Abstract
Dihydroorotate dehydrogenase (DHODH; EC 1.3.99.11) is a central enzyme of pyrimidine biosynthesis and catalyzes the oxidation of dihydroorotate to orotate. DHODH is an important target for antiparasitic and cytostatic drugs since rapid cell proliferation often depends on the de novo synthesis of pyrimidine nucleotides. We have cloned the pyr4 gene encoding mitochondrial DHODH from the basidiomycetous plant pathogen Ustilago maydis. We were able to show that pyr4 contains a functional mitochondrial targeting signal. The deletion of pyr4 resulted in uracil auxotrophy, enhanced sensitivity to UV irradiation, and a loss of pathogenicity on corn plants. The biochemical characterization of purified U. maydis DHODH overproduced in Escherichia coli revealed that the U. maydis enzyme uses quinone electron acceptor Q6 and is resistant to several commonly used DHODH inhibitors. Here we show that the expression of the human DHODH gene fused to the U. maydis mitochondrial targeting signal is able to complement the auxotrophic phenotype of pyr4 mutants. While U. maydis wild-type cells were resistant to the DHODH inhibitor brequinar, strains expressing the human DHODH gene became sensitive to this cytostatic drug. Such engineered U. maydis strains can be used in sensitive in vivo assays for the development of novel drugs specifically targeted at either human or fungal DHODH.
Pyrimidine de novo biosynthesis is an important biosynthetic pathway that is highly conserved among prokaryotic and eukaryotic organisms. The de novo pathway consists of six enzymes, which are encoded by single genes or are parts of larger multifunctional proteins (28). Dihydroorotate dehydrogenase (DHODH; EC 1.3.99.11) is the fourth enzyme of this pathway and catalyzes the conversion of dihydroorotate (DHO) to orotate. Although the enzymatic function of DHODH is conserved in all organisms, there is an interesting difference between the enzymes of prokaryotic and eukaryotic origins (43, 45). In most eukaryotes, DHODH is located at the inner mitochondrial membrane, facing the intermembrane space (48). For its activity, mitochondrial DHODH depends on a functional respiratory chain and requires ubiquinone as a direct electron acceptor (38). Strictly aerobic prokaryotes contain membrane-bound DHODH enzymes that resemble the mitochondrial enzymes of eukaryotes (27). In obligate or facultative anaerobic bacteria, a cytosolic form of DHODH (53), which uses fumarate or NAD as an electron acceptor, has been identified. Interestingly, the ascomycetous yeast Saccharomyces cerevisiae also contains a cytosolic fumarate-reducing DHODH, the product of the URA1 gene (43). The gene encoding this enzyme has most probably been recruited by horizontal gene transfer from a prokaryotic organism (19, 35). This unique feature allows for the anaerobic growth of Saccharomyces cerevisiae, since the DHODH-catalyzed oxidation of dihydroorotate in other species is the only step of all anabolic pathways of nucleotide and amino acid biosynthesis that depends strictly on the presence of oxygen (19, 22). Also, in the intracellular parasite Trypanosoma cruzi, which causes Chagas’ disease, a related fumarate-reducing cytosolic DHODH was identified, indicating a similar evolutionary pressure toward anaerobiosis (52). Phylogenetic analysis suggested that this horizontal gene transfer occurred when the kinetoplastid branch was separated from the Apicomplexa during evolution (3).
All enzymes of the pyrimidine de novo biosynthesis pathway have been considered as promising targets for the development of antiproliferative and immunosuppressive drugs (for a review, see reference 13). In multicellular organisms, the uptake of pyrimidine nucleosides and the generation of the corresponding nucleotides by the salvage pathway fulfill the pyrimidine requirement under normal circumstances (37). Rapidly growing cells and many parasites, however, depend on the de novo synthesis of pyrimidines for the efficient synthesis of nucleic acid precursors (16). Inhibitors of the mitochondrion-bound DHODH have been successfully tested as antiproliferative agents that interfere with neoplastic growth (12, 36) and suppress immunological reactions (20, 24). In addition, DHODH inhibitors have been considered as potential therapeutic agents for a wide range of parasitic infections (23, 26). The mutation of the DHODH gene and inhibition by RNA interference demonstrated that DHODH is required for the virulence of Toxoplasma gondii (18) and the malaria parasite Plasmodium falciparum (40). Therefore, malarial DHODH was used to search for selective inhibitors by high-throughput screening using a simple biochemical in vitro assay (5).
Here, we report the molecular characterization of the pyr4 gene encoding DHODH in the phytopathogenic basidiomycete Ustilago maydis. U. maydis is a dimorphic fungus which in its haploid form grows vegetatively by budding and is nonpathogenic. U. maydis is very amenable to genetic analysis and serves as a valuable model organism to study fungal development and pathogenicity (for reviews, see references 7, 10, and 17). The complete genome sequence of U. maydis has recently been determined (30) and is publicly available (http://www.broad.mit.edu/annotation/genome/ustilago_maydis/Home.html). Here we show that U. maydis DHODH contains a functional mitochondrial targeting signal and thus belongs to family 2 of the eukaryotic DHODHs (27). The deletion of pyr4 resulted in a loss of pathogenicity and pyrimidine auxotrophy, which could be relieved by the functional expression of human DHODH carrying the U. maydis pyr4-encoded mitochondrial targeting sequence. A biochemical analysis revealed that U. maydis DHODH is insensitive to commonly used DHODH inhibitors. Strains expressing human DHODH were rendered sensitive to the cytostatic drug brequinar. This property makes U. maydis a valuable in vivo assay system to validate potential DHODH inhibitors targeted specifically at either human or fungal DHODH.
MATERIALS AND METHODS
Materials.
Unless otherwise stated, all chemicals were obtained from Roche Diagnostics, Serva, Merck, or Sigma at the purest grade available. The following inhibitors were used: 2-hydroxyethylidene-cyanoacetic acid 4-trifluoromethyl anilide, A77-1726 (Aventis), anthranilic acid (Fluka), trans-2-[4-(chlorophenyl)cyclohexyl]-3-hydroxy-1,4-naphthoquinone, atovaquone, 566C80 (Wellcome Foundation), 6-fluoro-2-(2′-fluoro-1,1′-biphenyl-4yl)-3-methyl-4-quinoline carboxylic acid, brequinar sodium (NSC 368390; DuPont Pharma GmbH), coumarin (Sigma), 3,4-dihydroxybenzoic acid (3,4-DHB), 3,5-DHB, 5-fluorouracil, 5-fluoroorotate, flucytosine, carboxin, (2,2′-[3,3′-dimethoxy[1,1′-biphenyl]-4,4′-diyl]diimino)bis-benzoic acid (Redoxal; NSC-73735), dichloroallyl lawsone, lawsone, N-phenylanthranilic acid (Fluka), salicylhydroxamic acid, and 2-methyl-1,4-naphthoquinone (menadione, or vitamin K3).
Strains and growth conditions.
For all DNA manipulations, the Escherichia coli K-12 derivative DH5α (Bethesda Research Laboratories) was used. U. maydis strains FB1, FB2, and FBD11 have been described previously (8). U. maydis cells were grown at 28°C in liquid YEPS (1% yeast extract, 2% peptone, 2% sucrose), in liquid potato dextrose broth, or on solid potato dextrose agar that contained 1.5% (wt/vol) Bacto agar. The transformation of U. maydis was performed according to the procedure described in reference 50. For selection, potato dextrose agar plates containing 200 μg of hygromycin/ml were used. For the induction or repression of the carbon source-dependent crg promoter (11), cells were grown on yeast nitrogen base medium (Difco), pH 5.8, containing 0.5% ammonium sulfate and 2% arabinose or glucose. For the determination of nutrient requirements, cells were grown in yeast nitrogen base medium with and without uracil (1 mg/ml). To test whether U. maydis strains were susceptible to brequinar, strains were cultivated in 20 ml of minimal medium containing 2% glucose and 100 μM brequinar at 28°C. Growth was measured after 16 h.
Oligonucleotides.
For the amplification and subcloning of DHODH genes, the following oligonucleotides were used (restriction sites used for cloning are shown in italics): UmDHODHΔN-fwd, 5′-CTAGTCTAGATAACGAGGGCAAAAAATGTCTCGTTCGGCTATCCATCG-3′; UmDHODH-rev, 5′-CCGGAATTCAACAACGCGCACCTTGTCGATC-3′; A-fwd (U. maydis promoter), 5′-GCATGCCTTGTCAAGCTCGTGGTGTC-3′; E-rev (U. maydis promoter), 5′-GGTACCGACAGAGCTAGACAG-3′; D-Um-rev (U. maydis DHODH gene), 5′-GCGGCCGCTTAAACAACGCGCACCTT-3′; B-Um-rev (human DHODH gene fused to the U. maydis DHODH gene regions corresponding to the mitochondrial targeting sequence and the transmembrane domain), 5′-CTCATCTCCCGTGGCCATTGCATAGTAGGCAATGCC-3′; C-Hs-fwd (human DHODH gene lacking the sequence encoding the N terminus), 5′-GGCATTGCCTACTATGCAATGGCCACGGGAGATGAG-3′; F-Hs-fwd (human DHODH gene), 5′-GGTACCATGGCGTGGAGACACC-3′; D-Hs-rev (human DHODH gene), 5′-GCGGCCGCTCACCTCCGATGATCTGC-3′; GF1 (5′ green fluorescent protein [GFP] gene fusion), 5′-CCCGGGATGCTTGCCTCTCGTAGC-3′; GF2 (3′ fusion of the DHODH gene to the first 73 codons of the GFP gene), 5′-CCATGGGCTTGGTGCGCGCCAGAG-3′; and GF3 (3′ fusion of the DHODH gene to the first 110 codons of the GFP gene), 5′-CCATGGATGCTTTCAGAGCTGGAA-3′.
Cloning of the U. maydis pyr4 gene and construction of chimeric DHODH genes.
The pyr4 gene was isolated from a cosmid library derived from U. maydis strain FBD11. The sequence was determined by subcloning. A cDNA library of U. maydis FBD11 (49) was used to isolate pyr4 cDNA. The cDNA inserts were amplified by PCR and sequenced after subcloning into pCR2.1-Topo (Invitrogen).
For the deletion of pyr4, 1.5 kb of upstream and 1.1 kb of downstream sequences were fused to a hygromycin resistance cassette and strains FB1 and FB2 were transformed with the constructs. The correct replacement of the pyr4 gene by homologous recombination was verified by Southern analysis. The conditional pyr4 mutant was generated by replacing the endogenous promoter with the regulatable crg promoter (11), which is repressed in the presence of glucose and induced in the presence of arabinose.
For enhanced GFP (EGFP) fusion constructs, two DNA fragments including the first 73 or 110 codons of the GFP gene and the DHODH gene were amplified with PCR primer pairs GF1-GF2 and GF1-GF3, respectively, which introduced a SmaI site at the 5′ ends and a NcoI site at the 3′ ends. DNA fragments were fused to the EGFP gene in plasmid p123 (1), where the fusion constructs were expressed under the control of the constitutive etef promoter.
For complementation, the U. maydis full-length pyr4 gene including its own promoter was amplified with primers A-fwd and D-Um-rev. The full-length human DHODH gene was amplified from plasmid pASKMh (4) with primers F-Hs-fwd and D-Hs-rev. The human open reading frame was then fused at an introduced KpnI site to the native U. maydis pyr4 promoter, which had been generated by PCR by using primers A-fwd and E-rev. For the generation of the chimeric U. maydis-human DHODH gene, the U. maydis pyr4 promoter and the region corresponding to the N-terminal targeting sequence were amplified with primers A-fwd and B-Um-rev. The sequence corresponding to the catalytic domain of human DHODH was amplified with primers C-Hs-fwd and D-Hs-rev and fused to the U. maydis promoter and targeting signal by overlapping PCR using primers A-fwd and D-Hs-rev. All DNA fragments were introduced into plasmid p123 (1) and used for the transformation of U. maydis protoplasts.
Determination of radiation sensitivity.
U. maydis strains were grown overnight in potato-dextrose medium. Cells were washed and diluted with H2O to a final optical density at 600 nm of 0.5. Dilutions of 1:10, 1:100, and 1:1,000 (100 μl) were plated onto minimal medium agar supplemented with 2% glucose or 2% arabinose. Plates were irradiated in the UV Stratalinker 2400 (Stratagene) with UV radiation (254 nm), and the survival rate was determined after incubation for 2 to 5 days at 28°C.
Plant inoculations.
Maize seedlings (early golden bantam variety purchased from Olds Seed Company, Madison, WI) were grown in a growth chamber (14 h of light at 28°C and 10 h of darkness at 20°C at 60% humidity). The infections of 9-day-old maize seedlings were performed as described previously by the injection of ca. 300-μl volumes of cell suspensions (109 cells/ml) (33). Tumor formation in infected corn plants was scored 2 weeks after infection.
Gene expression in E. coli and protein purification.
For expression in E. coli, the region of pyr4 corresponding to the catalytic domain was amplified with primers UmDHODHΔN-fwd and UmDHODH-rev. During the amplification, BamHI and EcoRI restriction sites were introduced and these sites were used to clone the PCR fragment into the corresponding sites of pGEX-6P-3 (Amersham Bioscience). Recombinant U. maydis N-terminally truncated DHODH (DHODH-ΔN) was purified as glutathione S-transferase (GST) fusion protein from E. coli strain BL21 induced for 24 h in medium containing 0.1 mM flavin mononucleotide (FMN). Cells were harvested at 4,000 × g for 15 min; resuspended in buffer A (140 mM NaCl, 2.7 mM KCl, 0.1 mM FMN, 10 mM Na2HPO4, 1.8 mM KH2PO4, and 1% Triton X-100, pH 7.3); and disrupted by sonication. After centrifugation for 60 min at 15,000 × g, the supernatant was applied to a 1-ml GSTrap FF column (Amersham Bioscience). The column was washed with 10 volumes of buffer A and 10 volumes of a solution containing 50 mM Tris-HCl, 150 mM NaCl, 1 mM EDTA, 1 mM dithiothreitol, and 0.1% Triton X-100, pH 7. The GST tag was removed by digestion with PreScission protease (Amersham Bioscience) at 4°C and above. The purified proteins were transferred into a solution containing 50 mM Tris-HCl, 150 mM KCl, 10% (vol/vol) glycerol, and 0.1% Triton X-100 (vol/vol), pH 8, by using a PD-10 column (Amersham Bioscience).
Protein analysis.
For the fluorimetric determination of the flavin concentration, 0.5 to 1 μg of protein/ml was denatured by incubation at 100°C for 10 min. After centrifugation, the flavin concentration in the supernatant was determined using a spectrofluorimeter (SFM 25; Bio-Tek) at an excitation wavelength of 465 nm and an emission wavelength of 518 nm, with FMN (0 to 100 μM) as the standard marker.
Mitochondria were prepared from logarithmically growing U. maydis cells according to the protocol described in reference 29 and directly used for sodium dodecyl sulfate (SDS)-polyacrylamide gel electrophoresis (PAGE). For immunodetection, U. maydis DHODH-ΔN or the mitochondrial proteins were transferred from the SDS gel onto ImmobilonP (Millipore) by semidry blotting (1.5 h at 0.8 mA/cm2 of SDS gel). After blocking with 5% nonfat dried milk in 10 mM sodium phosphate buffer (pH 7.5)-150 mM NaCl, the membrane was exposed to affinity-purified rabbit anti-human DHODH antibodies (diluted 1:15,000) (14). As secondary antibodies, goat anti-rabbit horseradish peroxidase-conjugated immunoglobulin G antibodies (Sigma), diluted 1:10,000, were used. The detection of bound antibodies was done by ECL (Amersham Bioscience) detection.
Enzyme assays.
The Km values of U. maydis DHODH for DHO and the artificial electron acceptor 2,6-dichlorophenol-indophenol (DCIP) were determined by using DHODH reaction buffer (50 mM Tris-HCl, 150 mM KCl, 0.1% [vol/vol] Triton X-100, pH 8) at 30°C by monitoring the increase in the absorption of UV radiation by the product orotate (280 nm; ɛ = 7,500 M−1 cm−1) and the decrease in the absorption of UV radiation by DCIP (600 nm; ɛ = 18,800 M−1 cm−1), respectively. Kinetic data were determined in the DCIP chromogen reduction assay (57). The data were evaluated under initial velocity conditions (31) and fitted to the following Michaelis-Menten equation: v = V[S]/(Km + [S]), where v is the initial velocity, V is the limiting value at the saturating substrate concentration, and [S] is the substrate concentration.
The pH dependence of initial velocities was measured at a saturating substrate concentration (1 mM DHO) in different buffer systems (MES [morpholineethanesulfonic acid]-HCl, HEPES-HCl, and Tris-HCl) covering a pH range from 5 to 9 by using the chromogen reduction assay. Overlapping pH ranges were measured in two buffer systems to exclude salt effects. The following equation was fitted to the data: v = V/{[10−(pH)/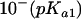 ] + [
] + [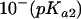 /10−(pH)] + 1}.
/10−(pH)] + 1}.
The comparison of various natural and artificial electron acceptors was done by using DHODH reaction buffer. The reduction of the following electron acceptors was measured at the indicated wavelengths: FeCy, 420 nm (ɛ = 1,020 M−1 cm−1), and NAD+, 340 nm (ɛ = 6,200 M−1 cm−1). The absorption of UV radiation by the product orotate was monitored at 280 nm (ɛ = 7,500 M−1 cm−1) for the electron acceptors fumarate and oxygen and at the appropriate isosbestic wavelengths for decylubiquinone QD (300 nm; ɛ = 2,950 M−1 cm−1), Q10 (300 nm; ɛ = 2,950 M−1 cm−1), Q6 (293 nm; ɛ = 4,700 M−1 cm−1), Q3 (287 nm; ɛ = 5,680 M−1 cm−1), and 2,3-dimethoxy-5-methyl-1,4-benzoquinone (Q0; 287 nm; ɛ = 5,680 M−1 cm−1).
To determine the inhibitory potencies of different compounds, the chromogen reduction assay was used with concentrations of up to 1 mM of the putative inhibitor. Stock solutions of all inhibitors were prepared freshly in DHODH reaction buffer without Triton X-100 or in dimethyl sulfoxide (DMSO). Only quinoxyfen was dissolved in ethyl alcohol, 96% pure. The appropriate controls were run in buffer, in ethyl alcohol, or in the presence of DMSO; 2% DMSO or ethyl alcohol in the assay mixtures was found not to interfere with the DHODH activity. All measurements were done in triplicate. Percentages of inhibition were determined relative to the control (100% activity).
Microscopy.
The intracellular localization of DHODH-EGFP fusion proteins was determined by confocal laser scanning microscopy analysis with a Leica instrument. U. maydis cells were grown overnight and mixed with glycerol (final concentration, 25%) to reduce mobility. The endocytosis dye FM4-64 (Molecular Probes) was added at 15 μM to visualize cellular membranes. EGFP fluorescence was detected at an excitation wavelength of 488 nm and an emission wavelength of 509 nm. FM4-64 was used for visualization at an excitation wavelength of 543 nm and an emission wavelength of 640 nm. The colocalization of DHODH-EGFP and mitochondria was monitored with cells stained with Mitotracker rhodamine B (excitation wavelength, 578 nm; emission wavelength, 556 nm) with a Zeiss Axiophot microscope.
Nucleotide sequence accession number.
The sequence of the pyr4 gene from U. maydis has been deposited in GenBank under accession number DQ869012.
RESULTS
The U. maydis pyr4 gene encodes DHODH with an unusually large carboxy-terminal extension.
We cloned and sequenced the pyr4 gene encoding DHODH (41) since no DHODH from basidiomycetes had yet been characterized on the molecular level. The integrity of the predicted intron-lacking open reading frame was confirmed by isolating cDNA clones (data not shown). The derived polypeptide of 677 amino acids contains the highly conserved domains for FMN and dihydroorotate binding that are characteristic of dihydroorotate dehydrogenases. Within the catalytic center of U. maydis DHODH, a highly conserved serine residue is present, which is characteristic of membrane-bound enzymes of family 2 that depend on a functional respiratory chain (9, 46). A database comparison using BLAST (2) revealed that U. maydis DHODH is most similar to PyrD from Schizosaccharomyces pombe (E value = 10−109). Remarkably, the polypeptide sequence of DHODH from U. maydis contains an unusually long extension at the C-terminal end which substantially exceeds the lengths of the C-terminal ends of all previously characterized DHODH proteins. Only the basidiomycetous yeast Cryptococcus neoformans, whose genome sequence has recently been determined (39), contains a DHODH that carries a C-terminal domain of comparable length.
DHODH is targeted to the mitochondria in U. maydis.
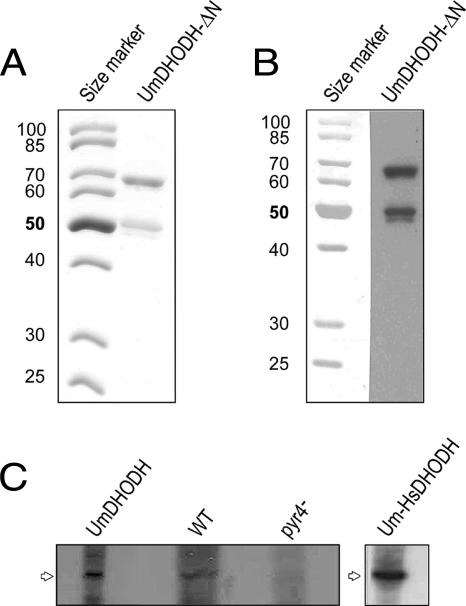
A putative N-terminal mitochondrial targeting signal with an adjacent transmembrane domain between amino acid residues 74 and 90 was detected by the program PSORT, which predicts the subcellular localization patterns of proteins (44). This result suggests the targeting of the protein to the inner mitochondrial membrane, which is characteristic of eukaryotic DHODHs that depend on a functional respiratory chain (28). To determine the subcellular localization of U. maydis DHODH, we fused the predicted N-terminal mitochondrial targeting sequence of DHODH to GFP. GFP was fused either directly to the predicted mitochondrial targeting signal or behind the putative transmembrane domain (Fig. 1B). The fusion constructs were expressed in haploid cells under the control of a constitutive promoter. Mitochondria were visualized by staining with Mitotracker rhodamine B (see Materials and Methods). A microscopic inspection of transformed cells revealed a clear colocalization of GFP fluorescence and the mitochondrion-specific dye (Fig. 1A). We could not detect any differences in the localization patterns of the GFP fusion proteins between constructs with and without the adjacent putative membrane-spanning domain (Fig. 1C). The accumulation of DHODH in mitochondria was also confirmed by the purification of mitochondrial proteins from logarithmically growing cells. The mitochondrial fraction was stained with cross-reacting antibodies directed against human DHODH, revealing a clear signal in the mitochondrial fraction (Fig. 2C). Together, these results indicate that U. maydis DHODH is targeted to the mitochondria, where the enzyme is supposed to be localized at the outer face of the inner mitochondrial membrane.
FIG. 1.
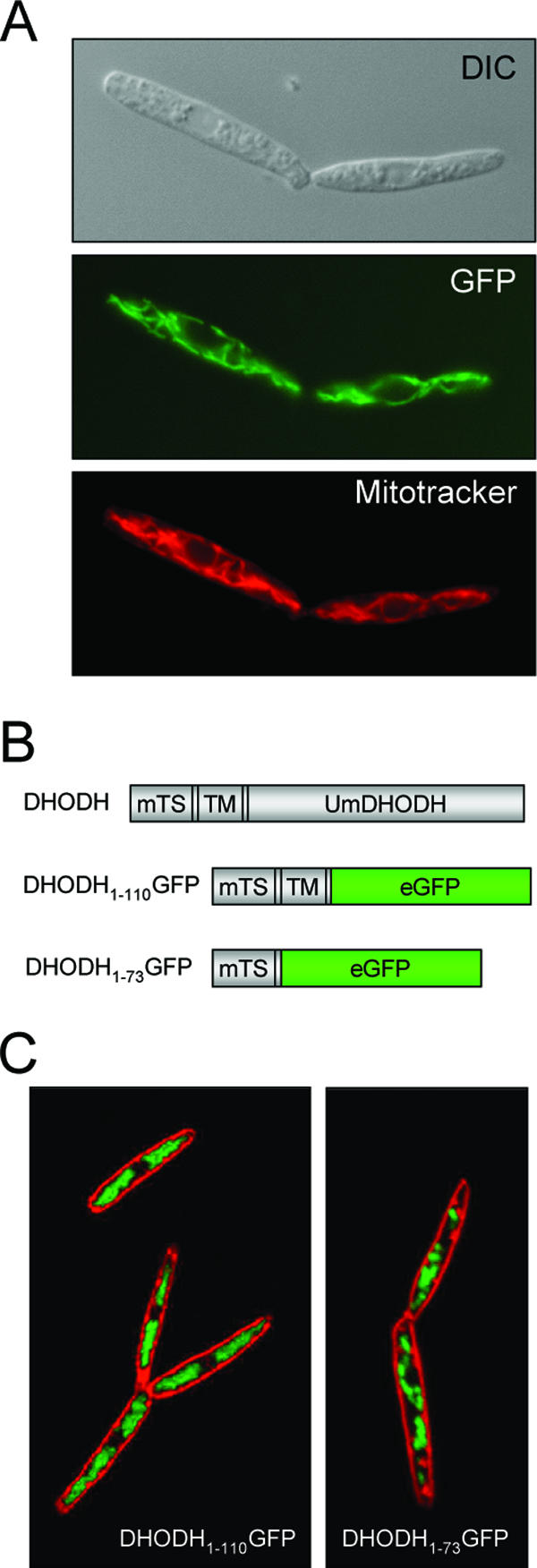
U. maydis DHODH is targeted to the mitochondria. The mitochondrial targeting sequence of U. maydis DHODH was fused to GFP and expressed in haploid U. maydis cells. (A) The colocalization of the U. maydis DHODH fused to amino acids 1 to 73 of GFP (DHODH1-73GFP) with mitochondria stained by Mitotracker rhodamine B indicates the targeting of U. maydis DHODH to the mitochondria. DIC, differential interference contrast. (B) Schematic drawing of different GFP constructs used for the localization of U. maydis DHODH (UmDHODH). The mitochondrial targeting sequences (mTS) and the transmembrane domains (TM) are indicated. (C) Confocal fluorescence micrographs of FM4-64-stained cells expressing DHODH fused to amino acids 1 to 110 of GFP (DHODH1-110GFP) or DHODH1-73GFP reveal no differences in localization.
FIG. 2.
Expression of recombinant U. maydis dihydroorotate dehydrogenase in E. coli and Western analysis of mitochondrial proteins. (A) The N-terminally truncated U. maydis DHODH (UmDHODH-ΔN) was expressed in E. coli and purified by affinity chromatography. Shown is a Coomassie-stained SDS gel of the purified protein (2 μg). (B) Western blot of purified U. maydis DHODH-ΔN. The purified protein (1 μg) was separated by SDS-PAGE and immunostained with rabbit polyclonal anti-human DHODH serum. (C) Mitochondria were prepared from an FB1 Δpyr4 mutant strain transformed with U. maydis DHODH (UmDHODH), the wild-type (WT) FB1 strain, an FB1 Δpyr4 mutant strain (pyr4−), and an FB1 Δpyr4 mutant strain expressing the chimeric U. maydis-human DHODH construct (Um-HsDHODH); separated by SDS-PAGE; and immunostained with rabbit polyclonal anti-human DHODH serum.
Expression and biochemical characterization of recombinant U. maydis DHODH.
To study the enzymatic activity in vitro, we expressed the pyr4 gene product in E. coli as a GST fusion protein. U. maydis DHODH-ΔN, which lacks both the bipartite mitochondrial targeting signal and the transmembrane domain, was purified by affinity chromatography on glutathione Sepharose. The recombinant U. maydis DHODH-ΔN, whose GST tag was cleaved off during elution, had the expected molecular mass of 63 kDa (Fig. 2A) and yielded approximately 1 mg of purified protein from 1,000 ml of E. coli BL21 cultures. The purified enzyme could be detected on a Western blot by using anti-human DHODH antibodies (Fig. 2B). The flavin/protein ratio (mol/mol) of the recombinant enzyme was estimated by fluorimetric cofactor analysis, which revealed a ratio of 0.7 to 0.8 flavin cofactor molecules per protein molecule.
The enzymatic activity of U. maydis DHODH-ΔN was measured with DCIP as the electron acceptor and tested in various buffers, which revealed a maximum of activity at pH 8.0. From the characteristic bell-shaped activity profile, two distinct pKa values could be calculated: pKa1, 6.5 ± 0.04, and pKa2, 9.4 ± 0.11. The specific activity was determined to be 6 U/mg, with kcat in the range around 1.6 s−1. The Km values for DCIP (37 ± 7 μM) and l-dihydroorotate (43 ± 7 μM) were comparable.
Next, we tested the activity of U. maydis DHODH-ΔN with a variety of natural and artificial electron acceptors. Of the latter, U. maydis DHODH-ΔN was able to use potassium hexacyanoferrate(III) (FeCy) and DCIP (Table 1). Of the naturally occurring acceptors, quinone Q6 was significantly better accepted than Q10, which is used by most higher eukaryotes as an endogenous electron acceptor (47) (Table 1). Fumarate, NAD+, and NADP+, which can be used by cytosolic DHODHs, turned out to be inadequate electron acceptors for U. maydis DHODH-ΔN. In the presence of atmospheric oxygen, a very low basal level of DHODH activity was observed, suggesting that molecular oxygen may serve as a poor direct acceptor of the reaction electrons. Together, these results indicate that U. maydis DHODH fulfills all the criteria of eukaryotic membrane-bound enzymes and displays a preference for the ubiquinone derivative Q6.
TABLE 1.
Electron-accepting substrates for recombinant U. maydis DHODH-ΔN
| Electron acceptor (concn, mM) | Enzyme activitya (%) |
|---|---|
| FeCy (1) | 100 |
| DCIP (1) | 90 ± 2 |
| QD (0.1) | 5 ± 1.9 |
| Q10 (0.1) | 2 ± 0.2 |
| Q6 (0.1) | 104 ± 19 |
| Q3 (0.1) | 8 ± 5 |
| Q0 (0.1) | 8 ± 9 |
| Fumarate (1) | 2 ± 0.3 |
| NAD+ (0.1) | 1 ± 0.2 |
| NADP+ (0.1) | 1 ± 0.3 |
| None | 2 ± 0.4 |
Enzyme activities were determined as relative initial velocities and are expressed as mean percentages (± standard errors of the means) of the activity with FeCy as the electron acceptor. All reaction mixtures contained 1 mM DHO as the substrate, and reactions were performed in the presence molecular oxygen at atmospheric pressure (equivalent to about 230 μM).
U. maydis DHODH is resistant to commonly used DHODH inhibitors and substrate analogues.
The purified U. maydis DHODH-ΔN was tested in vitro for its susceptibility to various known DHODH inhibitors and substrate analogues. Although brequinar, A77-1726, and atovaquone are highly efficient inhibitors of mitochondrially bound family 2 DHODHs (31, 32), none of these compounds reduced the activity of U. maydis DHODH-ΔN more than 25% (Table 2). This finding demonstrates that the mechanism of inhibition of these substances seems to be quite specific for mammalian enzymes. In addition, quinoxyfen, a compound highly similar to the antifungal substance LY214352 [8-chloro-4-(2-chloro-4-fluorophenoxy)-quinoline] (21), also did not interfere with U. maydis DHODH activity. A number of substance analogues that resemble dihydroorotate, the natural substrate of DHODH, were also tested as competitive inhibitors. As shown in Table 2, U. maydis DHODH was not affected by any of these compounds.
TABLE 2.
Activity of recombinant U. maydis DHODH-ΔN in the presence of putative inhibitors
| Compound (concn, mM) | Enzyme activitya (%) |
|---|---|
| Brequinar | 85 ± 15 |
| A77-1726 | 108 ± 8 |
| Atovaquone (0.5) | 117 ± 8 |
| Dichloroallyl lawsone | 75 ± 3 |
| Lawsone | 90 ± 4 |
| Menadione | 91 ± 7 |
| Salicylhydroxamic acid | 84 ± 7 |
| Anthranilic acid | 95 ± 7 |
| N-Phenylanthranilic acid (0.1) | 90 ± 3 |
| Coumarin (0.1) | 82 ± 3 |
| 5-Fluorocytosine | 91 ± 8 |
| 5-Fluoroorotate | 98 ± 4 |
| 5-Fluorouracil | 90 ± 3 |
| Quinoxyfen (0.1) | 94 ± 7 |
| Carboxin | 92 ± 2 |
| 3,4-DHB | 87 ± 3 |
| 3,5-DHB | 98 ± 3 |
Relative initial velocities were determined in chromogen reduction assays with 1 mM DHO as the substrate and 0.1 mM DCIP as the acceptor. The enzyme activity without an inhibitor was set at 100%, and values are expressed as mean percentages ± standard errors of the means. For compounds dissolved in DMSO or ethanol, the respective solvent was added to the control. If not otherwise stated, the concentration of tested compounds was 1 mM.
Depletion of pyr4 results in enhanced sensitivity to UV irradiation and loss of pathogenicity.
We generated a pyr4 deletion mutant by replacing the complete open reading frame with a cassette conferring hygromycin resistance. The disruption of the pyr4 gene was confirmed by Southern analysis (data not shown). While pyr4 mutant cells could grow in complete medium, they were completely unable to propagate in synthetic minimal medium lacking a pyrimidine source (Fig. 3B). The supplementation of the medium with uracil restored growth, confirming that U. maydis pyr4 mutants are dependent on pyrimidine bases. While the addition of orotate to the medium also cancelled the growth defect of pyr4 mutants, supplementation with DHO did not allow for the growth of pyr4 mutants (data not shown). This finding is in accordance with the predicted enzymatic function of DHODH, which is required for the conversion of DHO to orotate.
FIG. 3.
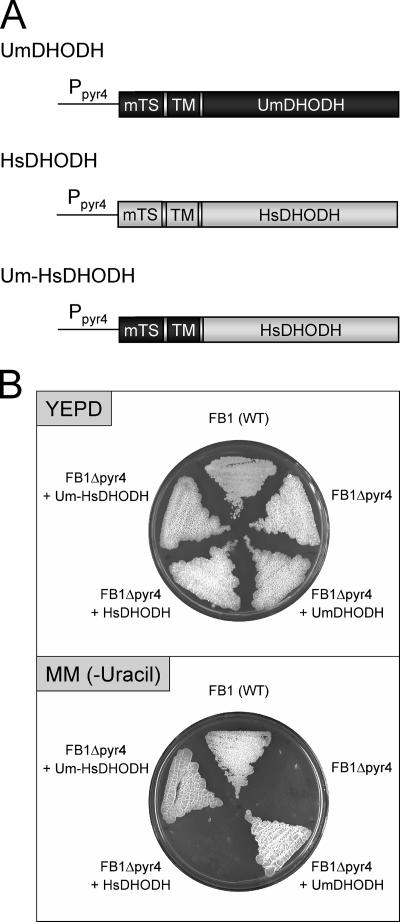
The functional expression of human dihydroorotate dehydrogenase in U. maydis complements the growth defect of Δpyr4 mutants. (A) Full-length and chimeric DHODH genes were expressed under the control of the U. maydis pyr4 (DHODH) promoter (Ppyr4). Dark gray, U. maydis DHODH (UmDHODH) open reading frame; light gray, human DHODH (HsDHODH) open reading frame; mTS, mitochondrial targeting sequence; TM, transmembrane domain; Um-HsDHODH, chimeric U. maydis-human DHODH construct. (B) Wild-type (WT) strain FB1, the FB1 Δpyr4 mutant (FB1Δpyr4), and transformed strains were grown on full medium (yeast extract-peptone-dextrose [YEPD]) and minimal medium without uracil [MM (−Uracil)].
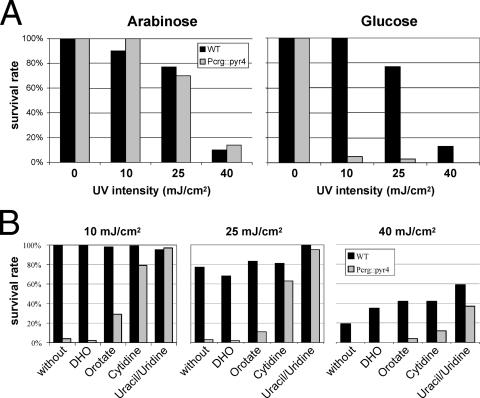
It has been reported previously that U. maydis mutants defective in pyrimidine biosynthesis are highly sensitive to UV irradiation (25, 41). This phenomenon has been ascribed to an imbalance in the deoxynucleoside triphosphate (dNTP) pools, which results in error-prone DNA repair by endogenous DNA polymerases (42). To test whether the complete deletion of U. maydis pyr4 also affects UV irradiation resistance, we determined the lethal dose of UV irradiation. Whereas wild-type strains were highly resistant to UV irradiation, pyr4 mutant strains displayed a significant reduction in the level of tolerance of UV irradiation (data not shown). To test whether UV irradiation sensitivity depends directly on the presence of DHODH, we constructed a conditional pyr4 mutant strain that expressed pyr4 under the control of the arabinose-inducible crg promoter (11). Under conditions in which the crg promoter was repressed (the presence of glucose), cells were highly sensitive to UV irradiation (Fig. 4A). However, if cells were grown on arabinose-containing medium, they displayed the same level of resistance to UV irradiation as wild-type cells (Fig. 4A). If the conditional mutants were grown on medium supplemented with uridine/uracil or cytidine, UV irradiation resistance was restored nearly to the wild-type level (Fig. 4B). While the addition of orotate resulted in the partial suppression of sensitivity to UV irradiation, DHO was unable to affect UV irradiation sensitivity (Fig. 4B).
FIG. 4.
The depletion of DHODH results in enhanced sensitivity to UV radiation. (A) Cells expressing the pyr4 open reading frame under the control of the arabinose-inducible crg promoter (Pcrg::pyr4) are highly sensitive to UV radiation if the expression of pyr4 is repressed in glucose-containing medium. If the medium contains arabinose, no difference from wild-type (WT) cells can be observed. (B) The UV radiation sensitivity of conditional pyr4 mutants (Pcrg::pyr4) grown under repressing conditions (in the presence of glucose) is alleviated if uridine/uracil, cytidine, or orotate is added to the medium. DHO is unable to restore the radiation resistance.
Next we tested whether the presence of pyr4 is required for pathogenic development in plants. To this end, wild-type and mutant cells of compatible mating types were mixed and coinoculated into maize seedlings. While the coinoculation of pyr4 mutants with wild-type cells resulted in normal pathogenic development, no disease symptoms could be observed when two compatible pyr4 mutants were used for infection. Although the loss of pathogenicity was not surprising, since many auxotrophic mutants of U. maydis are known to be nonvirulent, this finding indicates that DHODH is a potential target to prevent fungal infection.
Functional expression of human DHODH restores prototrophy but confers sensitivity to brequinar.
Next we asked whether human DHODH is able to abolish the pyrimidine biosynthesis defect of U. maydis pyr4 mutants. To this end, we expressed full-length human DHODH under the control of the endogenous U. maydis pyr4 promoter (Fig. 3A). Since it cannot be excluded that human DHODH may not be processed properly in U. maydis, we also generated a chimeric version of DHODH in which the N-terminal mitochondrial targeting and transmembrane domains of the human protein were replaced by the corresponding U. maydis domains (Fig. 3A). Both constructs were introduced into auxotrophic pyr4 mutants, and the resulting transformants were assayed for growth on minimal medium. Interestingly, only the chimeric DHODH protein was able to restore growth on minimal medium, while cells expressing full-length human DHODH were unable to grow without the addition of uracil (Fig. 3B). This result indicates that the catalytic domain of human DHODH is able to complement the biosynthesis defect of pyr4 mutants. At the same time, this finding indicates that the U. maydis targeting signal seems to be necessary for the correct localization of the fusion protein at the mitochondrial membrane of U. maydis cells.
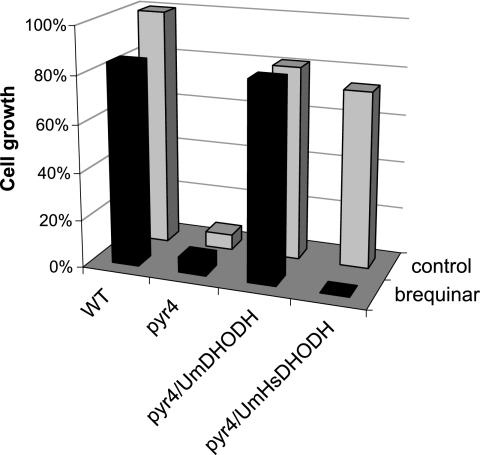
Next we tested whether U. maydis cells expressing a humanized version of DHODH are sensitive to known inhibitors of mammalian DHODH. The biphenyl quinoline-carboxylic acid derivative brequinar blocks the activity of human DHODH at nanomolar concentrations and thus is one of the most potent inhibitors of mammalian DHODH (12, 31). Inhibition was tested in vivo by incubating transformants in minimal medium containing various concentrations of brequinar. As shown in Fig. 5, recombinant U. maydis strains expressing human DHODH were sensitive to brequinar at a 10 μM concentration. In contrast, neither U. maydis wild-type cells nor pyr4 mutants transformed with the U. maydis DHODH were inhibited by brequinar at this concentration and these cells could grow normally on minimal medium without pyrimidine (Fig. 5). Together, these data indicate that upon correct targeting to the mitochondria, human DHODH is fully active in the basidiomycetous fungus U. maydis. While the auxotrophic phenotype of U. maydis pyr4 mutants was fully complemented by the expression of the chimeric DHODH, such strains displayed sensitivity to a commonly used inhibitor of the mammalian enzyme. This finding provides the opportunity to use such strains in bioassays for inhibitors specifically targeted at either the fungal or the human DHODH (see Discussion).
FIG. 5.
U. maydis strains expressing human DHODH are inhibited by brequinar. U. maydis wild-type cells (WT), pyr4 mutants (pyr4), and pyr4 mutants transformed with either full-length U. maydis DHODH (pyr4/UmDHODH) or the chimeric U. maydis-human DHODH (pyr4/Um-HsDHODH) were grown in minimal medium. Cell growth was determined 16 h after the addition of the inhibitor brequinar (10 μM).
DISCUSSION
We have characterized the U. maydis pyr4 DHODH-encoding gene on the molecular level. Although mutations affecting the de novo pyrimidine biosynthesis pathway in several fungi, including U. maydis, have been described previously (6, 41, 51), here we report the first complete knockout of a fungal mitochondrial DHODH. pyr4 mutants were unable to grow on minimal medium, but growth was completely restored if uracil or cytosine was added to the medium. As expected from the predicted function of DHODH, the supplementation of the medium with orotate but not with dihydroorotate alleviated the growth defect of pyr4 mutant cells.
It has been reported previously that pyrimidine auxotrophic mutants of U. maydis are highly sensitive to UV irradiation (25, 41). This effect, which has been observed only in U. maydis so far, has been explained by an imbalance in the dNTP pools, which is assumed to affect DNA repair processes (42). This hypothesis was supported by the observation that pyrimidine auxotrophic mutants contain an unusually low level of dTTP (42). It has been suggested that the repair polymerase is unable to work properly at these low dTTP levels (56) and, therefore, pyrimidine dimers induced by UV radiation are left uncorrected in large numbers. Why strains with mutations affecting pyrimidine biosynthesis display this dNTP pool imbalance even in the presence of exogenously supplied uracil or cytosine remains unclear. We have constructed a U. maydis strain in which pyr4 is expressed under the control of the arabinose-inducible crg promoter. This conditional UV radiation-sensitive mutant may help to elucidate the molecular basis of this interesting regulatory phenomenon. In addition, this strain can be used to screen for synthetic lethal mutants that grow only in the presence of pyr4. Such mutants are expected to be involved in systems required for maintaining DNA integrity.
The presence of pyr4 is also required for virulence on corn plants, demonstrating that U. maydis depends on de novo pyrimidine biosynthesis during pathogenic development. This makes DHODH an interesting target for the development of antifungal drugs for agricultural crop protection. For the experimental fungicide LY214352 [8-chloro-4-(2-chloro-4-fluorophenoxy) quinoline], it has already been shown that DHODH is the primary target of this compound (21). Since LY214352 is not commercially available, we tested quinoxyfen, a closely related compound, for the inhibition of U. maydis DHODH activity in vitro. Quinoxyfen was not able to inhibit U. maydis DHODH, which is in line with the observation that LY214352 does not exhibit growth inhibition when tested against basidiomycetes fungi (21).
The U. maydis DHODH protein contains an unusually long extension at its C terminus. Since a similar extension can also be found in the related basidiomycete Cryptococcus neoformans, this structural architecture may be characteristic of basidiomycetous fungi. The availability of several additional fungal genome sequences, in particular those of basidiomycetous origin, will surely clarify this issue. Interestingly, the heterologous expression of a C-terminally truncated DHODH in E. coli resulted in a nonfunctional protein (J. Warneboldt and M. Löffler, unpublished results). This finding may indicate that the C-terminal extension is required either for catalytic activity or for the correct folding of the complete protein, at least in vitro.
Our biochemical analysis of U. maydis DHODH revealed that the activity of the recombinant enzyme is significantly lower than that of mammalian DHODH (6 U/mg compared to 99 U/mg for the human enzyme) (55). The Km value of U. maydis DHODH is significantly higher (43 μM for DHO) than that of human DHODH (9.7 μM for DHO) (55). In both cases, these values were determined for recombinant proteins lacking their N-terminal transmembrane domains. Nevertheless, the activity of recombinant U. maydis DHODH is still low and its Km is still high compared to those of other fungal enzymes, e.g., Candida albicans DHODH (57). The enzymes of the ascomycetous yeasts Schizosaccharomyces pombe and Saccharomyces kluyveri have even lower specific activities (58), which, however, may be due to the low flavin contents observed in the heterologously expressed enzymes (58). However, the large amount of flavin in the recombinant U. maydis enzyme (molar content, 70 to 80%) supports the notion that in general fungal DHODHs display lower specific activities in vitro than mammalian enzymes.
Although we observed that in vitro the U. maydis enzyme preferentially uses Q6 as an electron acceptor, it is still unknown which ubiquinone derivative acts as the native electron acceptor for DHODH in vivo. In early studies, either no coenzyme Q at all has been found (34) or only Q10 and Q9 have been identified (15) in U. maydis. For the ascomycetous yeast Saccharomyces cerevisiae, coenzyme Q6 has been described as a physiological acceptor of the respiratory chain (54). Q6 was also detected in a number of other fungal species; in particular, it was also found in basidiomycetes, e.g., Suillus luteus (47). This may indicate that Q6 may also be the native electron acceptor in U. maydis.
We succeeded in showing that U. maydis DHODH is not inhibited by brequinar, a very efficient inhibitor of mammalian DHODH. Most inhibitors targeted at DHODH block the quinone binding site. For brequinar, this pattern has been shown convincingly by the determination of the three-dimensional structure of human DHODH in a complex with this inhibitor (36). We used DeepView Swiss-PdbViewer 3.7 to superimpose the amino acid sequence of U. maydis DHODH onto this structure (Protein Data Bank accession number, 1D3G). Comparison of the inhibitor binding sites revealed that many hydrophobic residues of the human protein known to be involved in the binding of the inhibitor are not present in the U. maydis enzyme. These structural differences may explain the different affinities of these enzymes both for quinone electron acceptors and for inhibitors.
We were able to express the human DHODH gene in U. maydis. The complementation of pyrimidine auxotrophy was observed only when the catalytic domain of the mammalian enzyme was fused to the mitochondrial targeting and the membrane-spanning domains derived from U. maydis DHODH. This finding indicates that differences in the protein-sorting machineries may prevent correct targeting of the full-length human enzyme to the U. maydis mitochondria. The expression of the chimeric human enzyme also complemented the UV radiation sensitivity of pyr4 mutants. This result indicates that this sensitivity phenotype is a direct consequence of the disruption of pyrimidine de novo biosynthesis.
U. maydis cells engineered to express human DHODH became sensitive to brequinar, a common inhibitor of mammalian DHODH. Thus, such strains can be used for the in vivo validation of compounds that selectively inhibit either the fungal or the human version of this important target enzyme. One can imagine labeling U. maydis cells that express either the wild-type or the human enzyme specifically by the coexpression of different fluorescent proteins that differ in the emission spectra (e.g., GFP and a red-shifted variant). If a mixture of such strains was incubated with a substance that specifically interferes with only one of the DHODH enzymes, the selective growth inhibition of one of the strains would result. This effect could be monitored by detecting a spectral shift of the emitted fluorescence. A major advantage of this system would be that compounds that are toxic for both strains would not result in a spectral shift because the growth of both strains would be affected. In principle, this system could also be used to screen for inhibitors directed against DHODHs of other important human fungal pathogens such as Candida albicans and Cryptococcus neoformans or parasites like Plasmodium falciparum and Toxoplasma gondii. In this case, a competitive inhibition screening of U. maydis cells expressing either the human DHODH gene or the DHODH gene derived from the respective human pathogen would be performed. This system may allow for the identification of novel compounds that selectively target the pathogen without major cytostatic or immunosuppressive side effects.
Acknowledgments
This study was supported by funds from the Deutsche Forschungsgemeinschaft, Graduiertenkolleg “Protein Function at the Atomic Level,” to M. Löffler.
Footnotes
Published ahead of print on 16 March 2007.
REFERENCES
- 1.Aichinger, C., K. Hansson, H. Eichhorn, F. Lessing, G. Mannhaupt, W. Mewes, and R. Kahmann. 2003. Identification of plant-regulated genes in Ustilago maydis by enhancer-trapping mutagenesis. Mol. Genet. Genomics 270:303-314. [DOI] [PubMed] [Google Scholar]
- 2.Altschul, S. F., T. L. Madden, A. A. Schaffer, J. Zhang, Z. Zhang, W. Miller, and D. J. Lipman. 1997. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 25:3389-3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Annoura, T., T. Nara, T. Makiuchi, T. Hashimoto, and T. Aoki. 2005. The origin of dihydroorotate dehydrogenase genes of kinetoplastids, with special reference to their biological significance and adaptation to anaerobic, parasitic conditions. J. Mol. Evol. 60:113-127. [DOI] [PubMed] [Google Scholar]
- 4.Bader, B., W. Knecht, M. Fries, and M. Löffler. 1998. Expression, purification, and characterization of histidine-tagged rat and human flavoenzyme dihydroorotate dehydrogenase. Protein Expr. Purif. 13:414-422. [DOI] [PubMed] [Google Scholar]
- 5.Baldwin, J., C. H. Michnoff, N. A. Malmquist, J. White, M. G. Roth, P. K. Rathod, and M. A. Phillips. 2005. High-throughput screening for potent and selective inhibitors of Plasmodium falciparum dihydroorotate dehydrogenase. J. Biol. Chem. 280:21847-21853. [DOI] [PubMed] [Google Scholar]
- 6.Banks, G. R., and S. Y. Taylor. 1988. Cloning of the PYR3 gene of Ustilago maydis and its use in DNA transformation. Mol. Cell. Biol. 8:5417-5424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Banuett, F. 1995. Genetics of Ustilago maydis, a fungal pathogen that induces tumors in maize. Annu. Rev. Genet. 29:179-208. [DOI] [PubMed] [Google Scholar]
- 8.Banuett, F., and I. Herskowitz. 1989. Different alleles of Ustilago maydis are necessary for maintenance of filamentous growth but not for meiosis. Proc. Natl. Acad. Sci. USA 86:5878-5882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Björnberg, O., A. C. Gruner, P. Roepstorff, and K. F. Jensen. 1999. The activity of Escherichia coli dihydroorotate dehydrogenase is dependent on a conserved loop identified by sequence homology, mutagenesis, and limited proteolysis. Biochemistry 38:2899-2908. [DOI] [PubMed] [Google Scholar]
- 10.Bölker, M. 2001. Ustilago maydis: a valuable model system for the study of fungal dimorphism and virulence. Microbiology 147:1395-1401. [DOI] [PubMed] [Google Scholar]
- 11.Bottin, A., J. Kämper, and R. Kahmann. 1996. Isolation of a carbon source-regulated gene from Ustilago maydis. Mol. Gen. Genet. 253:342-352. [DOI] [PubMed] [Google Scholar]
- 12.Chen, S. F., F. W. Perrella, D. L. Behrens, and L. M. Papp. 1992. Inhibition of dihydroorotate dehydrogenase activity by brequinar sodium. Cancer Res. 52:3521-3527. [PubMed] [Google Scholar]
- 13.Christopherson, R. I., S. D. Lyons, and P. K. Wilson. 2002. Inhibitors of de novo nucleotide biosynthesis as drugs. Acc. Chem. Res. 35:961-971. [DOI] [PubMed] [Google Scholar]
- 14.Dietz, C., E. Hinsch, and M. Löffler. 2000. Immunocytochemical detection of mitochondrial dihydroorotate dehydrogenase in human spermatozoa. Int. J. Androl. 23:294-299. [DOI] [PubMed] [Google Scholar]
- 15.Erickson, R. E., K. S. Brown, Jr., D. E. Wolf, and K. Folkers. 1960. Coenzyme Q. XX. Isolation of coenzymes Q9 and Q10 from two Basidiomycetes. Arch. Biochem. Biophys. 90:314-317. [DOI] [PubMed] [Google Scholar]
- 16.Fairbanks, L. D., M. Bofill, K. Ruckemann, and H. A. Simmonds. 1995. Importance of ribonucleotide availability to proliferating T-lymphocytes from healthy humans. Disproportionate expansion of pyrimidine pools and contrasting effects of de novo synthesis inhibitors. J. Biol. Chem. 270:29682-29689. [PubMed] [Google Scholar]
- 17.Feldbrügge, M., M. Bölker, G. Steinberg, J. Kämper, and R. Kahmann. 2006. Regulatory and structural networks orchestrating mating, dimorphism, cell shape, and pathogenesis in Ustilago maydis, p. 375-392. In U. Kües and R. Fischer (ed.), Mycota, 2nd ed., vol. 1. Springer, Berlin, Germany. [Google Scholar]
- 18.Fox, B. A., and D. J. Bzik. 2002. De novo pyrimidine biosynthesis is required for virulence of Toxoplasma gondii. Nature 415:926-929. [DOI] [PubMed] [Google Scholar]
- 19.Gojkovic, Z., W. Knecht, E. Zameitat, J. Warneboldt, J. B. Coutelis, Y. Pynyaha, C. Neuveglise, K. Møller, M. Löffler, and J. Piskur. 2004. Horizontal gene transfer promoted evolution of the ability to propagate under anaerobic conditions in yeasts. Mol. Genet. Genomics 271:387-393. [DOI] [PubMed] [Google Scholar]
- 20.Greene, S., K. Watanabe, J. Braatz-Trulson, and L. Lou. 1995. Inhibition of dihydroorotate dehydrogenase by the immunosuppressive agent leflunomide. Biochem. Pharmacol. 50:861-867. [DOI] [PubMed] [Google Scholar]
- 21.Gustafson, G., G. Davis, C. Waldron, A. Smith, and M. Henry. 1996. Identification of a new antifungal target site through a dual biochemical and molecular-genetics approach. Curr. Genet. 30:159-165. [DOI] [PubMed] [Google Scholar]
- 22.Hall, C., S. Brachat, and F. S. Dietrich. 2005. Contribution of horizontal gene transfer to the evolution of Saccharomyces cerevisiae. Eukaryot. Cell 4:1102-1115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Harder, A., and A. Haberkorn. 1989. Possible mode of action of toltrazuril: studies on two Eimeria species and mammalian and Ascaris suum enzymes. Parasitol. Res. 76:8-12. [DOI] [PubMed] [Google Scholar]
- 24.Herrmann, M. L., R. Schleyerbach, and B. J. Kirschbaum. 2000. Leflunomide: an immunomodulatory drug for the treatment of rheumatoid arthritis and other autoimmune diseases. Immunopharmacology 47:273-289. [DOI] [PubMed] [Google Scholar]
- 25.Holliday, R. 1965. Radiation sensitive mutants of Ustilago maydis. Mutat. Res. 2:557-559. [DOI] [PubMed] [Google Scholar]
- 26.Ittarat, I., W. Asawamahasakda, M. S. Bartlett, J. W. Smith, and S. R. Meshnick. 1995. Effects of atovaquone and other inhibitors on Pneumocystis carinii dihydroorotate dehydrogenase. Antimicrob. Agents Chemother. 39:325-328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Jensen, K. F., and O. Björnberg. 1998. Evolutionary and functional families of dihydroorotate dehydrogenases. Paths Pyrimidines 6:20-28. [Google Scholar]
- 28.Jones, M. E. 1980. Pyrimidine nucleotide biosynthesis in animals: genes, enzymes, and regulation of UMP biosynthesis. Annu. Rev. Biochem. 49:253-279. [DOI] [PubMed] [Google Scholar]
- 29.Juarez, O., G. Guerra, F. Martinez, and J. P. Pardo. 2004. The mitochondrial respiratory chain of Ustilago maydis. Biochim. Biophys. Acta 1658:244-251. [DOI] [PubMed] [Google Scholar]
- 30.Kämper, J., R. Kahmann, M. Bölker, L.-J. Ma, T. Brefort, B. J. Saville, F. Banuett, J. W. Kronstad, S. E. Gold, O. Muller, M. H. Perlin, H. A. B. Wosten, R. de Vries, J. Ruiz-Herrera, C. G. Reynaga-Pena, K. Snetselaar, M. McCann, J. Perez-Martin, M. Feldbrügge, C. W. Basse, G. Steinberg, J. I. Ibeas, W. Holloman, P. Guzman, M. Farman, J. E. Stajich, R. Sentandreu, J. M. Gonzalez-Prieto, J. C. Kennell, L. Molina, J. Schirawski, A. Mendoza-Mendoza, D. Greilinger, K. Münch, N. Rössel, M. Scherer, M. Vranes, O. Ladendorf, V. Vincon, U. Fuchs, B. Sandrock, S. Meng, E. C. H. Ho, M. J. Cahill, K. J. Boyce, J. Klose, S. J. Klosterman, H. J. Deelstra, L. Ortiz-Castellanos, W. Li, P. Sanchez-Alonso, P. H. Schreier, I. Häuser-Hahn, M. Vaupel, E. Koopmann, G. Friedrich, H. Voss, T. Schlüter, J. Margolis, D. Platt, C. Swimmer, A. Gnirke, F. Chen, V. Vysotskaia, G. Mannhaupt, U. Güldener, M. Münsterkötter, D. Haase, M. Oesterheld, H.-W. Mewes, E. W. Mauceli, D. DeCaprio, C. M. Wade, J. Butler, S. Young, D. B. Jaffe, S. Calvo, C. Nusbaum, J. Galagan, and B. W. Birren. 2006. Insights from the genome of the biotrophic fungal plant pathogen Ustilago maydis. Nature 444:97-101. [DOI] [PubMed] [Google Scholar]
- 31.Knecht, W., J. Henseling, and M. Löffler. 2000. Kinetics of inhibition of human and rat dihydroorotate dehydrogenase by atovaquone, lawsone derivatives, brequinar sodium and polyporic acid. Chem. Biol. Interact. 124:61-76. [DOI] [PubMed] [Google Scholar]
- 32.Knecht, W., and M. Löffler. 1998. Species-related inhibition of human and rat dihydroorotate dehydrogenase by immunosuppressive isoxazol and cinchoninic acid derivatives. Biochem. Pharmacol. 56:1259-1264. [DOI] [PubMed] [Google Scholar]
- 33.Kronstad, J. W., and S. A. Leong. 1989. Isolation of two alleles of the b locus of Ustilago maydis. Proc. Natl. Acad. Sci. USA 86:978-982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lester, R. L., and F. L. Crane. 1959. The natural occurrence of coenzyme Q. and related compounds. J. Biol. Chem. 234:2169-2175. [PubMed] [Google Scholar]
- 35.Liti, G., and E. J. Louis. 2005. Yeast evolution and comparative genomics. Annu. Rev. Microbiol. 59:135-153. [DOI] [PubMed] [Google Scholar]
- 36.Liu, S., E. A. Neidhardt, T. H. Grossman, T. Ocain, and J. Clardy. 2000. Structures of human dihydroorotate dehydrogenase in complex with antiproliferative agents. Structure 8:25-33. [DOI] [PubMed] [Google Scholar]
- 37.Löffler, M., L. D. Fairbanks, E. Zameitat, A. M. Marinaki, and H. A. Simmonds. 2005. Pyrimidine pathways in health and disease. Trends Mol. Med. 11:430-437. [DOI] [PubMed] [Google Scholar]
- 38.Löffler, M., J. Jöckel, G. Schuster, and C. Becker. 1997. Dihydroorotat-ubiquinone oxidoreductase links mitochondria in the biosynthesis of pyrimidine nucleotides. Mol. Cell. Biochem. 174:125-129. [PubMed] [Google Scholar]
- 39.Loftus, B. J., E. Fung, P. Roncaglia, D. Rowley, P. Amedeo, D. Bruno, J. Vamathevan, M. Miranda, I. J. Anderson, J. A. Fraser, J. E. Allen, I. E. Bosdet, M. R. Brent, R. Chiu, T. L. Doering, M. J. Donlin, C. A. D'Souza, D. S. Fox, V. Grinberg, J. Fu, M. Fukushima, B. J. Haas, J. C. Huang, G. Janbon, S. J. Jones, H. L. Koo, M. I. Krzywinski, J. K. Kwon-Chung, K. B. Lengeler, R. Maiti, M. A. Marra, R. E. Marra, C. A. Mathewson, T. G. Mitchell, M. Pertea, F. R. Riggs, S. L. Salzberg, J. E. Schein, A. Shvartsbeyn, H. Shin, M. Shumway, C. A. Specht, B. B. Suh, A. Tenney, T. R. Utterback, B. L. Wickes, J. R. Wortman, N. H. Wye, J. W. Kronstad, J. K. Lodge, J. Heitman, R. W. Davis, C. M. Fraser, and R. W. Hyman. 2005. The genome of the basidiomycetous yeast and human pathogen Cryptococcus neoformans. Science 307:1321-1324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.McRobert, L., and G. A. McConkey. 2002. RNA interference (RNAi) inhibits growth of Plasmodium falciparum. Mol. Biochem. Parasitol. 119:273-278. [DOI] [PubMed] [Google Scholar]
- 41.Moore, P. D. 1975. Radiation-sensitive pyrimidine auxotrophs of Ustilago maydis. I. Isolation and characterization of mutants. Mutat. Res. 28:355-366. [DOI] [PubMed] [Google Scholar]
- 42.Moore, P. D. 1975. Radiation-sensitive pyrimidine auxotrophs of Ustilago maydis. II. A study of repair mechanisms and UV recovery in pyr I. Mutat. Res. 28:367-380. [DOI] [PubMed] [Google Scholar]
- 43.Nagy, M., F. Lacroute, and D. Thomas. 1992. Divergent evolution of pyrimidine biosynthesis between anaerobic and aerobic yeasts. Proc. Natl. Acad. Sci. USA 89:8966-8970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Nakai, K., and P. Horton. 1999. PSORT: a program for detecting sorting signals in proteins and predicting their subcellular localization. Trends Biochem. Sci. 24:34-36. [DOI] [PubMed] [Google Scholar]
- 45.Nara, T., T. Hshimoto, and T. Aoki. 2000. Evolutionary implications of the mosaic pyrimidine-biosynthetic pathway in eukaryotes. Gene 257:209-222. [DOI] [PubMed] [Google Scholar]
- 46.Nørager, S., K. F. Jensen, O. Björnberg, and S. Larsen. 2002. E. coli dihydroorotate dehydrogenase reveals structural and functional distinctions between different classes of dihydroorotate dehydrogenases. Structure 10:1211-1223. [DOI] [PubMed] [Google Scholar]
- 47.Ramasarma, T. 1985. Natural occurrence and distribution of coenzyme Q, p. 67-81. In G. Lenaz (ed.), Coenzyme Q. John Wiley, London, United Kingdom.
- 48.Rawls, J., W. Knecht, K. Diekert, R. Lill, and M. Löffler. 2000. Requirements for the mitochondrial import and localization of dihydroorotate dehydrogenase. Eur. J. Biochem. 267:2079-2087. [DOI] [PubMed] [Google Scholar]
- 49.Schauwecker, F., G. Wanner, and R. Kahmann. 1995. Filament-specific expression of a cellulase gene in the dimorphic fungus Ustilago maydis. Biol. Chem. Hoppe-Seyler 376:617-625. [DOI] [PubMed] [Google Scholar]
- 50.Schulz, B., F. Banuett, M. Dahl, R. Schlesinger, W. Schäfer, T. Martin, I. Herskowitz, and R. Kahmann. 1990. The b alleles of U. maydis, whose combinations program pathogenic development, code for polypeptides containing a homeodomain-related motif. Cell 60:295-306. [DOI] [PubMed] [Google Scholar]
- 51.Spanos, A., N. Kanuga, D. W. Holden, and G. R. Banks. 1992. The Ustilago maydis pyr3 gene: sequence and transcriptional analysis. Gene 117:73-79. [DOI] [PubMed] [Google Scholar]
- 52.Takashima, E., D. K. Inaoka, A. Osanai, T. Nara, M. Odaka, T. Aoki, K. Inaka, S. Harada, and K. Kita. 2002. Characterization of the dihydroorotate dehydrogenase as a soluble fumarate reductase in Trypanosoma cruzi. Mol. Biochem. Parasitol. 122:189-200. [DOI] [PubMed] [Google Scholar]
- 53.Taylor, M. L., W. H. Taylor, D. F. Eames, and C. D. Taylor. 1971. Biosynthetic dihydroorotate dehydrogenase from Lactobacillus bulgaricus. J. Bacteriol. 105:1015-1027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Tsai, A. L., J. S. Olson, and G. Palmer. 1987. The kinetics of reoxidation of yeast complex III. An evaluation of the Q-cycle. J. Biol. Chem. 262:8677-8684. [PubMed] [Google Scholar]
- 55.Ullrich, A., W. Knecht, M. Fries, and M. Löffler. 2001. Recombinant expression of N-terminal truncated mutants of the membrane bound mouse, rat and human flavoenzyme dihydroorotate dehydrogenase. A versatile tool to rate inhibitor effects? Eur. J. Biochem. 268:1861-1868. [PubMed] [Google Scholar]
- 56.Yarranton, G. T., P. D. Moore, and A. Spanos. 1976. The influence of DNA binding protein on the substrate affinities of DNA polymerase from Ustilago maydis: one polymerase implicated in both DNA replication and repair. Mol. Gen. Genet. 145:215-218. [DOI] [PubMed] [Google Scholar]
- 57.Zameitat, E., Z. Gojkovic, W. Knecht, J. Piskur, and M. Löffler. 2006. Biochemical characterization of recombinant dihydroorotate dehydrogenase from the opportunistic pathogenic yeast Candida albicans. FEBS J. 273:3183-3191. [DOI] [PubMed] [Google Scholar]
- 58.Zameitat, E., W. Knecht, J. Piskur, and M. Löffler. 2004. Two different dihydroorotate dehydrogenases from yeast Saccharomyces kluyveri. FEBS Lett. 568:129-134. [DOI] [PubMed] [Google Scholar]