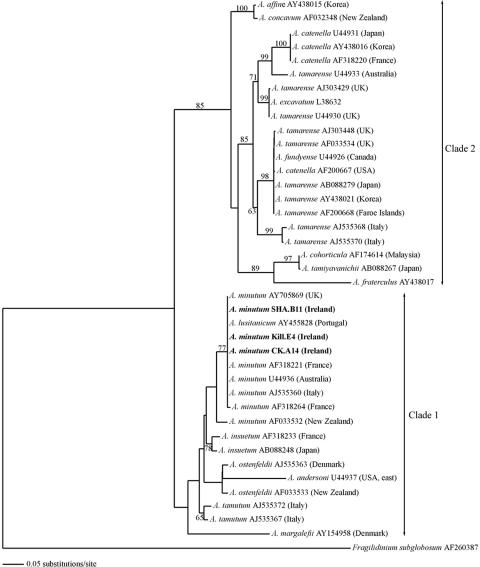
Partial sequences were obtained for the 28S subunit and complete ones for the ITS1-5.8S-ITS2 region. The expected size of the product (675 bp) was generated for all the LSU rDNA D1-D2 domain samples. A data matrix comprising 640 nucleotides was then compiled from 40 sequences in order to elucidate the phylogeny of strains of Alexandrium spp. and to determine where the Irish isolates fitted. No nucleotide differences were observed when comparing the sequences between the strains from the south and those from the west of Ireland, i.e., those that did produce PSP toxins and those that did not. The LSU rDNA sequences of the Irish strains were identical to those of A. minutum strains from Italy (AL9T), Portugal (181NT), the United Kingdom (3.9 h), and Australia (AMAD06). The divergence was greater with a strain from Brittany, France (95/4), and even greater still with a strain from New Zealand (Anakoha) (data not shown). Based on the D1-D2 domain nucleotide sequences, species of Alexandrium were separated into two major clades after the establishment of a phylogenetic tree inferred by maximum likelihood analysis, which had a score of −ln 4586.48 (Fig. 4). Clade 1 comprised A. minutum, A. tamutum, A. ostenfeldii, A. insuetum, A. andersoni, and A. margalefi; and Clade 2 included the A. tamarense species complex (A. tamarense/A. catenella/A. fundyense) together with separate groups of A. affine and A. concavum on the one hand and A. cohorticula, A. tamiyavanichi, and A. fraterculus on the other. The topology of the tree was consistent with previous findings (26, 42, 52, 53), with the exceptions of some branch positions in Clade 1. This may have resulted from the type of analysis used and/or the taxa included in the sequence alignment. Another explanation would be that, with the exception of the monophyly of A. minutum strains, the branch patterns were not well supported by bootstrap values in Clade 1, particularly for the positioning of A. andersoni and A. margalefi.
FIG. 4.
The most likely phylogenetic tree inferred from the maximum likelihood analysis of LSU rDNA sequences (D1-D2 domain). The optimal base substitution model derived from the Akaike information criterion in Modeltest 3.7 was a TIM+G model with the following constraining parameters for base frequencies, base substitution frequencies, proportion of invariable sites and gamma distribution shape parameter, respectively: A = 0.2569, C = 0.1639, G = 0.2543, T = 0.3249; A-C = 1.0000, A-G = 2.2110, A-T = 0.7666, C-G = 0.7666, C-T = 4.1370, G-T = 1.0000; I = 0; G = 0.6214. Numbers on the branches indicate branch frequency from 100 bootstrap replicates (values of <50% were not included).
The ITS1-5.8S-ITS2 rDNA sequence alignment of some species from Clade 1 showed that toxic and nontoxic forms of A. minutum from Ireland could not be differentiated and were also identical to strains from Italy and Portugal. The alignment showed that the 5.8S gene was very conservative among the taxa used for the analysis, the nucleotide variability being associated mainly with the intergenic spacers (data not shown). Surprisingly, the sequence divergence between European strains of A. minutum and the A. minutum ANAKOHA strain from New Zealand was greater than that with A. tamutum or A. insuetum (Table 4). The greatest divergence among the ITS1-5.8S-ITS2 rDNA sequences was found between A. margalefi and A. minutum from New Zealand, the former being the isolate most distantly related to any of the strains used for the comparative analysis.
TABLE 4.
Distance values between sequences of the ITS1-5.8S-ITS2 rDNA region obtained from nine strains of Alexandrium spp.a
| Taxa (strain or GenBank accession no.) | Distance between rDNA sequences (bp)
|
||||||||
|---|---|---|---|---|---|---|---|---|---|
| CK. A14 | Kill. E4 | ALAJ 5050 | AJ3 18460 | Anakoha | AB0 06996 | SHA. A11 | BY. K04 | AJ 251208 | |
| A. minutum (CK.A14) | 0 | 0 | 0 | 0.1504 | 0.1189 | 0.1097 | 0.1620 | 0.2350 | |
| A. minutum (Kill. E4) | 0 | 0 | 0 | 0.1504 | 0.1189 | 0.1097 | 0.1620 | 0.2350 | |
| A. lusitanicum (ALAJ5050) | 0 | 0 | 0 | 0.1504 | 0.1189 | 0.1097 | 0.1620 | 0.2350 | |
| A. minutum (AJ318460) | 0 | 0 | 0 | 0.1504 | 0.1189 | 0.1097 | 0.1620 | 0.2350 | |
| A. minutum (Anakoha) | 0.1685 | 0.1685 | 0.1685 | 0.1685 | 0.1466 | 0.1592 | 0.1712 | 0.2738 | |
| A. insuetum (AB006996) | 0.1301 | 0.1301 | 0.1301 | 0.1301 | 0.1636 | 0.0765 | 0.1353 | 0.2528 | |
| A. tamutum (SHA.A11) | 0.1187 | 0.1187 | 0.1187 | 0.1187 | 0.1796 | 0.0807 | 0.1288 | 0.2483 | |
| A. ostenfeldii (BY.K04) | 0.1833 | 0.1833 | 0.1833 | 0.1833 | 0.1948 | 0.1496 | 0.1417 | 0.2670 | |
| A. margalefi (AJ251208) | 0.2820 | 0.2820 | 0.2820 | 0.2820 | 0.3417 | 0.0926 | 0.3022 | 0.3306 | |
Distance values, computed with PAUP*, between sequences of the ITS1-5.8S-ITS2 rDNA region obtained from nine strains of Alexandrium spp., which grouped together in a cluster as a result of a phylogenetic analysis (Clade 1 in Fig. 3). Uncorrected distances are given above the diagonal and distance estimates based on the Kimura-2 parameter model are given below the diagonal.
RAPD comparison of A. minutum isolates from Ireland.
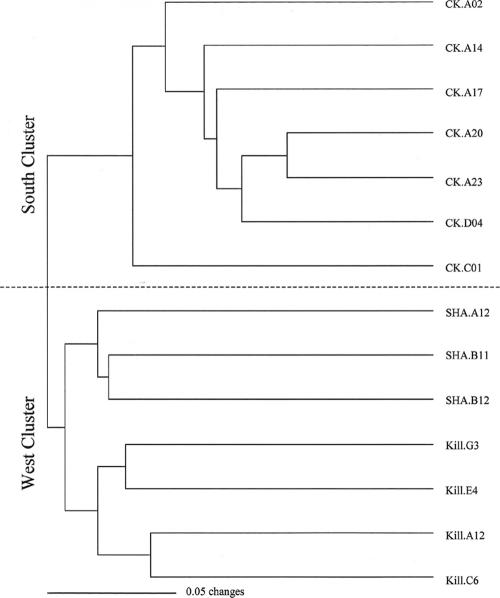
Successful amplifications were obtained for all strains and primers used for the analysis. All the primer sets produced different band profiles; a total of 555 bands were checked on a presence/absence basis, with an average of 106 ± 17 bands generated per strain. The RAPD reactions proved reproducible, and identical profiles were obtained from repeated amplifications with the same primer. One nontoxic strain of A. tamarense (CK.C01) isolated in the same location as the A. minutum strains from the south coast was also included in the RAPD analysis as a local, closely related out-group. The similarity dendrogram obtained with UPGMA showed two main clusters, one which included all the strains from the south coast and the other one comprising exclusively A. minutum strains from the west coast (Fig. 5). The analysis clustered the A. tamarense CK.C01 strain with the “south branch,” suggesting a relationship with the A. minutum strains from the area, as opposed to fitting it in a separate group as might have been expected. Nevertheless, it was clearly positioned alone on a separate branch within the cluster. For the “west branch,” two subgroups that could be clearly defined by geographical origin were identified. One subgroup contained strains isolated from Killary Harbor and the other contained strains isolated near the Shannon Estuary.
FIG. 5.
Similarity dendrogram obtained from the cluster analysis (UPGMA) of 14 strains of Alexandrium spp. collected from the south (CK.) and west (Kill. and SHA.) coasts of Ireland. Strains with the greatest number of shared RAPD bands cluster together. CK.C01, the A. tamarense strain initially selected as an out-group fits separately in the south cluster.
DISCUSSION
Alexandrium minutum is a cosmopolitan dinoflagellate found along the Atlantic, Mediterranean, and North Sea coasts of Europe. Elsewhere, it has been recorded along the coasts of Australia, New Zealand, Southeast Asia, and India but is apparently scarce along the coasts of the Americas (7, 22, 25, 62). Morphological variations within the species among isolates from diverse regions around the globe are mainly in the first apical plate region. These are manifested by the presence or absence of a ventral pore and the type of connection between that plate and the apical pore complex. No such morphological differences were recorded between strains isolated from the west and the south coasts of Ireland. These A. minutum isolates corresponded closely to Balech's description of the species (6), the main diagnostic features being a narrow 6′ and a wider-than-long posterior sulcal plate. Samples from both regions lacked a ventral pore, similar to strains from the United Kingdom and Denmark but unlike other European strains from Spain or Italy, as reported previously (26). Nevertheless, Hansen et al. (26) noticed that 6% of A. minutum vegetative cells in a preserved field sample collected off the west coast of Ireland in 2001 possessed a ventral pore. This indicates that there is a further A. minutum morphotype from the west of Ireland which has yet to be brought into culture.
The A. minutum strains studied during this investigation were collected from close geographic areas, but only those isolated from Cork Harbor produced PSP toxins. This result identifies A. minutum as the organism most likely responsible for the PSP events recorded in this area in the past, as the toxin profile (GTX2 and GTX3) is identical to that obtained in 1996 for contaminated shellfish from the region, on the only occasion for which detailed toxin analysis is available (19). At the time, A. tamarense was the suspected causative organism (19), but recent studies have shown that this species from Cork Harbor corresponds to the nontoxic Western European ribotype (27, 60a). The toxin profile is quite similar to that found in A. minutum cultures obtained from the Fal estuary in the United Kingdom (46a) and in a strain isolated from the Brittany coast of France (15), the latter differing by the additional presence of C-toxins. The most notable result, however, was that none of the cultured isolates of A. minutum from the west coast of Ireland synthesized PSP toxins. Considering the possible nondetection of very small quantities of toxins in those samples, the cellular PSP toxin quota would be less than 2.5 fg cell−1, which is 1,000 times inferior to the value obtained with the toxic strains from Cork Harbor. The only previous reports of naturally nontoxic A. minutum referred to in the literature concerned a strain obtained from Taiwan and two strains isolated from the Gulf of Trieste in Italian coastal waters (37, 60). Toxin composition results obtained with monoclonal cultures were also confirmed after analysis of field samples. No PSP toxins were detected in Killary Harbor in July 2004 when A. minutum cells were present in the water column, but GTX2 and GTX3 were observed in a net sample taken during a bloom of Alexandrium spp. in Cork Harbor in June 2004. The use of toxins to establish taxonomic relationships does not usually appear to be reliable because too many external parameters, such as bacterial activity, salinity, or nutrient concentrations, may influence both the toxin synthesis and the toxin profile (16, 29, 38). Within the Irish context, however, the presence of toxin could be a discriminative character, allowing strains isolated from the south coast to be distinguished from those isolated from the west coast, a distinction morphology does not allow.
At least two well-defined clades exist in the phylogeny of Alexandrium based on the sequence of the D1-D2 domain of the LSU rDNA (31, 42, 63). The Irish isolates of A. minutum fitted into Clade 1, showing 100% homology with strains from the United Kingdom, Italy, Portugal, and Australia. The nontoxic and toxic A. minutum strains from Ireland could not be resolved in the phylogenetic analysis based on the D1-D2 domain of the LSU rDNA. This was because no nucleotide variability existed between them in the rDNA sequences examined, even though the D1-D2 domain is one of the most rapidly evolving regions of the eukaryotic rDNA genes (63). As with the LSU rDNA, no differences were detected between strains from the west coast and those from the south coast of Ireland in the ITS1-5.8S-ITS2 genomic region, or even between Irish strains and two strains from Italy. The level of genetic resolution provided by this region is therefore not satisfactory to explain morphological or physiological differences between these strains. Similar results were obtained for several isolates of Gymnodinium catenatum (Graham) and Pfiesteria piscicida (Steidinger and Burkholder) (11, 59). The ribosome-encoding regions are usually a good choice for oligonucleotide probe design because ribosomes are produced in high numbers within cells, making the detection of a bright fluorescent signal possible after binding of probes to their targets. However, such probes could not discriminate between PSP toxin-producing and nontoxic isolates of Irish A. minutum; to do so would require the development of alternative genetic markers.
RAPD was used as a final attempt to determine genetic polymorphism between A. minutum strains from the south (toxic) and west (nontoxic) coasts of Ireland. The technique has been widely used for fingerprinting and genetic mapping studies or even for population structure analysis (54, 64). It has already been applied to the study of microalgae such as Chattonella or Gymnodinium to compare isolates of the same genus or species from different locations (2, 41). In the present investigation, the similarity dendrogram revealed assemblages which reflected the geographic origin of the strains. In other studies, strains from the same location have clustered in different groups (10, 13). In these cases the hypothesis advanced suggested that genetic variation was partitioned mainly within populations of the same species, compared with the variation between regions and between populations within regions (13). It is commonly accepted that microalgae are associated with bacteria which can be either free in the surrounding phycosphere, attached externally on the surface membrane, or within cells in small inclusions (reference 55 and references therein). This was also demonstrated for some dinoflagellates by transmission electron microscopy (35), by fluorescent in situ hybridization, or through bacterial isolation and culturing (8, 16, 20). As a result, native bacteria are probably always included in cultures when cultures are initiated from resting cysts or vegetative cells. Often, contaminant bacteria appear when nonaxenic conditions are applied, particularly when dealing with a large number of cultures. When DNA extractions from the monoclonal cell cultures were performed, DNA belonging to the bacteria flora within or exteriorly associated with the Alexandrium vegetative cells was also extracted. As random primers are by definition nonspecific, the genetic information coming from bacteria would have been incorporated in the results of the analysis. The motivation for the use of RAPD in this study was to allow the comparison of integrated isolates, i.e., A. minutum with the associated bacterial flora. The possibility of forcing bacteria on the relationships between isolates was purposefully favored by selecting a single closely related out-group from the location where some of the isolates of interest originated. As a result, A. tamarense and A. minutum from Cork Harbor, which were clearly genetically separated regarding the LSU or ITS regions of the rDNA, grouped together in the southern assemblage. Thus, the clustering of A. tamarense with the toxic A. minutum from the south coast probably reflected a combination of the intrinsic polymorphism of both species, whereby they appeared on different branches, and the sharing of bacteria typical of the harbor area, which is an embayed retention zone influenced by anthropogenic inputs. This suggestion would be supported by the fact that the “West” isolates also clustered by geographic origin. Killary Harbor is a fjord subjected to high freshwater input and usually has a distinct halocline, whereas the Shannon Estuary is well mixed, and west of the mouth, the salinities are nearly always high (<34 practical salinity units) (40, 49). To investigate the likelihood that different bacterial profiles would be associated with different A. minutum isolates would require using techniques such as denaturant gradient gel electrophoresis. This could also help in determining whether the same bacteria are associated with the toxic and nontoxic strains. A recent study that focused on the genetic variability between A. minutum and A. tamarense strains from various locations was based on RAPD results (39). Interestingly, the authors included in their analysis A. tamarense and A. minutum strains from the same location in conjunction with strains from other areas. The influence of bacteria was probably attenuated in this case as the final dendrogram clearly separated both species, independently of their geographical origins. These results underline the fact that caution is necessary when interpreting data derived from fingerprinting analyses as the analysis orientates the selection of out-groups. Comparison of integrated samples of one species from close geographical areas should favor the use of local out-groups. On the other hand, the analysis of genetic structures among globally distributed strains and species facilitates the dilution of a possible bacterial influence. Undoubtedly, the best way to reveal strict genetic polymorphism between various species or strains is to use axenic cultures. This however is a technically constraining procedure which does not reflect ecological reality. Finally, the concomitant use of RAPD with several fingerprinting techniques, such as amplified fragment length polymorphism and microsatellite markers, which do not theoretically guarantee the elimination of bacterial interference, appears to be the most elegant approach for population genetic studies.
One remaining question is whether the supposedly toxic and nontoxic A. minutum populations are separated by biological and/or by physical barriers. Two scenarios are possible. The first scenario would suppose that the two populations of A. minutum sometimes share the same ecological niche, but because of reproduction incompatibilities, such as pre- or postzygotic barriers (D. Kulis and D. M. Anderson, unpublished data), competition for nutrients or allelopathy, they remain separated. The other scenario relies on the lack of physical contact between the two populations. The North Channel of Cork Harbor, where A. minutum blooms originate within the estuary, is a highly retentive zone that harbors the toxic form, while the shelf seas along the southwest and west coasts support the nontoxic form. A previous study (48) has shown that under prevailing winds from the southwest quarter, water in the northern Celtic Sea is separated from that over the Atlantic Shelf at the southwest of Ireland. Populations on the west coast could therefore develop separately from those off the south coast. It is unlikely however that these two regions remain geographically distinct as the coastal current which flows clockwise around the southwest corner of Ireland is known to operate between them under suitable meteorological conditions (48).
In conclusion, the divergence in toxin profiles among the A. minutum strains isolated from Irish waters was not associated with differences in the LSU and ITS regions of rDNA. Furthermore, the morphology of A. minutum did not allow an explanation for the divergence in the toxin profile between the toxic and nontoxic strains of this species. Therefore, the use of morphology or genetic information of rDNA does not appear suitable enough to distinguish between toxin-producing and nontoxic forms of this species. The RAPD analysis revealed, nevertheless, a clear distinction between the southern and western coast isolates, a segregation which also coincided with the toxic and nontoxic character of the strains analyzed. This has important implications for monitoring some toxigenic species for public health reasons, as it would appear that prevention of harmful outbreaks must rely on the chemical analysis of the biotoxins alone. The presence of Alexandrium spp. observed in samples following examination by conventional microscopy methods, however, would still be a convenient trigger with which to initiate toxin analysis, and results would provide a basis for determining if the geographic distribution of populations of the nontoxic A. minutum form is expanding.
Acknowledgments
This work was supported by the Higher Education Authority of Ireland (Department of Education and Science) (PRTLI Cycle III).
We thank the skipper and crew of the RV Celtic Voyager and Aoife Ní Rathaille for assistance in field sampling. We are grateful to Pilar Riobó Agulla and Beatriz Paz Pino (IEO, Vigo) for assistance with toxin analysis.
Footnotes
Published ahead of print on 2 March 2007.
REFERENCES
- 1.Adachi, M., Y. Sako, and Y. Ishida. 1996. Analysis of Alexandrium (Dinophyceae) species using sequences of the 5.8S ribosomal DNA and internal transcribed spacer regions. J. Phycol. 32:424-432. [Google Scholar]
- 2.Adachi, M., Y. Sako, and Y. Ishida. 1997. Analysis of Gymnodinium catenatum Dinophyceae using sequences of the 5.8S rDNA-ITS regions and random amplified polymorphic DNA. Fish Sci. 63:701-707. [Google Scholar]
- 3.Anderson, D. M. 1997. Bloom dynamics of toxic Alexandrium species in the northeastern U.S. Limnol. Oceanogr. 42:1009-1022. [Google Scholar]
- 4.Anderson, D. M., D. M. Kulis, G. J. Doucette, J. C. Gallagher, and E. Balech. 1994. Biogeography of toxic dinoflagellates in the genus Alexandrium from the northeastern United States and Canada. Mar. Biol. 120:467-478. [Google Scholar]
- 5.Anderson, D. M., D. M. Kulis, and B. A. Keafer. 1999. Detection of the toxic dinoflagellate Alexandrium fundyense (Dinophyceae) with oligonucleotide and antibody probes; variability in labelling intensity with physiological condition. J. Phycol. 35:870-883. [Google Scholar]
- 6.Balech, E. 1989. Redescription of Alexandrium minutum Halim (Dinophyceae) type species of the genus Alexandrium. Phycologia 28:206-211. [Google Scholar]
- 7.Balech, E. 1995. The genus Alexandrium Halim (Dinoflagellata). Sherkin Island Marine Station, Sherkin Island Co., Cork, Ireland.
- 8.Biegala, I. C., G. Kennaway, E. Alverca, J.-F. Lennon, D. Vaulot, and N. Simon. 2002. Identification of bacteria associated with dinoflagellates (Dinophyceae) Alexandrium spp. using tyramide signal amplification-fluorescent in situ hybridization and confocal microscopy. J. Phycol. 38:404-411. [Google Scholar]
- 9.Boelens, R. G. V., D. M. Maloney, A. P. Parsons, and A. R. Walsh. 1999. Ireland's marine and coastal areas and adjacent sea: an environmental assessment. Marine Institute, Dublin, Ireland.
- 10.Bolch, C. J. S., S. I. Blackburn, G. M. Hallegraeff, and R. E. Vaillancourt. 1999. Genetic variation among strains of the toxic dinoflagellate Gymnodinium catenatum (Dinophyceae). J. Phycol. 35:356-367. [DOI] [PubMed] [Google Scholar]
- 11.Bolch, C. J. S., A. P. Negri, and G. M. Hallegraeff. 1999. Gymnodinium microreticulatum sp. nov. (Dinophyceae): a naked, microreticulate cyst-producing dinoflagellate, distinct from Gymnodinium catenatum and Gymnodinium nolleri. Phycologia 38:301-313. [Google Scholar]
- 12.Bornet, B., E. Antoine, E. Francoise, and C. Marcaillou-Le Baut. 2005. Development of sequence characterized amplified region markers from intersimple sequence repeat fingerprintings for the molecular detection of toxic phytoplankton Alexandrium catenella (Dinophyceae) and Pseudo-nitzschia pseudodelicatissima (Bacillariophyceae) from French coastal waters. J. Phycol. 41:704-711. [Google Scholar]
- 13.Camino Ordas, M., S. Fraga, J. M. Franco, A. Ordas, and A. Figueras. 2004. Toxin and molecular analysis of Gymnodinium catenatum (Dinophyceae) strains from Galicia (NW Spain) and Andalucia (S Spain). J. Plankton Res. 26:341-349. [Google Scholar]
- 14.Cembella, A. D., J. J. Sullivan, G. L. Boyer, F. J. R. Taylor, and R. J. Andersen. 1987. Variation in paralytic shellfish toxin composition within the Protogonyaulax tamarensis/catenella species complex; red tide dinoflagellates. Biochem. Syst. Ecol. 15:171-186. [Google Scholar]
- 15.Chang, F. H., D. M. Anderson, D. M. Kulis, and D. J. Till. 1997. Toxin production of Alexandrium minutum (Dinophyceae) from the Bay of Plenty, New Zealand. Toxicon 35:393-409. [DOI] [PubMed] [Google Scholar]
- 16.Dantzer, W. R., and R. E. Levin. 1997. Bacterial influence on the production of paralytic shellfish toxins by dinoflagellated algae. J. Appl. Microbiol. 83:464-469. [DOI] [PubMed] [Google Scholar]
- 17.Franco, J. M., and P. Fernandez Vila. 1993. Separation of paralytic shellfish toxins by reversed phase high performance liquid chromatography with postcolumn reaction and fluorimetric detection. Chromatographia 35:613-620. [Google Scholar]
- 18.Fritz, L., and R. E. Triemer. 1985. A rapid simple technique utilizing Calcofluor White M2R for the visualization of dinoflagellate thecal plates. J. Phycol. 21:662-664. [Google Scholar]
- 19.Furey, A., K. J. James, and I. R. Sherlock. 1998. First report of paralytic shellfish poisoning toxins in the Republic of Ireland, p. 70-71. In B. Reguera, J. Blanco, M. L. Fernandez, and T. Wyatt (ed.), Harmful algae. Xunta de Galicia and IOC of UNESCO, Santiago de Compostela, Spain.
- 20.Gallacher, S., K. J. Flynn, J. M. Franco, E. E. Brueggemann, and H. B. Hines. 1997. Evidence for production of paralytic shellfish toxins by bacteria associated with Alexandrium spp. (Dinophyta) in culture. Appl. Environ. Microbiol. 63:239-245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Galluzzi, L., A. Penna, E. Bertozzini, M. Vila, E. Garces, and M. Magnani. 2004. Development of a real-time PCR assay for rapid detection and quantification of Alexandrium minutum (a dinoflagellate). Appl. Environ. Microbiol. 70:1199-1206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Godhe, A., S. K. Otta, H. A. S. Rehnstam, I. Karunasagar, and I. Karunasagar. 2001. Polymerase Chain Reaction in detection of Gymnodinium mikimotoi and Alexandrium minutum in field samples from Southwest India. Mar. Biotechnol. 3:152-162. [DOI] [PubMed] [Google Scholar]
- 23.Guillard, R. 1975. Culture of phytoplankton for feeding marine invertebrates, p. 29-60. In W. Smith and M. H. Chanley (ed.), Culture of marine invertebrate animals. Plenum Press, New York, NY.
- 24.Hallegraeff, G. M. 1993. A review of harmful algal blooms and the apparent global increase. Phycologia 32:79-99. [Google Scholar]
- 25.Hallegraeff, G. M., D. A. Steffensen, and R. Wetherbee. 1988. Three estuarine dinoflagellates that can produce paralytic shellfish toxins. J. Plankton Res. 10:533-541. [Google Scholar]
- 26.Hansen, G., N. Daugbjerg, and J. M. Franco. 2003. Morphology, toxin composition and LSU rDNA phylogeny of Alexandrium minutum (Dinophyceae) from Denmark, with some morphological observations on other European strains. Harmful Algae 2:317-335. [Google Scholar]
- 27.Higman, W. A., D. M. Stone, and J. M. Lewis. 2001. Sequence comparison of toxic and non-toxic Alexandrium tamarense (Dinophyceae) isolates from UK waters. Phycologia 40:256-262. [Google Scholar]
- 28.Hosoi-Tanabe, S., and Y. Sako. 2004. Rapid detection of natural cells of Alexandrium tamarense and A. catenella (Dinophyceae) by fluorescence in situ hybridization. Harmful Algae 4:319-328. [Google Scholar]
- 29.Hwang, D. F., and Y. H. Lu. 2000. Influence of environmental and nutritional factors on growth, toxicity, and toxin profile of dinoflagellate Alexandrium minutum. Toxicon 38:1491-1503. [DOI] [PubMed] [Google Scholar]
- 30.John, U., A. Cembella, C. Hummert, M. Elbrachter, R. Groben, and L. Medlin. 2003. Discrimination of the toxigenic dinoflagellates Alexandrium tamarense and A. ostenfeldii in co-occurring natural populations from Scottish coastal waters. Eur. J. Phycol. 38:25-40. [Google Scholar]
- 31.John, U., R. A. Fensome, and L. K. Medlin. 2003. The application of a molecular clock based on molecular sequences and the fossil record to explain biogeographic distributions within the Alexandrium tamarense “species complex” (Dinophyceae). Mol. Biol. Evol. 20:1015-1027. [DOI] [PubMed] [Google Scholar]
- 32.John, U., R. Groben, B. Beszteri, and L. Medlin. 2004. Utility of amplified fragment length polymorphisms (AFLP) to analyze genetic structures within the Alexandrium tamarense species complex. Protist 155:169-179. [DOI] [PubMed] [Google Scholar]
- 33.Kao, C. Y. 1993. Paralytic shellfish poisons, p. 75-86. In I. R. Falconer (ed.), Algal toxins in seafood and drinking waters. Academic Press, London, United Kingdom.
- 34.Lenaers, G., L. Maroteaux, B. Michot, and M. Herzog. 1989. Dinoflagellates in evolution. A molecular phylogenetic analysis of large subunit ribosomal RNA. J. Mol. Evol. 29:40-51. [DOI] [PubMed] [Google Scholar]
- 35.Lewis, J., G. Kennaway, S. Franca, and E. Alverca. 2001. Bacterium-dinoflagellate interactions: investigative microscopy of Alexandrium spp. (Gonyaulacales, Dinophyceae). Phycologia 40:280-285. [Google Scholar]
- 36.Lilly, E. L., D. M. Kulis, P. Gentien, and D. M. Anderson. 2002. Paralytic shellfish poisoning toxins in France linked to a human-introduced strain of Alexandrium catenella from the western Pacific: evidence from DNA and toxin analysis. J. Plankton Res. 24:443-452. [Google Scholar]
- 37.Lilly, E. L., K. M. Halanych, and D. M. Anderson. 2005. Phylogeny, biogeography, and species boundaries within the Alexandrium minutum group. Harmful Algae 4:1004-1020. [Google Scholar]
- 38.Lippemeier, S., D. M. F. Frampton, S. I. Blackburn, S. Geier, and A. P. Negri. 2003. Influence of phosphorus limitation on toxicity and photosynthesis of Alexandrium minutum (Dinophyceae) monitored by in-line detection of variable chlorophyll fluorescence. J. Phycol. 38:320-331. [Google Scholar]
- 39.Martinez, R., C. Anibarro, and S. Fernandez. 2006. Genetic variability among Alexandrium tamarense and Alexandrium minutum strains studied by RAPD banding pattern analysis. Harmful Algae 5:599-607. [Google Scholar]
- 39a.McDermott, G., and R. Raine. 2006. The dinoflagellate genus Ceratium in Irish shelf seas. The Martin Ryan Institute, Galway, Ireland.
- 40.McMahon, T. G., R. C. T. Raine, T. Fast, L. Kies, and J. W. Patching. 1992. Phytoplankton biomass, light attenuation and mixing in the Shannon Estuary, Ireland. J. Mar. Biol. 72:709-720. [Google Scholar]
- 41.Murayama-Sadaaki, E., S. Yoshimatsu, T. Kayano, T. Nishio, H. Ueda, and T. Nagamune. 1998. Application of the random amplified polymorphic DNA (RAPD) technique to distinguishing species of the red tide phytoplankton Chattonella (Raphydophyceae). J. Ferment. Bioeng. 85:343-345. [Google Scholar]
- 42.Montresor, M., U. John, A. Beran, and L. K. Medlin. 2004. Alexandrium tamutum sp. nov. (Dinophyceae): a new nontoxic species in the genus Alexandrium. J. Phycol. 40:398-411. [Google Scholar]
- 43.Nagai, S., C. Lian, M. Hamaguchi, Y. Matsuyama, S. Itakura, and T. Hogetsu. 2004. Development of microsatellite markers in the toxic dinoflagellate Alexandrium tamarense (Dinophyceae). Mol. Ecol. Notes 4:83-85. [Google Scholar]
- 44.Nascimento, S. M., D. A. Purdie, E. L. Lilly, J. Larsen, and S. Morris. 2005. Toxin profile, pigment composition, and large subunit rDNA phylogenetic analysis of an Alexandrium minutum (Dinophyceae) strain isolated from the Fleet Lagoon, United Kingdom. J. Phycol. 41:343-353. [Google Scholar]
- 45.Oshima, Y., K. Sugino, and T. Yasumoto. 1989. Latest advances in HPLC analysis of paralytic shellfish toxins, p. 319-326. In S. Natori, K. Hashimoto, and Y. Ueno (ed.), Mycotoxins and phycotoxins ′88. Elsevier, Amsterdam, The Netherlands.
- 46.Penna, A., E. Garces, M. Vila, M. G. Giacobbe, S. Fraga, A. Luglie, I. Bravo, E. Bertozzini, and C. Vernesi. 2005. Alexandrium catenella (Dinophyceae), a toxic ribotype expanding in the NW Mediterranean Sea. Mar. Biol. 148:13-23. [Google Scholar]
- 46a.Percy, L., W. Higman, D. Stone, S. Morris, and J. Lewis. 2004. A detailed study of phytoplankton in the Fal Estuary, U.K., and the relationship between Alexandrium species and PSP toxin profiles of Mytilus edulis, p. 12. Abstr. 5th Int. Conf. Molluscan Shellfish Safety, Galway, Ireland, 14 to 18 June 2004.
- 47.Posada, D., and K. A. Crandall. 1998. Modeltest: testing the model of DNA substitution. Bioinformatics 14:817-818. [DOI] [PubMed] [Google Scholar]
- 48.Raine, R., and T. McMahon. 1998. Physical dynamics on the continental shelf off southwestern Ireland and their influence on coastal phytoplankton blooms. Cont. Shelf Res. 18:883-914. [Google Scholar]
- 49.Roden, C. M., P. G. Rodhouse, M. P. Hensey, T. McMahon, T. H. Ryan, and J. P. Mercer. 1987. Hydrography and the distribution of phytoplankton in Killary Harbour: a fjord in western Ireland. J. Mar. Biol. 67:359-371. [Google Scholar]
- 50.Sako, Y., C. M. Adachi, and Y. Ishida. 1993. Preparation of characterization of monoclonal antibodies to Alexandrium species, p. 87-93. In T. J. Smayda and Y. Shimizu (ed.), Toxic phytoplankton blooms in the sea. Elsevier, New York, NY.
- 51.Sako, Y., H. Kim, H. Ninomiya, M. Adachi, and Y. Ishida. 1990. Isozyme and cross analysis of mating populations in the Alexandrium catenella/tamarense species complex, p. 320-323. In E. Graneli, E. Sunderstrom, L. Edler, and D. M. Anderson (ed.), Toxic marine phytoplankton. Elsevier, New York, NY.
- 52.Scholin, C. A., G. M. Hallegraeff, and D. M. Anderson. 1995. Molecular evolution of the Alexandrium tamarense species complex (Dinophyceae): dispersal in the North American and west Pacific regions. Phycologia 34:472-485. [Google Scholar]
- 53.Scholin, C. A., M. Herzog, M. Sogin, and D. M. Anderson. 1994. Identification of group- and strain-specific genetic markers for globally distributed Alexandrium (Dinophyceae). II. Sequence analysis of a fragment of the LSU rRNA gene. J. Phycol. 30:999-1011. [Google Scholar]
- 54.Shrestha, M. K., A. Golan-Goldhirsh, and D. Ward. 2002. Population genetic structure and the conservation of isolated populations of Acacia raddiana in the Negev Desert. Biol. Conserv. 108:119-127. [Google Scholar]
- 55.Simon, N., I. C. Biegala, E. A. Smith, and D. Vaulot. 2002. Kinetics of attachment of potentially toxic bacteria to Alexandrium tamarense. Aquat. Microb. Ecol. 28:249-256. [Google Scholar]
- 56.Simon, N., L. Campbell, E. Ornolfsdottir, R. Groben, L. Guillou, M. Lange, and L. K. Medlin. 2000. Oligonucleotide probes for the identification of three algal groups by dot blot and fluorescent whole-cell hybridization. J. Eukaryot. Microbiol. 47:76-84. [DOI] [PubMed] [Google Scholar]
- 57.Spalter, R. A., D. Walsh, R. A. Reeves, D. J. Saul, R. D. Gray, P. L. Bergquist, L. McKenzie, and P. R. Bergquist. 1997. Sequence heterogeneity of the ribosomal RNA intergenic region Alexandrium species. Biochem. Syst. Ecol. 25:231-239. [Google Scholar]
- 58.Swofford, D. L. 2002. PAUP* phylogenetic analysis using parsimony, version 4.0b10. Sinauer Associates, Sunderland, MA.
- 59.Tengs, T., H. A. Bowers, H. B. Glasgow, Jr., J. M. Burkholder, and D. W. Oldach. 2003. Identical ribosome DNA sequence data from Pfiesteria piscicida (Dinophyceae) isolates with different toxicity phenotypes. Environ. Res. 93:88-91. [DOI] [PubMed] [Google Scholar]
- 60.Tillmann, U., and U. John. 2002. Toxic effects of Alexandrium spp. on heterotrophic dinoflagellates: an allelochemical defense mechanism independent of PSP-toxin content. Mar. Ecol. Prog. Ser. 230:47-58. [Google Scholar]
- 60a.Touzet, N., B. Paz Pino, P. Riobó Agulla, J. M. Franco, and R. Raine. 2004. Abstr. 5th Int. Conf. Molluscan Shellfish Safety, abstr. 120.
- 61.Usup, G., L. C. Pin, A. Ahmad, and L. P. Teen. 2002. Phylogenetic relationship of Alexandrium tamiyavanichii (Dinophyceae) to other Alexandrium species based on ribosomal RNA gene sequences. Harmful Algae 1:59-68. [Google Scholar]
- 62.Usup, G., L. C. Pin, A. Ahmad, and L. P. Teen. 2002. Alexandrium (Dinophyceae) species in Malaysia waters. Harmful Algae 1:265-275. [Google Scholar]
- 63.Walsh, D., R. A. Reeves, D. J. Saul, R. D. Gray, L. Mackenzie, P. R. Berquist, and P. L. Bergquist. 1998. Heterogeneity of SSU and LSU rDNA sequences of Alexandrium species. Biochem. Syst. Ecol. 26:495-509. [Google Scholar]
- 64.Wang, X. L., Y. X. Yang, Y. Z. Cong, and D. L. Duan. 2004. DNA fingerprinting of selected Laminaria (Phaeophyta) gametophytes by RAPD markers. Aquaculture 238:143-153. [Google Scholar]