Abstract
“Photobacterium mandapamensis” (proposed name) and Photobacterium leiognathi are closely related, phenotypically similar marine bacteria that form bioluminescent symbioses with marine animals. Despite their similarity, however, these bacteria can be distinguished phylogenetically by sequence divergence of their luminescence genes, luxCDAB(F)E, by the presence (P. mandapamensis) or the absence (P. leiognathi) of luxF and, as shown here, by the sequence divergence of genes involved in the synthesis of riboflavin, ribBHA. To gain insight into the possibility that P. mandapamensis and P. leiognathi are ecologically distinct, we used these phylogenetic criteria to determine the incidence of P. mandapamensis as a bioluminescent symbiont of marine animals. Five fish species, Acropoma japonicum (Perciformes, Acropomatidae), Photopectoralis panayensis and Photopectoralis bindus (Perciformes, Leiognathidae), Siphamia versicolor (Perciformes, Apogonidae), and Gadella jordani (Gadiformes, Moridae), were found to harbor P. mandapamensis in their light organs. Specimens of A. japonicus, P. panayensis, and P. bindus harbored P. mandapamensis and P. leiognathi together as cosymbionts of the same light organ. Regardless of cosymbiosis, P. mandapamensis was the predominant symbiont of A. japonicum, and it was the apparently exclusive symbiont of S. versicolor and G. jordani. In contrast, P. leiognathi was found to be the predominant symbiont of P. panayensis and P. bindus, and it appears to be the exclusive symbiont of other leiognathid fishes and a loliginid squid. A phylogenetic test for cospeciation revealed no evidence of codivergence between P. mandapamensis and its host fishes, indicating that coevolution apparently is not the basis for this bacterium's host preferences. These results, which are the first report of bacterial cosymbiosis in fish light organs and the first demonstration that P. leiognathi is not the exclusive light organ symbiont of leiognathid fishes, demonstrate that the host species ranges of P. mandapamensis and P. leiognathi are substantially distinct. The host range difference underscores possible differences in the environmental distributions and physiologies of these two bacterial species.
“Photobacterium mandapamensis” (proposed name) is a widely distributed marine luminous bacterium found in coastal seawater (1, 18). Originally described as a separate species, P. mandapamensis overlaps in various taxonomic traits with Photobacterium leiognathi, a luminous bacterial symbiont of leiognathid fishes. This similarity led to the synonymization of these two species as P. leiognathi (3, 18, 34). Phylogenetic criteria, however, based on the luminescence genes luxAB(F)E separates these bacteria into two evolutionarily distinct lineages that match up with the original species descriptions (1). These contrasting views suggest that P. mandapamensis may be either a subspecies of P. leiognathi (1) or a separate species. Ecological and further phylogenetic evidence distinguishing these bacteria is presented here, and we therefore refer to them using the original species names, P. mandapamensis (18) and P. leiognathi (3).
The ability to distinguish P. mandapamensis from P. leiognathi on the basis of molecular phylogenetic criteria provides the means to determine whether these closely related, phenotypically similar bacteria are ecologically distinct. With respect to bioluminescent symbioses, we hypothesized that P. mandapamensis might associate with species of host animals different from those colonized by P. leiognathi (1). A difference in host species ranges might reflect other ecological or physiological differences between these bacteria. Photobacterium leiognathi has repeatedly been identified as the specific, exclusive light organ symbiont of leiognathid fishes (Perciformes, Leiognathidae), a well-studied group of marine bioluminescent fishes (e.g., see references 1, 3, 6, 9, 36, and 38), and P. mandapamensis has not previously been reported to be isolated from leiognathid light organs. Consistent with our hypothesis, a recent report indicates that bacteria provisionally identified as P. mandapamensis are light organ symbionts of two other fishes, Acropoma japonicum (Perciformes, Acropomatidae) and Siphamia versicolor (Perciformes, Apogonidae) (41).
The criteria used for previous identifications of these bacteria, however, i.e., comparisons of phenotypic traits and limited sequence analyses (e.g., see references 6, 9, 36, and 41), lack phylogenetic resolution and therefore leave the actual identities of these bacteria substantially in doubt. Specifically, the light organ bacteria from A. japonicum, S. versicolor, and certain other Siphamia species have been identified variously in the past either as P. leiognathi or as P. mandapamensis (12, 14, 16, 19, 22, 33, 41, 44). Furthermore, except for leiognathid fishes, relatively few species of host animals have been sampled for luminous bacteria. As a consequence, the host range of P. mandapamensis and the extent to which its host preferences and other aspects of its ecology may differ from those of P. leiognathi are indefinitely and incompletely known.
To begin investigating these issues, we used a multigene phylogenetic approach and examined a wide diversity of deep-water- and shallow-water-dwelling bioluminescent animals for the presence of P. mandapamensis as a light organ symbiont. Multiple bacterial strains were isolated from the light organs of each host animal to obtain a sampling of the bacterial diversity present. The strains were then screened using repetitive extragenic palindromic PCR (rep-PCR) genome profiling and sequence-based methods to identify genetically distinct strain types. Representatives of the different types from each light organ were identified phylogenetically based on sequences of the gyrB, luxAB(F)E, and ribBHA genes, the analysis of which provides unambiguous resolution between P. mandapamensis and P. leiognathi. We also reconstructed a phylogeny of the host animals found to harbor P. mandapamensis to test for cospeciation between the fish and bacterial strains, a possible indication of host-symbiont coevolution. The results reveal a host range for P. mandapamensis that is both unexpectedly wide and substantially distinct from that of P. leiognathi. They also demonstrate the effectiveness of molecular phylogenetic criteria for examining the ecological specificity of closely related, phenotypically similar bacteria.
MATERIALS AND METHODS
Collection of host specimens.
Fishes and squids bearing bacterial light organs were obtained from catches from various shallow coastal and deeper continental shelf locations in the Pacific, Atlantic, and Indian oceans. Specimens were collected at regional wholesale markets on the morning of capture, directly from trawls or trap nets, with handheld lines, and by hand using scuba. When possible, multiple specimens of each species were collected and examined. Results for host species not reported here are described by Dunlap et al. (10). In the present study, specimens of Acropoma japonicum and Gadella jordani were collected at Dahsi Fish Market on the east coast of Taiwan and at Tungkang Fish Market on the southern coast of Taiwan, respectively, from commercial catches of deepwater benthic trawls at an ∼200- to 600-m depth in the northwest portion of the Philippine Sea, east of Taiwan (Table 1). Additional specimens of A. japonicum were collected at Saga Market, Shikoku, Japan, from a commercial benthic trawl at a 100- to 300-m depth in Tosa Bay and from a commercial surface set net in Suruga Bay, Honshu, Japan, approximately 1 km from Yumachi. Specimens of Photopectoralis panayensis were taken by surface drop net 1 to 2 km from shore in the Panay Gulf (Visayan Sea), the Philippines, in the vicinity of Tigbauan, Panay (Table 1), and specimens of Photopectoralis bindus were taken by trap net from Nakagusuku Bay, Okinawa, Japan, landed at Hama Fish Market. Specimens of Siphamia versicolor were collected from their protective association with the longspine urchin Diadema setosum at a 3- to 5-m depth along coral reefs at Sesoko Island, Motobu, Okinawa, Japan (Table 1). Fishes were identified to the species level by reference to Nakabo (24), Kimura et al. (21), and Sparks et al. (38). Ichthyological nomenclature follows the work of Nelson (27) and Sparks et al. (38). Fish specimen designations follow the work of Dunlap et al. (9). Samples for fish mitochondrial DNA extraction were flank muscle tissue, which was excised free of skin and stored in 90% ethanol at −20°C. DNA was extracted from small chunks of the tissue using the QIAGEN QIAquick tissue extraction kit, according to the manufacturer's protocol. After fish specimens were sampled for muscle tissue and their light organs dissected (see below), they were tagged individually, preserved, and later deposited into the laboratory's permanent specimen collection.
TABLE 1.
Bacterially bioluminescent fishes harboring P. mandapamensis as a light organ symbiont
| Host family and species | Depth; habitata | Collection location; source | Specimen | Cosymbiont |
|---|---|---|---|---|
| Moridae | ||||
| Gadella jordani | 400-760 m; bathydemersal | Tungkang, Taiwan; Philippine Sea | Gjord.1 | |
| Acropomatidae | ||||
| Acropoma japonicum | 100-500 m; bathydemersal | Dahsi, Taiwan; Philippine Sea | Ajapo.2 | P. leiognathi |
| Saga, Shikoku, Japan; Tosa Bay | Ajapo.3 | |||
| Ajapo.4 | P. leiognathi | |||
| Ajapo.5 | P. leiognathi | |||
| Yui, Honshu, Japan; Suruga Bay | Ajapo.6 | |||
| Ajapo.7 | ||||
| Ajapo.8 | ||||
| Apogonidae | ||||
| Siphamia versicolor | 0-68 m; tropical reef | Sesoko Island, Okinawa, Japan | Svers.1 | |
| Svers.3 | ||||
| Svers.4 | ||||
| Svers.9 | ||||
| Leiognathidae | ||||
| Photopectoralis bindus | 10-110 m; coastal demersal | Hama, Okinawa, Japan; Nakagusuku Bay | Pbind.5 | P. leiognathi |
| Photopectoralis panayensis | 10-110 m; coastal demersal | Tigbauan, Panay, The Philippines; Visayan Sea | Ppana.1 | |
| Ppana.2 | P. leiognathi | |||
| Ppana.3 | P. leiognathi |
Reported depth and habitat data are from R. Froese and D. Pauly (FishBase [www.fishbase.org, version 03/2006]), interspersed with data for P. panayensis and P. bindus for Leiognathidae in general. Various leiognathid species, e.g., Leiognathus fasciatus, Leiognathus nuchalis, and Leiognathus stercorarius, however, can be caught as adult specimens at 0- to 1-m depths in coastal areas (P. V. Dunlap, personal observation), whereas P. panayensis is thought to be an atypically deep-dwelling species (J. Ledesma, personal communication).
Isolation of bacterial strains.
The bacterial strains used in this study are listed in Table 2. Bacteria newly reported here were isolated from the light organs of fishes essentially as previously described (2, 5, 7, 10). Specifically, fish were kept chilled on ice until dissection of the light organ, usually within an hour or two of collection. The ventral light organ (A. japonicum, S. versicolor, G. jordani) or circumesophageal light organ (P. panayensis, P. bindus) was aseptically dissected from the fish and homogenized in 0.5 ml or 1.0 ml of sterile artificial 70% seawater containing 25 mM HEPES buffer (pH 7.25) (BSW-70) in a sterile handheld Tenbroeck glass tissue grinder. The light organ homogenates were then serially diluted in BSW-70, and portions of one or more of the end dilutions were spread on plates of LSW-70 agar, which contained, per liter, 10 g tryptone, 5 g yeast extract, 350 ml double-strength artificial seawater (25), 650 ml deionized water, and 15 g agar. Dilutions of light organ homogenates, generally to 10−5, and plating volumes, generally 25 to 100 μl, were based on a typical light organ population size of approximately 108 cells (5). Plates were incubated for 18 to 24 h at room temperature (typically 18 to 27°C, depending on the season, location, and availability of air conditioning). This plating procedure typically gave rise to approximately 50 to 500 well-isolated luminous colonies per plate. Nonluminous colonies were not observed. Previous comparisons of counts of viable cells and direct counts of bacteria from light organs handled in this way indicated a 100% plating efficiency (5). Ten to 40 colonies (i.e., individual strains) were then picked at random from the plates for each fish specimen, purified on LSW-70 agar plates, and stored as viable cultures at −75°C in cryoprotective medium (8) in the laboratory's permanent strain collection. Bacterial strain designations report the host fish species and specimen number of this laboratory (9); e.g., ajapo.2.16 indicates strain number 16 from specimen number 2 of A. japonicum. Strain AJ-1a, isolated from the light organ of a specimen of A. japonicum collected at Kochi, Shikoku, Japan (14), was a gift of S. Fukasawa. Genomic DNA was purified from 1-ml cultures of strains grown overnight in LSW-70 broth by using the QIAGEN DNeasy tissue extraction kit and by following the manufacturer's protocol for gram-negative bacteria.
TABLE 2.
Bacterial species and strains used in this study
| Species | Strain | Ecological sourcea | Reference(s) |
|---|---|---|---|
| Photobacterium angustum | ATCC 25915T | SW | 35 |
| Photobacterium damselae subsp. damselae | ATCC 33539T | Skin ulcer, Chromis punctipinnus | 15 |
| Photobacterium iliopiscarium | ATCC 51760T | Pyloric cecum, Clupea harengus | 2, 31 |
| Photobacterium kishitanii | ahane.1.1 | LO, Acropoma hanedai, Ahane.1 | 10 |
| pjapo.1.1T | LO, Physiculus japonicus, Pjapo.1 | 2, 7 | |
| Photobacterium leiognathi | ATCC 25521T | LO, Leiognathus splendens | 3 |
| ATCC 25587 | LO, L. splendens | 3 | |
| lequu.1.1 | LO, Leiognathus equulus, Lequu.1 | 9 | |
| lleuc.1.1 | LO, Leiognathus leuciscus, Lleuc.1 | 9 | |
| ajapo.2.16 | LO, Acropoma japonicum, Ajapo.2 | This study | |
| ajapo.4.22 | LO, A. japonicum, Ajapo.4 | This study | |
| ajapo.5.37 | LO, A. japonicum, Ajapo.5 | This study | |
| lbind.5.1, lbind.5.3 | LO, Photopectoralis bindus, Pbind.5 | This study | |
| ppana.1.1-ppana.1.20 | LO, Photopectoralis panayensis, Ppana.1 | This study | |
| ppana.2.2, ppana.2.3, ppana.2.5-ppana.2.12 | LO, P. panayensis, Ppana.2 | This study | |
| ppana.3.2, ppana.3.6, ppana.3.9 | LO, P. panayensis, Ppana.3 | This study | |
| Photobacterium mandapamensis | ATCC 27561T | SW | 1, 18 |
| ATCC 33981 | SW | 1, 18 | |
| PL-721 | Skin, Coccorella sp. | 1, 26 | |
| AJ-1a | LO, A. japonicum | 14 | |
| ajapo.2.1-ajapo.2.15, ajapo.2.17-ajapo.2.20 | LO, A. japonicum, Ajapo.2 | This study | |
| ajapo.3.1, ajapo.3.7 | LO, A. japonicum, Ajapo.3 | This study | |
| ajapo.4.1, ajapo.4.5, ajapo.4.10, ajapo.4.11, ajapo.4.31, ajapo.4.40 | LO, A. japonicum, Ajapo.4 | This study | |
| ajapo.5.1, ajapo.5.7, ajapo.5.21 | LO, A. japonicum, Ajapo.5 | This study | |
| ajapo.6.1-ajapo.6.20 | LO, A. japonicum, Ajapo.6 | This study | |
| ajapo.7.1-ajapo.7.23 | LO, A. japonicum, Ajapo.7 | This study | |
| ajapo.8.1-ajapo.8.24 | LO, A. japonicum, Ajapo.8 | This study | |
| gjord.1.1, gjord.1.3, gjord.1.5 | LO, Gadella jordani, Gjord.1 | This study | |
| lbind.5.10 | LO, P. bindus, Pbind.5 | This study | |
| ppana.2.1 | LO, P. panayensis, Ppana.2 | This study | |
| ppana.3.1, ppana.3.3-ppana.3.5, ppana.3.7, ppana.3.10, ppana.3.14 | LO, P. panayensis, Ppana.3 | This study | |
| seafl.1.1, seafl.1.3, seafl.1.4 | SW | 1 | |
| svers.1.1, svers.1.2, svers.1.11 | LO, Siphamia versicolor, Svers.1 | This study | |
| svers.3.2, svers.3.7 | LO, S. versicolor, Svers.3 | This study | |
| svers.9.9 | LO, S. versicolor, Svers.9 | This study | |
| Photobacterium phosphoreum | ATCC 11040T | Skin, marine fish | 2, 18 |
| Photobacterium profundum | JCM 10084T | SW | 29 |
| Vibrio fischeri | ATCC 7744T | SW | 18 |
Abbreviations: SW, seawater; LO, light organ.
Amplification of the gyrB, luxAB(F)E, and ribBHA genes.
The gyrB, luxAB(F)E, and ribBHA genes code for DNA gyrase subunit B, the α and β subunits of luciferase, a nonfluorescent flavoprotein, a protein involved in the synthesis of aldehyde (a luciferase substrate), and three proteins involved in riboflavin synthesis, respectively. These genes were amplified by PCR, using Taq polymerase along with reagents from either the Eppendorf (Hamburg, Germany) MasterTaq kit or the Promega (Madison, WI) Taq DNA polymerase kit, essentially as described previously (1). For details of the amplification protocols and specific primers, see the supplemental material.
Strain typing and phylogenetic screening.
DNA genomic profile analysis was carried out with rep-PCR on purified genomic DNA of individual strains using primers REP1R-I (5′-IIIICGICGICATCIGGC-3′) and REP2-I (5′-ICGICTTATCIGGCCTAC-3′) (40) according to the methods of Di Meo et al. (4) and essentially as described previously (7). For sequence-based screening, PCR products generated with the luxA primers CWLAf and CWLAr (43) or CWLAforPl and CWLArevPl (see the supplemental material) or the luxA-luxB primers luxAforPlPm (5′-TACAATGARRTTGCRGCWGARCATGG-3′) and luxBrevPhoto (5′-TCRTARCANGCTTCRAATTGYSGYTGYTG-3′) were sequenced. The template for the reactions was prepared directly from cell pellets resuspended in 50 μl of sterile deionized water and repelleted to remove the supernatant. The amplification protocol was as described for the amplification of gyrB, with primer annealing at 50°C and a 1-min extension time. Provisional species identifications were based on 90% or greater identity to sequences of the luxA amplicon (approximately 650 nucleotides) or the luxA-luxB amplicon (approximately 800 nucleotides) of P. mandapamensis or P. leiognathi from GenBank.
Sequencing and phylogenetic analyses.
PCR products were sequenced by staff of the University of Michigan Sequencing Core using the respective amplification primers and dye terminator cycle sequencing on a Perkin-Elmer (Wellesley, MA) ABI 3730 DNA analyzer. The gyrB, luxAB(F)E, and ribBHA sequences were aligned manually by inferred amino acid sequence. Bacterial phylogeny was reconstructed by parsimony analysis performed with PAUP* (39), using 1,000 heuristic search replicates with tree bisection reconnection branch swapping. Jackknife support was calculated with PAUP* using 1,000 replicates (with 10 heuristic searches per replicate) and 34% deletion, emulating Jac resampling. For strains bearing an insertion in luxF (ajapo.3.1, ajapo.3.7, ppana.3.1, and ppana.3.14), the portion of luxF downstream of the insertion was excluded from the analysis. For nonluminous taxa, luxABFE sequences were treated as missing data.
To construct the fish phylogeny, the mitochondrial genes encoding 16S rRNA and cytochrome oxidase subunit I (COI) were amplified and sequenced, using the primer sequences and PCR protocols described by Sparks et al. (38). Sequence data were analyzed by direct optimization as implemented in OY (POY without parallel options) (42). The 100 OY replicates included randomizing outgroup and taxon order input (-replicates 100 -randomizeoutgroup -nooneasis), with retention of a maximum of two trees per initial build (-buildmaxtrees 2), all changes set to a cost of 1 (-change 1 -gap 1 -extensiongap 1), two iterations of ratcheting with 30% data perturbation saving one tree per iteration (-ratchettbr 2 -rachettrees 1 -ratchetpercent 30), and precise calculations on the optimization down-pass (-exact). Suboptimal trees were evaluated during each iteration using slop commands (-slop 5 -checkslop 10). Jackknife percentages were calculated in PAUP* using 1,000 branch and bound replicates.
Nucleotide sequence accession numbers.
Accession numbers for the gyrB sequences of newly sequenced P. leiognathi and P. mandapamensis strains are DQ371341 through DQ371367 and DQ790866 through DQ790884, respectively, and that for pbind.5.1 is EF372600. The luxAB(F)E sequences of newly sequenced P. leiognathi and P. mandapamensis strains have the numbers DQ371368 through DQ371394 and DQ790849 through DQ790865, respectively; EF372601 is the number for the luxABE sequence of pbind.5.1, and EF372602 is the number for luxAB of pbind.5.10. The gyrB and luxABFE sequences of “Photobacterium kishitanii” (proposed name) strain ahane.1.1 are DQ648287 and DQ648331. Sequences for the rib genes (ribB, ribH, and ribA) of Photobacterium strains are DQ988873 through DQ988881. For fishes, the accession numbers for the mitochondrial 16S gene and COI, respectively, are as follows: for Acropoma hanedai, DQ648414 and DQ648436; for Acropoma japonicum, DQ790843 and DQ790845; for Gadella jordani, DQ648427 and DQ648449; for Physiculus japonicus, DQ648431 and DQ648453, and for Siphamia versicolor, DQ790844 and DQ790846. Sequences for Photopectoralis panayensis (38) were downloaded from GenBank.
For the previously reported lux sequences in the following strains used in this study, the GenBank accession numbers are as indicated in parentheses: Vibrio fischeri ATCC 7744T (AY341062); Photobacterium phosphoreum ATCC 11040T (AY341063); P. kishitanii pjapo.1.1T (AY341065); P. leiognathi ATCC 25521T (M63594); P. leiognathi ATCC 25587 (AY456750); P. mandapamensis ATCC 27561T (AY341067); P. mandapamensis ATCC 33981 (AY341068), lequu.1.1 (AY341069), and lleuc.1.1 (AY341070); and P. mandapamensis PL-721 (AY341066), seafl.1.3 (AY456752), seafl.1.1 (AY456751), and seafl.1.4 (AY456753). For the previously reported gyrB sequences in the following strains used in this study, the GenBank accession numbers are as indicated in parentheses: V. fischeri ATCC 7744T (AY455874); Photobacterium damselae subsp. damselae ATCC 33539T (AY455889); Photobacterium angustum ATCC 25915T (AY455890); Photobacterium profundum JCM 10084T (AY455892); Photobacterium iliopiscarium ATCC 51760T (AY455878); P. phosphoreum ATCC 11040T (AY455875); P. kishitanii pjapo.1.1T (AY455877); P. leiognathi ATCC 25521T (AY455879); P. leiognathi ATCC 25587 (AY455880); P. mandapamensis ATCC 27561T (AY455883); P. mandapamensis ATCC 33981 (AY455884), lequu.1.1 (AY455881), lleuc.1.1 (AY455882), seafl.1.1 (AY455886), seafl.1.3 (AY455887), and seafl.1.4 (AY455888); and P. mandapamensis PL-721 (AY455885).
RESULTS
Symbiotic host range of P. mandapamensis.
Light organs of a diversity of deep-water- and shallow-water-dwelling bioluminescent animals were examined for the presence of P. mandapamensis and other luminous bacteria. Sequence-based screening and preliminary phylogenetic analyses (see Materials and Methods) presumptively identified P. mandapamensis as a light organ symbiont of five species of marine fishes, Acropoma japonicum, Photopectoralis panayensis, Photopectoralis bindus, Siphamia versicolor, and Gadella jordani (Tables 1 and 2). These fishes, which represent four families in two teleost orders, occur at various depths in a variety of different marine habitats (Table 1). Other animals examined were found to harbor P. kishitanii, P. leiognathi, or V. fischeri (10).
Diversity and cosymbiosis of the light organ symbionts of Acropoma japonicum.
To conclusively identify the bacterial symbionts of A. japonicum, we first examined a representative strain, AJ-1a, from an earlier study in which the bacteria were identified phenotypically as P. leiognathi (14). Analysis of the gyrB and luxAB(F)E genes identified AJ-1a as a strain of P. mandapamensis (Fig. 1). The presence of luxF, which discriminates P. mandapamensis from P. leiognathi (1), in the lux operon of AJ-1a confirmed this finding. These results correct an earlier misidentification of the bacteria that are symbiotic with A. japonicum (14).
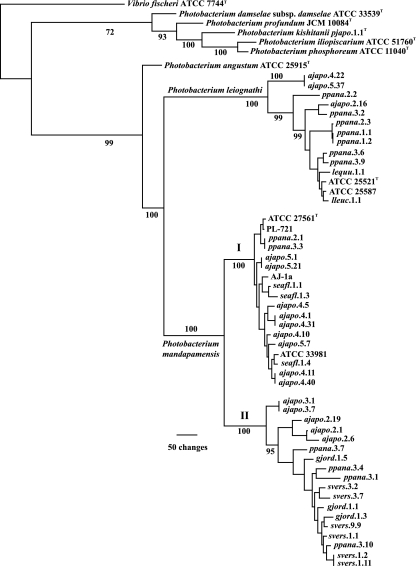
FIG. 1.
Phylogram representing 1 of 56 equally most parsimonious hypotheses resulting from combined multilocus analysis of gyrB and luxAB(F)E gene sequences of Photobacterium species (1,393 informative characters; length, 3,491; consistency index [CI], 0.578; retention index [RI], 0.879). A strict consensus of the 56 trees reduces resolution only among smaller clades near the tips of the tree. The separation of P. leiognathi and P. mandapamensis results primarily from differences in lux operon sequences, as described previously (1), and was confirmed here through analysis of ribBHA sequences (Fig. 3). Numbers at the nodes indicate Jackknife resampling values; some values were omitted for clarity. Representative strains from different specimens of S. versicolor were used here. Strains whose gyrB and luxAB(F)E sequences were identical and that were therefore excluded from this analysis are gjord.1.4, gjord.1.7, and gjord.1.9 (identical to gjord.1.1); gjord.1.8 (identical to gjord.1.3); gjord.1.10 (identical to gjord.1.5); and ppana.3.14 (identical to ppana.3.1 [see the text]). Strains with identical rep-PCR profiles and therefore not sequenced or, if sequenced, not used in this analysis due to sequence identity are gjord.1.2 (identical to gjord.1.1), gjord.1.6 (identical to gjord.1.4), ppana.1.3 through ppana.1.20 (identical to ppana.1.1 and ppana.1.2), ppana.2.5 through ppana.2.12 (identical to ppana.2.3), and ppana.3.5 (identical to ppana.3.3). For strains from specimen Ajapo.2, representatives of each strain type, ajapo.2.1, ajapo.2.6, ajapo.2.16, and ajapo.2.19, were sequenced and used in this analysis. Roman numerals I and II refer to P. mandapamensis clade I and clade II, respectively (see the text).
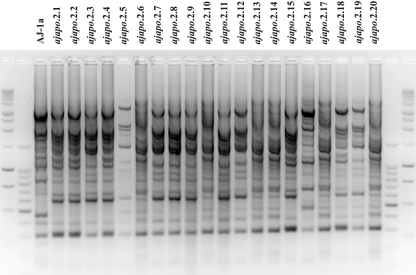
To determine whether other specimens of this fish also harbor P. mandapamensis as a symbiont, we isolated bacteria from the light organs of several recently collected specimens of A. japonicum (Table 1). The bacteria were screened by rep-PCR genomic profiling or by analysis of luxA or luxA-luxB sequences to identify genetically distinct types from each light organ, representatives of which were then examined phylogenetically (see Materials and Methods). Substantial bacterial diversity was present in light organs of specimens of A. japonicum. For example, at least three genetically distinct strain types, distinguished by rep-PCR profiling, were present among the light organ bacteria of specimen Ajapo.2, collected in Taiwan (Fig. 2). The gyrB and luxAB(F)E genes of strains representative of each strain type (ajapo.2.1, ajapo.2.6, ajapo.2.16, and ajapo.2.19) were sequenced. Strains ajapo.2.1, ajapo.2.6, and ajapo.2.19 were identified as P. mandapamensis. These strains formed a clade within P. mandapamensis, designated clade II, that is phylogenetically distinct from clade I, formed by AJ-1a and previously identified strains of this species, e.g., ATCC 27561T, ATCC 33981, and PL-721 (Fig. 1). In contrast, ajapo.2.16 was identified as P. leiognathi (Fig. 1). Analysis of the luxF region confirmed these results; the lux operons of ajapo.2.1, ajapo.2.6, and ajapo.2.19 contained luxF, whereas luxF was absent from the lux operon of ajapo.2.16. Sequence analysis of the ribBHA genes of ajapo.2.16 further confirmed this identification (Fig. 3). These results suggest that P. mandapamensis is a common light organ symbiont of A. japonicum. They also demonstrate that P. leiognathi can cooccur with P. mandapamensis in light organs of A. japonicum. This is the first example of two bacterial species cooccurring in the light organ of a fish, a situation we term cosymbiosis.
FIG. 2.
rep-PCR genomic profiling of strains from A. japonicum specimen Ajapo.2. Three distinct strain types were identified: (i) strains ajapo.2.1 to ajapo.2.5, ajapo.2.7 to ajapo.2.9, ajapo.2.11, ajapo.2.12, ajapo.2.15, ajapo.2.18, and ajapo.2.19 (strains ajapo.2.1 and ajapo.2.19 [identified as P. mandapamensis] represent the minor genetic variation among these 13 strains); (ii) ajapo.2.6, ajapo.2.10, ajapo.2.13, ajapo.2.14, ajapo.2.17, ajapo.2.20 (with identical rep-PCR profiles, represented by ajapo.2.6 [P. mandapamensis]); and (iii) ajapo.2.16 (P. leiognathi). Included for comparison is strain AJ-1a from the study of Fukasawa et al. (14), identified here as P. mandapamensis. Flanking unlabeled lanes are 1-kb and 100-bp DNA size standard ladders.
FIG. 3.
Phylogram of the single most parsimonious tree resulting from analysis of the genes ribB, ribH, and ribA (868 informative characters; length, 1,603; CI, 0.812; RI, 0.861). Sequences were aligned by inferred amino acid sequences and analyzed using the branch-and-bound algorithm as implemented in PAUP* (39). Jackknife resampling values appear at the nodes and were calculated using 1,000 branch-and-bound replicates with 34% deletion, emulating Jac resampling. Roman numerals I and II refer to P. mandapamensis clade I and clade II, respectively. Compare the figure with Fig. 1.
To investigate how commonly P. mandapamensis and P. leiognathi occur as cosymbionts of A. japonicum and to further assess the extent of phylogenetic diversity within P. mandapamensis, we examined the light organ bacteria of several additional specimens of this fish species (Table 1). The sequence of luxA or luxA-luxB was used to provisionally identify strains from each fish (see Materials and Methods). Of the 120 strains examined from three specimens of A. japonicum from Tosa Bay, Shikoku, Japan, Ajapo.3, Ajapo.4, and Ajapo.5, two strains, ajapo.4.22 and ajapo.5.37, were provisionally identified as P. leiognathi. The remaining 118 strains were identified as P. mandapamensis. All 40 strains from specimen Ajapo.3 were members of P. mandapamensis clade II, whereas all 78 P. mandapamensis strains from specimens Ajapo.4 and Ajapo.5 were members of clade I. Of the 67 strains examined from three specimens of A. japonicum from Suruga Bay, Honshu, Japan, Ajapo.6, Ajapo.7, and Ajapo.8, all were identified as P. mandapamensis. All 20 strains from Ajapo.6 were members of clade II, and all 23 strains from Ajapo.7 were members of clade I. Of the 24 strains from specimen Ajapo.8, 5 were members of clade I and 19 were members of clade II. The provisional identifications were confirmed by sequence analysis of the gyrB and luxAB(F)E genes for 13 representative strains from specimens Ajapo.3, Ajapo.4, and Ajapo.5 (Fig. 1). Furthermore, analysis of the ribBHA genes of ajapo.3.1 and ajapo.4.11 confirmed the placement of these strains in clade II and clade I, respectively, of P. mandapamensis (Fig. 3). We note parenthetically here that the luxF genes of ajapo.3.1 and ajapo.3.7 contained insertions that apparently inactivate this gene (see the supplemental material), the first demonstration of nonsense mutations in the lux genes of bacteria from nature.
These results reveal the presence of substantial phylogenetic diversity among the symbionts of A. japonicum, with strains representing three phylogenetically distinct clades, P. mandapamensis clade I and clade II and P. leiognathi. Furthermore, they demonstrate that cosymbiosis of P. mandapamensis and P. leiognathi, while not frequently observed, might not be rare.
Diversity and cosymbiosis of the light organ symbionts of Photopectoralis panayensis and Photopectoralis bindus.
Cosymbiosis of P. mandapamensis and P. leiognathi was also found in the light organs of two leiognathid fishes, P. panayensis and P. bindus. rep-PCR analysis identified the presence of a single strain type among the 20 strains of specimen Ppana.1, three types among the 11 strains of Ppana.2, eight among the 10 strains of Ppana.3, and three among the 10 strains of Pbind.5 (data not shown). The gyrB and luxAB(F)E genes of representatives of these strain types were sequenced for identification. Strains identified as P. leiognathi were ppana.1.1 and ppana.1.2; ppana.2.2 and ppana.2.3; ppana.3.2, ppana.3.6, and ppana.3.9; and, pbind.5.1 and pbind.5.3. Strains identified as P. mandapamensis were ppana.2.1 (clade I); ppana.3.1 (clade II), ppana.3.3 (clade I), ppana.3.4 (clade II), ppana.3.5 (clade I), ppana.3.7 (clade II), ppana.3.10 (clade II), and ppana.3.14 (clade II); and pbind.5.10 (clade II) (Fig. 1). We note parenthetically here a rare instance of different fish specimens harboring the same strain type (7, 9); strain ppana.2.3 was identical in rep-PCR profile and in gyrB and luxABE sequence to ppana.1.1 and other strains from specimen Ppana.1 (Fig. 1). Furthermore, strains ppana.3.1 and ppana.3.14 were found to bear a transposase-containing insertion in luxF that inactivates the gene (see the supplemental material).
The identification of P. mandapamensis as a light organ symbiont of P. panayensis and P. bindus, 9 of 43 strains examined at the sequence level, provides the first demonstration that P. leiognathi is not the exclusive bioluminescent symbiont of leiognathid fishes. Furthermore, these results confirm and extend to two additional fish species the finding that P. mandapamensis and P. leiognathi occur as cosymbionts.
Phylogenetic clustering of the light organ symbionts of Siphamia versicolor.
In contrast to the diversity described above, a high degree of phylogenetic clustering was found for the light organ symbionts of S. versicolor. All of the 92 bacterial strains isolated from light organs of the four examined specimens of this fish were presumptively identified by luxA or luxA-luxB sequence screening as P. mandapamensis. Phylogenetic analysis based on sequences of the gyrB and luxAB(F)E genes of six representative strains unambiguously identified these strains as P. mandapamensis, and all were members of clade II (Fig. 1). The presence of luxF in the lux operons of these strains confirmed this identification, as did sequence analysis of the ribBHA genes of svers.1.2 (Fig. 3).
Identification of the light organ bacteria of Gadella jordani.
Phylogenetic clustering was found also for the light organ bacteria of G. jordani. Screening by rep-PCR of 10 representatives of the light organ population of specimen Gjord.1, the single specimen obtained, revealed the presence of four different strain types (data not shown). Sequence analysis of gyrB and luxAB(F)E and the presence of luxF identified the eight examined strains as P. mandapamensis, and all were members of clade II (Fig. 1). Sequence analysis of the ribBHA genes of gjord.1.3 confirmed this identification (Fig. 3).
Test of symbiont-host cospeciation between P. mandapamensis and luminous fishes.
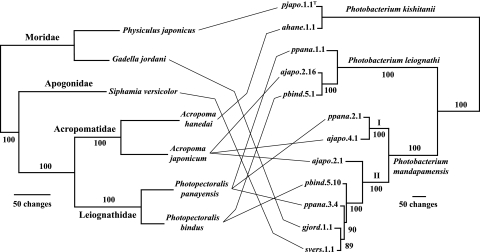
The identification of five fish species that harbor P. mandapamensis as a light organ symbiont and the ability to discriminate phylogenetically between this bacterium and other luminous bacteria allowed us to test the hypothesis that P. mandapamensis and its hosts have cospeciated. Cospeciation, visualized as a pattern of matching host and symbiont phylogenies, can reflect host-symbiont coevolution (32). Therefore, to test for cospeciation and ask whether coevolution might explain the host preferences of P. mandapamensis, we used mitochondrial 16S rRNA and COI gene sequences to reconstruct a phylogeny of the fishes harboring P. mandapamensis and compared it with a phylogeny of representative bacterial strains from these fishes.
The relationships among the fishes were unambiguously resolved, and a robust clade structure was evident both for the fishes and for the bacteria (Fig. 4). However, the clade structure of the bacteria did not topologically match that of the fish. Specifically (i) the distantly related fishes A. japonicum (Perciformes: Acropomatidae) and G. jordani (Gadiformes: Moridae) harbor the same species of bacteria as light organ symbionts; (ii) closely related fishes, i.e., in the same genus or family, namely, A. japonicum and A. hanedai and G. jordani and P. japonicus, harbor different bacterial species; and (iii) specimens of some fishes, A. japonicum, P. panayensis, and P. bindus, harbor members of two or three different bacterial clades. The absence of topological congruence between host and symbiont phylogenies refutes the hypothesis that P. mandapamensis and its host fishes have cospeciated. We conclude that factors other than symbiont-host coevolution account for the host preferences of P. mandapamensis.
FIG. 4.
Test of cospeciation between symbiotic luminous bacteria and their fish hosts. Fish specimens from seven species in four families from two teleost orders and bacterial strains representative of the symbionts of these fishes (Fig. 1) were tested for phylogenetic congruence. Genes analyzed were mitochondrial 16S rRNA and COI genes for the fish and gyrB and luxAB(F)E for the bacteria. The fish data were analyzed with OY (see Materials and Methods for settings), which resulted in a single most parsimonious hypothesis (330 informative characters; length, 829; CI, 0.813; RI, 0.678). The data for bacteria (1,407 informative characters) were analyzed by an exhaustive search in PAUP in which all possible tree topologies were evaluated; the single most parsimonious hypothesis (length, 2,001; CI, 0.870; RI, 0.911) is shown. Strains of P. kishitanii were isolated from the morid fish Physiculus japonicus (7) and the acropomatid fish Acropoma hanedai (10). Numbers at major nodes are jackknife support values (see Materials and Methods), and Roman numerals I and II refer to P. mandapamensis clades I and II, respectively.
DISCUSSION
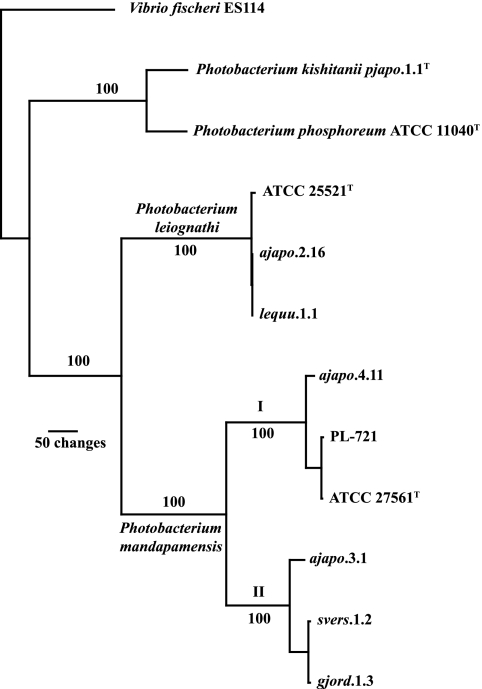
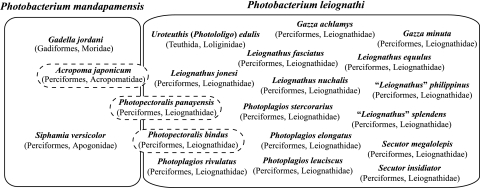
Despite their phenotypic similarity and close evolutionary relationship, P. mandapamensis and P. leiognathi can be distinguished by molecular phylogenetic criteria. Using these criteria, we demonstrate here that their symbiotic host species ranges, a major aspect of the ecology of these bacteria, are substantially distinct (Fig. 5). To date, five fishes, Acropoma japonicum (Perciformes: Acropomatidae), Photopectoralis panayensis and Photopectoralis bindus (Perciformes: Leiognathidae), Siphamia versicolor (Perciformes: Apogonidae), and Gadella jordani (Gadiformes: Moridae) harbor P. mandapamensis as a light organ symbiont. In specimens of two of these fishes, S. versicolor and G. jordani, P. mandapamensis appears to be the sole bacterial species present, whereas in light organs of the other fishes, P. mandapamensis occurs together with P. leiognathi as a cosymbiont. Molecular phylogenetic analysis also revealed the presence of two distinct clades of P. mandapamensis, the possible significance of which for bioluminescent symbioses or other aspects of the ecology of this bacterium is not yet obvious. In contrast, P. leiognathi occurs as the apparently exclusive light organ symbiont of most leiognathid fishes and the loliginid squid Uroteuthis (Photololigo) edulis (Fig. 5) (13).
FIG. 5.
Host species ranges of the light organ symbionts P. mandapamensis and P. leiognathi. Species of fish that harbor P. mandapamensis and species of fish and squid that harbor P. leiognathi are shown with host family and order in parentheses. Hosts reported are those for which detailed analysis of bacteria has been carried out using methods that discriminate between P. mandapamensis and P. leiognathi. Most leiognathids harbor apparently only P. leiognathi, whereas all examined specimens of S. versicolor and the one available specimen of G. jordani harbored only P. mandapamensis. In light organs of some fishes (A. japonicum, P. panayensis, and P. bindus), P. mandapamensis and P. leiognathi occur as cosymbionts. Nonetheless, the host ranges of these two bacterial species are substantially distinct.
Bacterial symbionts of G. jordani and P. panayensis previously had not been examined, and for P. bindus only a single bacterial strain from the light organ of a single specimen had been identified (9). Furthermore, for the symbionts of A. japonicum and S. versicolor, only phenotypic and limited sequence analysis had been carried out (14, 16, 19, 22, 33, 41, 44), leaving the identities of these bacteria in substantial doubt. In contrast, the extensive sampling and multigene phylogenetic analysis described here establishes P. mandapamensis as a light organ symbiont of these fishes, either as the apparently exclusive symbiont or as a cosymbiont with P. leiognathi. Therefore, five species of luminous bacteria, P. kishitanii, P. leiognathi, P. mandapamensis, V. fischeri, and Vibrio logei, have been documented to date as bioluminescent symbionts of marine animals (Fig. 1) (8, 10).
The difference in the host ranges of P. mandapamensis and P. leiognathi indicate that these bacteria might differ in other ways as well, such as in their environmental distributions or physiologies. Such differences presumably would relate in some way to the hosts that they preferentially colonize. Phenotypically, in their growth and luminescence responses to salinity and the color of light produced, these bacteria also differ (1). These ecological and phenotypic differences are underscored at the phylogenetic level by the robust and unambiguous divergence of the sequences of their lux and rib genes, as well as by the presence of luxF in all tested strains of P. mandapamensis and its absence in all tested strains of P. leiognathi. Despite the large number of strains examined here and previously (1, 9), no phylogenetically intermediate strains have been found. Together, these lines of evidence indicate that P. mandapamensis and P. leiognathi, despite the many phenotypic and genotypic traits they share (34), are biologically distinct. The ecological and phylogenetic differences described here appear consistent with their original descriptions as separate species (3, 18).
The presence of two bacterial species within an animal's light organ, termed here cosymbiosis, was first reported for sepiolid squids, certain specimens of which harbor both V. fischeri and V. logei in their light organs (11, 28). Cosymbiosis indicates that symbiotic associations between luminous bacteria and their fish and squid hosts are more accommodating of bacterial diversity than previously thought (8, 17). A strict host family-bacterial species specificity therefore does not characterize bioluminescent symbioses (10). Nonetheless, these associations do exhibit a high degree of symbiont specificity, with different bacterial species exhibiting substantially different symbiotic host ranges.
The use here and previously of methods to screen for genetic diversity in populations of light organ bacteria, rep-PCR genomic profiling and sequence analysis of the luxA gene or luxA-luxB genes, has led to the identification of unexpected intraspecies genetic diversity within light organ populations (Fig. 2; 7, 9) and to the finding that multiple bacterial species occur cosymbiotically in fish light organs. This diversity can easily be missed when only single strains are analyzed from a host specimen (e.g., see references 9 and 41). However, it is important to note that no obvious correlation was found between rep-PCR DNA fingerprints and luxAB(F)E sequences. Strains with generally similar DNA fingerprints, such as AJ-1a and ajapo.2.16 (Fig. 2), were found by sequence analysis to be different species, and strains with very different fingerprints often were found here to be the same species. The exceptions to this lack of correlation are those strains with the same DNA fingerprint, which we consistently find have identical or very nearly identical sequences for multiple genetic loci. Therefore, DNA fingerprint analysis, while highly effective for typing strains and for grouping them as genetically identical or distinct in ecological surveys of bacterial populations and communities, does not appear to reliably distinguish between or group strains phylogenetically.
The identification of nonsense mutations in luxF as small insertions or the presence of a putative transposase gene (see Fig. S1 in the supplemental material) in some strains of P. mandapamensis is the first example of nonsense mutations in the lux operons of luminous bacteria from nature. The mutations indicate that the LuxF protein is not necessary for the survival or reproduction of P. mandapamensis or for its ability to form bioluminescent symbioses. The mutations provide a natural test of the functional role of LuxF in luminescence. LuxF, a nonfluorescent flavoprotein (FP390), exhibits substantial amino acid sequence similarity to the α and β subunits of luciferase, especially to the β subunit, and it is coexpressed with LuxA, LuxB, and other lux operon proteins, suggesting that LuxF plays a role in light emission (20, 23, 30, 37). We found that strains carrying nonsense mutations in luxF are luminous, but less strongly so than other strains, and that the lower production of light probably is not due to a limitation for aldehyde, a substrate for the luminescence reaction (see the supplemental material). It therefore appears that while luxF is not required for luminescence in P. mandapamensis, it might contribute in some way to luminescence intensity.
The diversity of bacteria within light organs of A. japonicum, P. panayensis, and P. bindus has implications for the ecological interactions of these host animals with their luminous symbionts. First, it indicates that strains of both clades of P. mandapamensis and of P. leiognathi were likely to be present together in the environments where light organs of aposymbiotic juveniles of these fishes became colonized. This interpretation is based on the assumption that colonization occurs only during a short period early in the fish's development; however, later secondary colonization, as the fish matures, may be a possibility and could account for the observed diversity. The ability to discriminate between these bacteria using molecular phylogenetic criteria provides a means now to begin defining their specific ecological distributions and relative numbers in other habitats, such as seawater. Second, this diversity demonstrates that these fishes do not strictly discriminate among closely related but phylogenetically distinct bacteria. It is possible, therefore, that genetic and physiological differences between P. mandapamensis and P. leiognathi are unimportant for symbiosis; alternatively, bacterial attributes important for symbiosis might be shared by these bacteria. The predominance of P. mandapamensis in light organs of A. japonicum and of P. leiognathi in light organs of P. panayensis and P. bindus, however, introduces the possibility of competitive interactions that, over the life of the host animal, could result in changes in the presence and in numbers of one bacterial species over the other within individual light organs. The apparent lack of bacterial diversity in light organs of S. versicolor and G. jordani presents a potentially interesting contrast to the situation found for the other fishes, but it may simply be a consequence of limited sampling.
The identification of five fish species that harbor P. mandapamensis as a light organ symbiont and the ability to discriminate phylogenetically between this bacterium and other luminous bacteria allowed us test the hypothesis that P. mandapamensis and its hosts have cospeciated. The topological congruence expected for fish and bacterial phylogenies of symbiotic partners that have cospeciated, however, was not observed (Fig. 4). The results clearly contradict the suggestion, based on limited data, that the symbionts of certain of these fishes have diverged in a host-dependent manner (41). Instead of identifying a matchup between host and symbiont phylogenies, this analysis indicates that P. mandapamensis may be more of a generalist as a light organ symbiont, as reflected by the phylogenetic diversity of its host fishes. In colonizing host species in four families of two teleost orders, P. mandapamensis apparently is second only to P. kishitanii among light organ symbiotic bacteria in the breadth of its host species range (10). Furthermore, because coevolution, a process of reciprocal genetic adaptation between host and symbiont, is difficult to envision in the absence of cospeciation (32), the results presented here are consistent with the view that coevolution apparently is not the basis for this bacterium's host preferences. The factors that allow P. mandapamensis to affiliate with phylogenetically divergent hosts that inhabit widely different marine habitats are not known at this time. An intriguing possibility, however, is that of host selection for luminous bacteria (36) together with an overlap in the ecological distributions of P. mandapamensis and the host animals whose aposymbiotic juveniles this bacterium colonizes (2, 10).
Supplementary Material
Acknowledgments
We thank S. Fukasawa for the gift of strain AJ-1a. Assistance in collecting fishes was provided in Taiwan by T. Yoshino and H. Imai (University of the Ryukyus) and Y. Iwatsuki (University of Miyazaki); in Okinawa, Japan, by S. Nakamura and Y. Kojima (Tropical Biosphere Research Center) and T. Yoshino and H. Ishimori (University of the Ryukyus); in Shikoku, Japan, by Y. Machida (Kochi University); in Honshu, Japan, by A. Fukui, H. Ichikawa, and H. Kuge (Tokai University); and in the Philippines by C. Lavilla-Pitogo (SEAFDEC) and J. Ledesma (Tigbauan). We thank J. Paxton and J. Leis for helpful advice on Siphamia and T.-I. Chen and J.-H. Cheng for accommodations at the Tungkang Marine Laboratory, Taiwan Fisheries Research Institute. DNA sequencing was carried out by staff of the University of Michigan Sequencing Core.
This study is a contribution from Sesoko Station, Tropical Biosphere Research Center, University of the Ryukyus.
This work was supported by grant DEB 0413441 from the National Science Foundation.
Footnotes
Published ahead of print on 16 March 2007.
Supplemental material for this article may be found at http://aem.asm.org/.
REFERENCES
- 1.Ast, J. C., and P. V. Dunlap. 2004. Phylogenetic analysis of the lux operon distinguishes two evolutionarily distinct clades of Photobacterium leiognathi. Arch. Microbiol. 181:352-361. [DOI] [PubMed] [Google Scholar]
- 2.Ast, J. C., and P. V. Dunlap. 2005. Phylogenetic resolution and habitat specificity of the Photobacterium phosphoreum species group. Environ. Microbiol. 7:1641-1654. [DOI] [PubMed] [Google Scholar]
- 3.Boisvert, H., R. Chatelain, and J. M. Bassot. 1967. Étude d'un Photobacterium isolé de l'organe lumineux des poissons Leiognathidae. Ann. Inst. Pasteur (Paris) 112:520-524. [PubMed] [Google Scholar]
- 4.Di Meo, C. A., A. E. Wilbur, W. E. Holben, R. A. Feldman, R. C. Vrijenhoek, and S. C. Cary. 2000. Genetic variation among endosymbionts of widely distributed vestimentiferan tubeworms. Appl. Environ. Microbiol. 66:651-658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Dunlap, P. V. 1984. Physiological and morphological state of the symbiotic bacteria from light organs of ponyfish. Biol. Bull. 167:410-425. [DOI] [PubMed] [Google Scholar]
- 6.Dunlap, P. V. 1985. Osmotic control of luminescence and growth in Photobacterium leiognathi from ponyfish light organs. Arch. Microbiol. 141:44-50. [DOI] [PubMed] [Google Scholar]
- 7.Dunlap, P. V., and J. C. Ast. 2005. Genomic and phylogenetic characterization of the luminous bacteria symbiotic with the deep-sea fish Chlorophthalmus albatrossis (Aulopiformes: Chlorophthalmidae). Appl. Environ. Microbiol. 71:930-9399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Dunlap, P. V., and K. Kita-Tsukamoto. 2006. Luminous bacteria, p. 863-892. In M. Dworkin, S. Falkow, E. Rosenberg, K.-H. Schleifer, and E. Stackebrandt (ed.), The prokaryotes, a handbook on the biology of bacteria, 3rd ed., vol. 2. Ecophysiology and biochemistry. Springer, New York, NY. [Google Scholar]
- 9.Dunlap, P. V., A. Jiemjit, J. C. Ast, M. M. Pearce, R. R. Marques, and C. R. Lavilla-Pitogo. 2004. Genomic polymorphism in symbiotic populations of Photobacterium leiognathi. Environ. Microbiol. 6:145-158. [DOI] [PubMed] [Google Scholar]
- 10.Dunlap, P. V., J. C. Ast, S. Kimura, A. Fukui, T. Yoshino, and H. Endo. Phylogenetic analysis of host-symbiont specificity and codivergence in bioluminescent symbioses. Cladistics, in press.
- 11.Fidopiastis, P. M., S. von Boletzky, and E. G. Ruby. 1998. A new niche for Vibrio logei, the predominant light organ symbiont of squids in the genus Sepiola. J. Bacteriol. 180:59-64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Fitzgerald, J. M. 1979. Studies on the taxonomy and bioluminescence of some luminous marine bacteria. Ph.D. thesis. Monash University, Victoria, Australia.
- 13.Fukasawa, S., and P. V. Dunlap. 1986. Identification of luminous bacteria isolated from the light organs of the squid, Doryteuthis kensaki. Agric. Biol. Chem. 50:1645-1646. [Google Scholar]
- 14.Fukasawa, S., T. Suda, and S. Kubota. 1988. Identification of luminous bacteria isolated from the light organ of the fish, Acropoma japonicum. Agric. Biol. Chem. 52:285-286. [Google Scholar]
- 15.Gauthier, G., B. Lafay, R. Ruimy, V. Breittmayer, J. L. Nicolas, M. Gauthier, and R. Christen. 1995. Small-subunit rRNA sequences and whole DNA relatedness concur for the reassignment of Pasteurella piscicida (Snieszko et al.) Janssen and Surgalla to the genus Photobacterium as Photobacterium damsela subsp. piscicida. Int. J. Syst. Bacteriol. 45:139-144. [DOI] [PubMed] [Google Scholar]
- 16.Haneda, Y. 1966. Luminous apogonid fish from the Moreton Bay, Brisbane. Sci. Rep. Yokosuka City Mus. 12:1-3. [Google Scholar]
- 17.Hastings, J. W., and K. H. Nealson. 1981. The symbiotic luminous bacteria, p. 1332-1345. In M. P. Starr, H. Stolp, H. G. Trüper, A. Balows, and H. G. Schlegel (ed.), The prokaryotes: a handbook on habitats, isolation, and identification of bacteria. Springer-Verlag, Berlin, Germany.
- 18.Hendrie, M. S., W. Hodgkiss, and J. M. Shewan. 1970. The identification, taxonomy and classification of luminous bacteria. J. Gen. Microbiol. 64:151-169. [Google Scholar]
- 19.Herring, P. J., and J. G. Morin. 1978. Bioluminescence in fishes, p. 273-329. In P. J. Herring (ed.), Bioluminescence in action. Academic Press, London, England.
- 20.Karatani, H., T. Konaka, and C. Katsukawa. 2000. Properties of the bimodal fluorescent protein produced by Photobacterium phosphoreum. Photochem. Photobiol. 71:230-236. [DOI] [PubMed] [Google Scholar]
- 21.Kimura, S., P. V. Dunlap, T. Peristiwady, and C. R. Lavilla-Pitogo. 2003. The Leiognathus aureus complex (Perciformes: Leiognathidae) with the description of a new species. Ichthyol. Res. 50:221-232. [Google Scholar]
- 22.Leis, J. M., and S. Bullock. 1986. The luminous cardinalfish Siphamia (Pisces, Apogonidae): development of larvae and the luminous organ, p. 703-714. In T. Uyeno, R. Arai, T. Taniuchi, and K. Matsuura (ed.), Indo-Pacific fish biology: proceedings of the second international conference on Indo-Pacific fishes. Ichthyological Society of Japan, Tokyo, Japan.
- 23.Meighen, E. A., and P. V. Dunlap. 1993. Physiological, biochemical and genetic control of bacterial bioluminescence. Adv. Microbial. Physiol. 34:1-67. [DOI] [PubMed] [Google Scholar]
- 24.Nakabo, T. (ed.). 2002. Fishes of Japan with pictorial keys to the species, vol. 1 and 2 (English ed.). Tokai University Press, Tokyo, Japan.
- 25.Nealson, K. H. 1978. Isolation, identification and manipulation of luminous bacteria. Methods Enzymol. 57:153-166. [Google Scholar]
- 26.Nealson, K. H., and J. W. Hastings. 1977. Low oxygen is optimal for luciferase synthesis in some bacteria. Arch. Microbiol. 112:9-16. [DOI] [PubMed] [Google Scholar]
- 27.Nelson, J. S. 2006. Fishes of the world, 4th ed. John Wiley & Sons, New York, NY.
- 28.Nishiguchi, M. K. 2000. Temperature affects species distribution in symbiotic populations of Vibrio spp. Appl. Environ. Microbiol. 66:3550-3555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Nogi, Y., N. Masui, and C. Kato. 1998. Photobacterium profundum sp. nov., a new, moderately barophilic bacterial species isolated from a deep-sea sediment. Extremophiles 2:1-7. [DOI] [PubMed] [Google Scholar]
- 30.O'Kane, D. J., and D. C. Prasher. 1992. Evolutionary origins of bacterial bioluminescence. Mol. Microbiol. 6:443-449. [DOI] [PubMed] [Google Scholar]
- 31.Onarheim, A. M., R. Wiik, J. Burghardt, and E. Stackebrandt. 1994. Characterization and identification of two Vibrio species indigenous to the intestine of fish in cold sea water; description of Vibrio iliopiscarius sp. nov. Syst. Appl. Microbiol. 17:370-379. [Google Scholar]
- 32.Page, R. D. M. 2003. Introduction, p. 1-21. In R. D. M. Page (ed.), Tangled trees: phylogeny, cospeciation and coevolution. University of Chicago Press, Chicago, IL.
- 33.Reichelt, J. L., and P. Baumann. 1973. Taxonomy of the marine, luminous bacteria. Arch. Mikrobiol. 94:283-330. [DOI] [PubMed] [Google Scholar]
- 34.Reichelt, J. L., and P. Baumann. 1975. Photobacterium leiognathi subsp. mandapamensis Hendrie et al., a later subjective synonym of Photobacterium leiognathi Boisvert et al. Int. J. Syst. Bacteriol. 25:208-209. [Google Scholar]
- 35.Reichelt, J. L., P. Baumann, and L. Baumann. 1976. Study of genetic relationships among marine species of the genera Beneckea and Photobacterium by means of in vitro DNA/DNA hybridization. Arch. Microbiol. 110:101-120. [DOI] [PubMed] [Google Scholar]
- 36.Reichelt, J. L., K. Nealson, and J. W. Hastings. 1977. The specificity of symbiosis: pony fish and luminescent bacteria. Arch. Microbiol. 112:157-161. [Google Scholar]
- 37.Soly, R. R., J. A. Mancini, S. R. Ferri, M. Boylan, and E. A. Meighen. 1988. A new lux gene in bioluminescent bacteria codes for a protein homologous to the bacterial luciferase subunits. Biochem. Biophys. Res. Commun. 155:351-358. [DOI] [PubMed] [Google Scholar]
- 38.Sparks, J. S., P. V. Dunlap, and W. L. Smith. 2005. Evolution and diversification of a sexually dimorphic luminescent system in ponyfishes (Teleostei: Leiognathidae), including diagnoses for two new genera. Cladistics 21:305-327. [DOI] [PubMed] [Google Scholar]
- 39.Swofford, D. L. 2003. PAUP*: phylogenetic analysis using parsimony (*and other methods), version 4.0 B10. Sinauer Associates, Sunderland, MA.
- 40.Versalovic, J., J. Schneider, F. J. de Bruijn, and J. R. Lupski. 1994. Genomic fingerprinting of bacteria using repetitive sequence-based polymerase chain reaction. Methods Mol. Cell. Biol. 5:25-40. [Google Scholar]
- 41.Wada, M., A. Kamiya, N. Uchiyama, et al. 2006. luxA gene of light organ symbionts of the bioluminescent fish Acropoma japonicum (Acropomatidae) and Siphamia versicolor (Apogonidae) forms a lineage closely related to that of Photobacterium leiognathi ssp. mandapamensis. FEMS Microbiol. Lett. 260:186-192. [DOI] [PubMed] [Google Scholar]
- 42.Wheeler, W., D. Gladstein, and J. De Laet. 2003. POY, phylogeny reconstruction via optimization of DNA and other data, version 3.0.11. American Museum of Natural History, New York, NY. ftp://ftp.amnh.org/pub/people/wheeler/poy/version3-current/poy.3.0.11.pdf.
- 43.Wimpee, C. F., T.-L. Nadeau, and K. H. Nealson. 1991. Development of species-specific hybridization probes for marine luminous bacteria using in vitro DNA amplification. Appl. Environ. Microbiol. 57:1319-1324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Yoshiba, S., and Y. Haneda. 1967. Bacteriological study on the symbiotic luminous bacteria cultivated from the luminous organ of the apogonid fish, Siphamia versicolor and the Australian pine cone fish, Cleidopus gloria-maris. Sci. Rep. Yokosuka City Mus. 13:82-84. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.