Abstract
Stable isotope probing (SIP) of nucleic acids is a powerful tool that can identify the functional capabilities of noncultivated microorganisms as they occur in microbial communities. While it has been suggested previously that nucleic acid SIP can be performed with 15N, nearly all applications of this technique to date have used 13C. Successful application of SIP using 15N-DNA (15N-DNA-SIP) has been limited, because the maximum shift in buoyant density that can be achieved in CsCl gradients is approximately 0.016 g ml−1 for 15N-labeled DNA, relative to 0.036 g ml−1 for 13C-labeled DNA. In contrast, variation in genome G+C content between microorganisms can result in DNA samples that vary in buoyant density by as much as 0.05 g ml−1. Thus, natural variation in genome G+C content in complex communities prevents the effective separation of 15N-labeled DNA from unlabeled DNA. We describe a method which disentangles the effects of isotope incorporation and genome G+C content on DNA buoyant density and makes it possible to isolate 15N-labeled DNA from heterogeneous mixtures of DNA. This method relies on recovery of “heavy” DNA from primary CsCl density gradients followed by purification of 15N-labeled DNA from unlabeled high-G+C-content DNA in secondary CsCl density gradients containing bis-benzimide. This technique, by providing a means to enhance separation of isotopically labeled DNA from unlabeled DNA, makes it possible to use 15N-labeled compounds effectively in DNA-SIP experiments and also will be effective for removing unlabeled DNA from isotopically labeled DNA in 13C-DNA-SIP applications.
The vast majority of microorganisms continue to resist cultivation in the laboratory, and even when cultivation can be achieved, the traits expressed by a microorganism in culture may not be representative of those expressed when the organism is present in its natural habitat. Stable isotope probing (SIP) of nucleic acids offers a means to study the metabolic activity of microorganisms as they occur in the environment and to characterize novel organisms that may have escaped detection previously (35). SIP has been used in a range of applications to follow the assimilation of isotopically labeled compounds into the nucleic acids of microbial communities (for a review, see references 6, 9, 22, 28, 36, and 42). While SIP using DNA (DNA-SIP) has been performed primarily with 13C-labeled compounds, it is also possible to use 15N to selectively label DNA from pure cultures, as was originally demonstrated by Meselson and Stahl (29).
While nucleic acid SIP provides a powerful tool for characterizing microbial activity under in situ conditions, the method has notable limitations (5, 19, 22, 25, 26, 35, 36). One consideration that can complicate the interpretation of DNA-SIP experiments is the effect that genome G+C content has on the buoyant density of DNA (39). This relationship between DNA G+C content and buoyant density in CsCl gradients is described by the equation ρ = (0.098[G+C]) + 1.66, where ρ represents density in g ml−1 and [G+C] is the mole fraction G+C content (1). Thus, the native buoyant density of DNA in a CsCl gradient can differ by as much as 0.05 g ml−1 over the range of genome G+C contents that can occur in complex communities. This natural variation in DNA density explains why DNA from complex communities can occur as a “smear” extending over a range of densities in a CsCl gradient (35). Since fully 13C-labeled DNA increases in buoyant density by 0.036 g ml−1 relative to unlabeled DNA (1), there arises the very real possibility that “heavy” fractions will contain isotopically labeled DNA, as well as unlabeled DNA, from organisms with a high genome G+C content (19, 25, 36). This problem can be compounded if the incubation conditions used for DNA-SIP experiments result in the unintended growth of high-G+C organisms as a result of changes in soil moisture, temperature, bottle effects, or other artifacts associated with substrate amendment.
While the G+C problem is a potential concern in any DNA-SIP experiment, it represents a fundamental obstacle to the successful application of DNA-SIP with 15N as an isotopic label. The change in DNA buoyant density for fully 15N-labeled DNA (0.016 g ml−1 [1]) is less than half of the change in density that occurs naturally as a result of natural variation in the genome G+C content. Thus, there is a considerable potential for 15N-labeled DNA to cooccur with unlabeled DNA. This problem has been emphasized in recent attempts to use 15N-labeled compounds in DNA-SIP that have found significant overlap in the buoyant densities of 15N-labeled and unlabeled DNA in CsCl gradients (2, 3), but the extent to which variation in the genome G+C content poses a problem in complex communities has not been systematically examined.
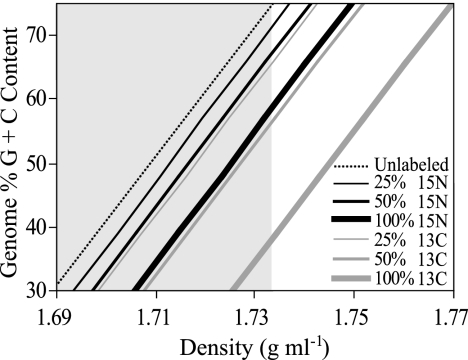
To illustrate the nature of this problem, calculations were performed to examine the theoretical effects of genome G+C content and isotope incorporation on DNA density in a CsCl gradient (Fig. 1). The buoyant density of unlabeled DNA with a G+C content between 30% and 80% was calculated to range from 1.689 to 1.738 g ml−1, respectively (Fig. 1). It was calculated that 100% 13C-labeled DNA which has less than 40% G+C content will have the same buoyant density as unlabeled DNA which has more than 65% G+C content (Fig. 1). As expected, considerably more overlap between unlabeled and labeled DNA was calculated to occur for 100% 15N-labeled DNA and for DNA that is only partially labeled (Fig. 1). For example, Fig. 1 predicts that 100% 15N-labeled DNA from an organism with a 51% genome G+C content, such as Escherichia coli, will have the same buoyant density as unlabeled DNA from an organism with a 67% genome G+C content, such as Pseudomonas aeruginosa.
FIG. 1.
Expected relationship between genome G+C content and buoyant density in a CsCl gradient represented for unlabeled DNA and DNA that is partially or completely labeled with either 13C or 15N. The shaded region of the chart represents the range of densities over which unlabeled DNA would be expected to occur based on biologically meaningful values of genome G+C content (30% to 80% G+C content).
To address the problem that variation in DNA G+C content poses to DNA-SIP experiments, we have developed a method that makes it possible to disentangle the effects of isotope incorporation and genome G+C content on DNA buoyant density in CsCl gradients. As illustrated in Fig. 1, isotopically labeled DNA must have a lower genome G+C content than any unlabeled DNA that cooccurs in a given primary gradient fraction. Thus, this difference in genome G+C content can be used to separate labeled DNA from unlabeled DNA. This is accomplished by performing secondary centrifugation of gradient fractions in the presence of an intercalating agent, such as bis-benzimide, which alters DNA buoyant density in a G+C-dependent manner. Bis-benzimide intercalates into DNA at A-T base pairs, altering the hydration state of DNA and causing a decrease in buoyant density that is inversely proportional to the DNA G+C content (12). This approach makes it possible to perform DNA-SIP experiments with 15N-labeled substrates and should also prove useful during 13C-DNA-SIP experiments as a quality control step to remove unlabeled DNA from isotopically labeled “heavy” fractions. The objective of this study was to develop and verify a method to enable DNA-SIP of complex communities with 15N-labeled compounds. To accomplish this goal, it was necessary to do the following: (i) develop a protocol that maximizes the resolution of CsCl density gradient fractionation, (ii) characterize the extent of the problem that variation in genome G+C content poses for DNA-SIP experiments involving complex communities, and (iii) develop a technique to disentangle the effects of genome G+C content and isotope incorporation on DNA buoyant density.
MATERIALS AND METHODS
CsCl gradient calculations.
The time (in hours) required for a given particle to reach equilibrium in a density gradient can be approximated by the following expression: [1.13 × 1014 × β° × (ρ − 1)]/(v4 × r2 × S), where ρ is particle density in g ml−1, β° is the density gradient proportionality constant (1.14 × 109 for 1.7 g ml−1 CsCl at 20°C), v is rotor velocity in rpm, r is the expected radius that the particle will occupy at equilibrium, and S is the sedimentation coefficient of the particle (1). It is apparent that the time required for a particle to reach equilibrium can be reduced greatly by increasing the rotor velocity. However, higher rotor speeds result in steeper density gradients, which are not optimal for separating particles having small differences in density. Thus, it is important to consider the effect of the DNA molecular weight on the time required to reach equilibrium. We used the above expression to calculate the impact that the DNA molecular weight has on the time required for a particle of 1.71 g ml−1 to reach equilibrium in a CsCl density gradient formed at 55,000 rpm (164,000 × g maximum) in a TLA110 rotor (see Fig. S1 in the supplemental material). The sedimentation coefficient of DNA was estimated as 2.8 + (0.00834 × M0.479), where M is the molecular weight of a linear DNA fragment (43). Based on the results of these calculations, we chose to use DNA of no less than 4,000 bp for all CsCl gradient experiments.
CsCl gradient formation.
High-molecular-weight genomic DNA for use in CsCl gradients was extracted from E. coli (ATCC 9637) and P. aeruginosa (ATCC 10145) by spooling as described previously (40). 15N labeling of E. coli DNA was accomplished by growth in minimal M9 medium (40) to which either NH4Cl or 98 atom% 15NH4Cl (Aldrich) was added in defined proportions to obtain different amounts of 15N label incorporation into DNA. Cultures were transferred twice in medium containing the desired proportion of 15NH4Cl, and the inocula were kept small (1/100 volume) to minimize 14N carryover from starter cultures.
Primary CsCl gradients were formed by filling 4.7-ml polyallomer Optiseal tubes (Beckman) with 4.3 ml of 1.762-g ml−1 CsCl in gradient buffer (15 mM Tris-HCl, 15 mM KCl, 15 mM EDTA, pH 8.0) and 0.45 ml of DNA in TE buffer (50 mM Tris-HCl, 15 mM EDTA, pH 8.0). CsCl concentrations were determined by using a Reichert AR200 handheld digital refractometer. The CsCl concentration was calculated from the refractive index using the equation ρ = anc − b, as described previously (1), where ρ is the density in g ml−1, nc is the corrected refractive index, and a and b are coefficients of values 10.927 and 13.593, respectively, for CsCl at 20°C. The corrected refractive index of CsCl in gradient buffer was calculated as nc = nobserved − (nbuffer − nwater), where nbuffer is the refractive index of the gradient buffer and nwater is that of water. Unless explicitly stated, CsCl gradients were loaded with 10 μg of DNA. Tubes were balanced and then sealed and mixed to obtain a homogeneous CsCl density of 1.69 g ml−1. Centrifugation was carried out for 66 h at 55,000 rpm (164,000 × g maximum) and 20°C in an Optima Max-E tabletop ultracentrifuge (Beckman-Coulter) equipped with a TLA110 rotor.
Secondary CsCl gradients were used to disentangle the effects of isotope incorporation from genome G+C content. These secondary CsCl gradients were prepared as described above with the exception that 8 μl of 10-mg/ml bis-benzimide (Hoechst no. 33258; Sigma-Aldrich) was added to the DNA sample.
Gradient fractionation.
Immediately following centrifugation, a fraction recovery system (Beckman) was used to collect 45 fractions of 100 μl from each CsCl gradient. The fraction collection method has been described previously (25, 26) with the exception that in the current experiment, 4.7-ml centrifuge tubes and 100-μl fractions were used to enhance the resolution of gradient fractionation. Fractions were displaced by using a syringe pump (KD Scientific) to dispense mineral oil into the tops of sealed tubes at a rate of 200 μl min−1. Fractions were collected through a side port needle inserted through the bottom of the tube. The density of each fraction was determined immediately by measurement of the refractive index using an AR200 digital refractometer (Reichert) which was modified to allow measurement from 5-μl volumes by covering the prism surface with a mask fashioned from black electrical tape with a 1.5-mm-diameter hole. CsCl was removed from DNA by ethanol precipitation and resuspended in 25 μl of 50 mM Tris-HCl, pH 8.0, and stored at −20°C.
Quantification of DNA in gradient fractions.
The distribution of DNA in primary CsCl gradients was determined by using the Quant-iT PICO Green dsDNA assay (Invitrogen) as per the manufacturer's instructions. A total of 5 μl of purified DNA from each fraction was used in each assay, and the assays were carried out in optically clear 96-well plates (Costar); fluorescent intensity was quantified by using a Fluor-S Multimager (Bio-Rad). Secondary CsCl gradients generally contained less DNA than primary gradients, and so quantitative PCR was used to determine the number of 16S rRNA genes in each fraction of these gradients. Quantitative PCR was conducted with primers Bact519F (5′-CAG CMG CCG CGG TAA NWC-3′) and Bact907R (5′-CCG TCA ATT CMT TTR AGT T-3′), which target bacterial 16S rRNA genes (41). Reactions were carried out in 50-μl volumes containing 1 μl of template DNA with each primer at a concentration of 0.3 μM, each deoxynucleoside triphosphate at a concentration of 50 μM, and 1× iTaq SYBR Green Supermix with ROX (Bio-Rad). Each PCR consisted of a 50°C hold for 2 min and a 95°C hold for 10 min, followed by 40 cycles of 15 s at 95°C and 60 s at 60°C. Reactions were carried out in an Applied Biosystems 7900 HT sequence detection system (ABI). E. coli 16S rRNA gene sequences were amplified by PCR and gel purified for use as a standard to relate threshold cycle values to the number of 16S rRNA genes present in each reaction.
Analysis of soil community DNA.
A soil sample was collected from a plot on Caldwell Field (Ithaca, NY) that has been maintained as a fallow for more than 30 years. DNA was extracted from four samples of 0.25 g soil using the UltraClean soil DNA extraction kit (MoBio, Inc.) as per the manufacturer's instructions, and these DNA extracts were subsequently pooled. DNA was further purified by electrophoresis through a 1% low-melting-temperature agarose gel to remove DNA fragments smaller than 4 kbp. DNA of greater than 4 kbp was excised from the gel, agarose removed by digestion with agarase (New England Biolabs) as per the manufacturer's instructions, and DNA obtained by ethanol precipitation. Analysis of a subsample of the DNA extract by Pico Green assay (Invitrogen) revealed total recovery of 1.8 μg of DNA per g of soil. This DNA sample was equilibrated in a primary CsCl gradient as described above, and the gradient was fractionated and gradient fractions analyzed by 16S rRNA gene terminal restriction fragment length polymorphism (T-RFLP) analysis.
T-RFLP analysis of 16S rRNA genes was carried out using the PCR primers Bact8F (5′-AGA GTT TGA TCM TGG CTC AG-3′), labeled at the 5′ end with the dye 6-carboxy-fluorescein, and the primer Univ1390R (5′-GAC GGG CGG TGT GTA CAA-3′) (15). Reactions were carried out in 100-μl volumes containing 2 μl of template DNA with the 6-carboxy-fluorescein-labeled primer at a concentration of 0.5 μM and unlabeled primer at 0.3 μM, each deoxynucleoside triphosphate at a concentration of 50 μM, 2.5 mM MgCl2, 5 U of AmpliTaq Gold DNA polymerase (Applied Biosystems, Foster City, CA), and 1× PCR buffer (supplied with the Taq enzyme). Each PCR consisted of a 95°C hold for 5 min followed by 35 cycles of 45 s at 95°C, 30 s at 55°C, and 30 s at 72°C. Following amplification, PCR products were desalted using Micro Bio-Spin P-30 Tris chromatography columns (Bio-Rad) and concentrated using an evaporative centrifuge (Eppendorf). PCR products were resuspended in 50 mM Tris-HCl, pH 8.0, and 250 to 400 ng of this DNA was digested with MspI (New England Biolabs) in 30-μl reaction volumes as per the manufacturer's instructions; the enzyme was subsequently inactivated by incubation at 65°C for 20 min. The digested PCR products were desalted and concentrated again and then resuspended in formamide loading buffer and resolved on an Applied Biosystems Automated 3730 DNA analyzer.
T-RFLP data were used to determine the density profiles of individual terminal restriction fragments (TRFs) in primary CsCl gradients. In addition, to determine whether genome G+C content influences the distribution of community DNA in CsCl gradients, overall similarity in T-RFLP patterns was determined with respect to gradient density. Similarity in T-RFLP profiles was determined by using the Sorenson index of similarity: S = (2 × ab)/(a + b), where a and b are the numbers of TRFs in any two samples and ab is the number of TRFs shared between those samples (23).
RESULTS
Precision of gradient fractionation.
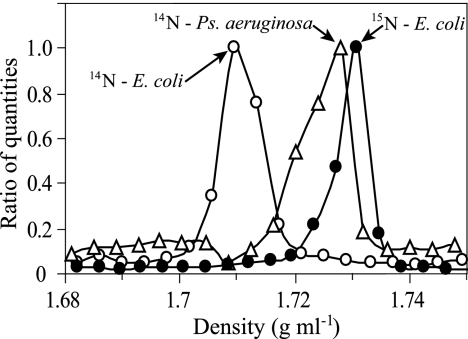
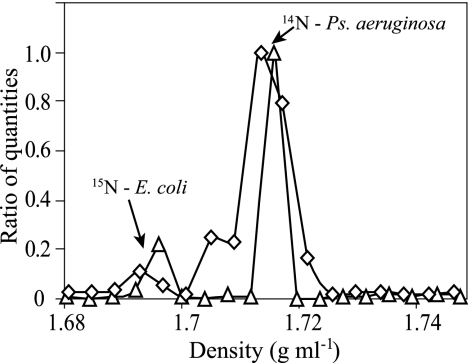
The buoyant density of unlabeled E. coli DNA in CsCl was determined to be 1.707 ± 0.001 g ml−1 (mean ± standard error [SE]; n = 4), which agreed with expectations (1, 38). In addition, 98% atom enriched 15N-DNA from E. coli was determined to have a buoyant density of 1.724 ± 0.002 g ml−1 (mean ± SE; n = 5), corresponding to a density change of 0.017 ± 0.002 g ml−1 (mean ± SE; n = 4) in response to 15N labeling (Fig. 2), which also matched predictions (1). The analytical error for determining DNA buoyant density was 0.0025 ± 0.0004 g ml−1 (mean ± SE; n = 15). As expected, buoyant density of DNA changed linearly in response to the degree of 15N incorporation (see Fig. S2 in the supplemental material). Differences between observed buoyant densities and fitted values (see Fig. S2 in the supplemental material) were used to calculate 95% confidence intervals of ±0.001 g ml−1 that define predictions for DNA atom% 15N incorporation made from buoyant density measurements. This result indicates that it is possible to use a shift in DNA buoyant density to predict the degree of 15N incorporation to within ±5 atom% 15N and that a particular DNA species must have at least 10 atom% 15N to be conclusively resolved from its unlabeled counterpart. When the distribution of completely 15N-labeled E. coli DNA in a CsCl gradient was compared to that of unlabeled E. coli DNA, it was apparent that 100% 15N-labeled DNA is easily resolved from its unlabeled counterpart (Fig. 2). However, the buoyant density of unlabeled P. aeruginosa DNA was 1.729 ± 0.001 g ml−1 (mean ± SE; n = 4), confirming that the buoyant density of unlabeled DNA with a 67% G+C content overlaps considerably with that of DNA that has a 51% G+C content and is 100% 15N labeled (Fig. 2).
FIG. 2.
Effect of 15N enrichment and genome G+C content on DNA distribution in CsCl density gradients. Lines indicate the density of unlabeled E. coli DNA (○), 100% 15N-labeled E. coli DNA (•), or unlabeled P. aeruginosa DNA (▵). The genome G+C contents of E. coli and P. aeruginosa are approximately 51% and 67%, respectively.
Effect of genome G+C content on buoyant density of DNA from complex communities.
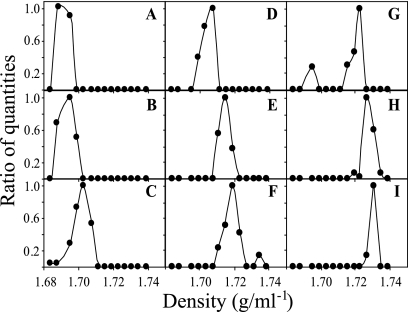
An experiment was conducted to determine whether DNA G+C content affects the buoyant density of complex DNA mixtures in the same way that it affects the buoyant density of DNA obtained from pure cultures. DNA extracted from soil was equilibrated in a density gradient, and T-RFLP analysis of 16S rRNA genes was conducted on DNA from each gradient fraction. The fluorescence intensity of each TRF was then plotted as a function of its buoyant density. A total of 1,415 fluorescence peaks were observed across all gradient fractions, corresponding to 322 unique TRFs. By plotting of the fluorescence intensities of TRFs in relation to gradient density, it was found that TRFs have distinct density profiles, consistent with the expectation that genome G+C content is an independent variable from TRF size (Fig. 3). TRFs could be identified that had distinct buoyant densities, ranging between 1.687 and 1.731 g ml−1 (Fig. 3). Individual TRFs in many cases were observed to have multiple peaks in fluorescence intensity with respect to buoyant density (e.g., Fig. 3F and G). This result is consistent with the expectation that different microorganisms that share a TRF can have different genome G+C contents and different buoyant densities. By using buoyant density to identify TRFs shared by multiple organisms, it was possible to identify 731 distinct microorganisms in the soil sample. To further confirm that differences in genome G+C content influenced the distribution of DNA in density gradients, pairwise similarity comparisons were made between T-RFLP profiles from different gradient fractions with respect to fraction density (see Fig. S3 in the supplemental material). The similarity between T-RFLP profiles was observed to decrease as the difference in density between fractions increased (see Fig. S3 in the supplemental material). The similarity between T-RFLP profiles of adjacent fractions (separated by less than 0.004 g ml−1) was significantly greater than that between fractions separated by an average of 0.008 g ml−1 (P < 0.05, Mann-Whitney U test) (see Fig. S3 in the supplemental material). Thus, the taxonomic composition of gradient fractions was shown to vary as a function of gradient density.
FIG. 3.
Fluorescence intensities of TRFs from soil DNA equilibrated in a CsCl density gradient and subjected to T-RFLP of 16S rRNA genes in gradient fractions. TRFs were selected to demonstrate that the buoyant densities of individual TRFs can vary between 1.69 g ml−1 and 1.73 g ml−1 in density gradients. Fluorescence intensity is shown as a ratio of the maximum value of fluorescence intensity that was observed for each TRF. The TRFs shown for panels A through I are 102 bp, 242 bp, 512 bp, 500 bp, 372 bp, 288 bp, 144 bp, 121 bp, and 177 bp, respectively.
Disentangling isotope incorporation from genome G+C content.
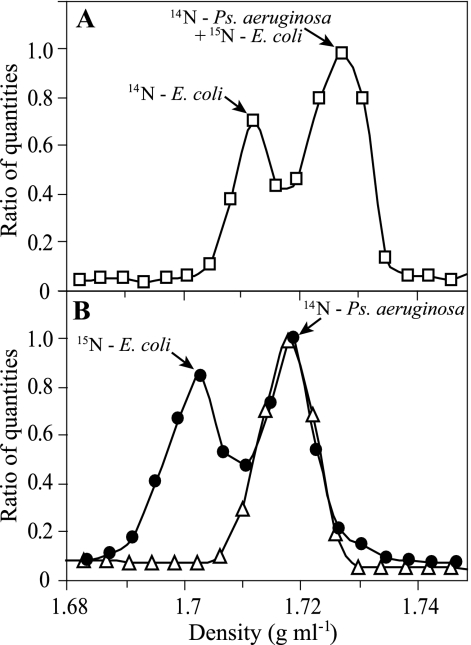
In a conventional CsCl gradient, there was significant overlap in buoyant density between unlabeled DNA with a 67% G+C content and 15N-labeled DNA with a 51% G+C content whether the DNA samples were run in separate gradients (Fig. 2) or were combined in a single gradient (Fig. 4A). When combined in a CsCl gradient containing bis-benzimide, however, these two types of DNA were easily resolved (Fig. 4B). In the presence of bis-benzimide, the buoyant density of P. aeruginosa DNA was 1.716 ± 0.001 g ml−1 (mean ± SE; n = 4), while that of 15N-labeled E. coli DNA was 1.701 ± 0.004 g ml−1 (mean ± SE; n = 4). Thus, bis-benzimide caused a 0.023 ± 0.002-g ml−1 (mean ± SE; n = 4) reduction in the buoyant density of DNA with a 51% G+C content and a 0.013 ± 0.001-g ml−1 (mean ± SE; n = 4) reduction in the density of DNA with a 67% G+C content (Fig. 2 and 4).
FIG. 4.
Ability of bis-benzimide to disentangle effects of stable isotope incorporation and genome G+C content on DNA density in CsCl gradients. (A) In the absence of bis-benzimide, it is impossible to resolve 100% 15N-labeled E. coli DNA from unlabeled P. aeruginosa DNA in a mixture of unlabeled E. coli DNA, 100% 15N-labeled E. coli DNA, and unlabeled P. aeruginosa DNA (□). (B) In the presence of bis-benzimide, it is possible to resolve 100% 15N-labeled E. coli DNA from unlabeled P. aeruginosa DNA in a mixture of the two (•). The density distribution of a pure sample of unlabeled P. aeruginosa DNA from a second gradient is depicted for reference (▵).
To determine the ability of this technique to resolve small quantities of target DNA in a background of nontarget DNA, mixtures of 10 μg of unlabeled P. aeruginosa DNA and either 2 μg (high) or 200 ng (low) of 15N-labeled E. coli DNA were run in CsCl primary gradients. The DNA from the 1.725-g ml−1 fraction of these primary CsCl gradients was then run in secondary gradients containing bis-benzimide. The total DNA transferred to the secondary gradients was 1.0 μg and 0.8 μg for the high and low samples, respectively, and from these values and the original proportions of labeled and unlabeled DNA loaded on primary gradients, the secondary gradients were expected to contain 167 ng and 17 ng of 15N-labeled E. coli DNA from the high and low samples, respectively. Quantitative PCR was used to determine the number of 16S rRNA genes in each fraction of the secondary gradients (Fig. 5). From these quantitative PCR results, it was possible to detect 232 ng and 22 ng of 15N-labeled DNA in the secondary gradients from the high and low samples, respectively (calculated using the number of 16S rRNA copies detected in the E. coli peak with a molecular weight of 3.1 × 109 and rrn operon copy number of 7 for E. coli genomes (31). This result confirms that effectively no DNA is being lost during gradient fractionation.
FIG. 5.
Secondary CsCl gradients containing bis-benzimide allow detection of small amounts of 15N-labeled DNA in the presence of excess unlabeled DNA. In separate experiments, either 2 μg (▵) or 200 ng (⋄) of 15N-labeled E. coli DNA was mixed with 10 μg of unlabeled P. aeruginosa DNA, and following primary centrifugation, 0.8 to 1.0 μg of this DNA with density of 1.725 g ml−1 was resolved in secondary CsCl gradients containing bis-benzimide. The peaks corresponding to 15N-labeled E. coli DNA and unlabeled P. aeruginosa DNA are labeled for clarity.
DISCUSSION
While 15N has been proposed as a suitable label for nucleic acid SIP experiments (35, 36), the successful use of 15N-DNA-SIP to identify microorganisms that assimilate 15N-labeled compounds in complex communities has not yet been achieved due to technical constraints (2, 3). Experiments with 15N-labeled DNA from pure cultures have demonstrated that it is possible to resolve 15N-labeled DNA from unlabeled DNA provided that label incorporation exceeds 40% (2, 29) but do not address the fundamental obstacle that variation in genome G+C content poses to the application of 15N-DNA-SIP in complex communities. Variation in DNA G+C content also presents a potential concern for the interpretation of 13C-DNA-SIP experiments (21, 25, 36), and this is particularly a problem when less than 100% of the C in DNA has been isotopically labeled (Fig. 1). For example, DNA-SIP experiments can be used to track the movement of isotopically labeled compounds into microbial food webs over time (5, 13, 21, 24, 28, 30, 45), but when incubation times are long or when environmentally relevant amounts of substrate are added to communities, it is likely that partial labeling of nucleic acids will be common (25, 28, 35). Thus, it is desirable to have a method that enables the accurate resolution of partially labeled DNA from unlabeled DNA in mixtures of DNA that vary widely in G+C content. We demonstrate a relatively straightforward approach to resolving this problem which takes advantage of the ability of DNA-intercalating agents, such as bis-benzimide, to change the buoyant density of DNA as a function of G+C content.
Variability of genome G+C content within microbial communities has previously been suggested as a factor that needs to be considered during the interpretation of DNA-SIP experiments (36), but the magnitude of this problem has not before been demonstrated empirically. The use of DNA from pure cultures reveals the extent to which genome G+C content can affect DNA buoyant density (1, 3), and microbial communities have been shown to contain a complex mixture of genomes with a range of G+C contents (11, 12, 32, 33). However, in complex mixtures of DNA (and in the absence of intercalating agents that enhance differences in DNA buoyant density), interactions between DNA molecules could affect the apparent buoyant density of DNA fragments, thereby reducing the ability of DNA G+C content to affect buoyant density. For example, changes in DNA conformation can influence DNA buoyant density in CsCl density gradients, since native and denatured DNA from E. coli can differ in buoyant density by 0.015 g ml−1 and circular and linear forms of the same DNA molecule will also differ in buoyant density (1). Thus, it could be hypothesized that DNA fragments from a complex mixture would be normally distributed within a density gradient, with a mean buoyant density corresponding to the mean G+C content of the community. However, Fig. 3 and Fig. S3 in the supplemental material provide separate lines of evidence that clearly demonstrate that this hypothesis is false. First, individual TRFs with distinct buoyant densities can be identified from soil DNA equilibrated in a density gradient (Fig. 3), and the range of buoyant densities spanned by these TRFs matches the range over which variation in genome G+C content is expected to influence DNA buoyant density based on theoretical predictions (Fig. 1). Second, analysis of T-RFLP profiles from soil DNA equilibrated in a density gradient reveals that the taxonomic composition of gradient fractions varies as a function of gradient density, consistent with the expectation that differences in genome G+C content affect the buoyant densities of DNA fragments within complex mixtures (see Fig. S3 in the supplemental material). The extent of the problem that variation in G+C content poses to DNA-SIP experiments can be easily seen, since the 102-bp TRF in Fig. 3A has a mean buoyant density of approximately 1.69 g ml−1 and if the DNA from this organism was fully 13C labeled it would be expected to have a buoyant density of 1.726 g ml−1, which overlaps considerably with unlabeled DNA from organisms with TRFs of 144 bp, 121 bp, and 177 bp (Fig. 3G, H, and I). These results, when considered in conjunction with the observation that the distribution of G+C contents in a microbial community can change in response to environmental characteristics (11, 12, 32, 33), emphasize the necessity of the proper use of controls in DNA-SIP experiments (22).
Our calculations show that DNA molecular weight is also a concern that must be considered when designing DNA-SIP experiments (see Fig. S1 in the supplemental material). Small DNA fragments below 2 kbp in length are commonly present in DNA preparations that use bead beating to achieve cell lysis (34), and this method of lysis is commonly employed for the extraction of DNA from environmental studies. These small DNA fragments may not reach equilibrium under the conditions typically used for establishing density gradients (see Fig. S1 in the supplemental material). An additional concern is that G+C content can vary locally within a genome, such that the G+C content of individual DNA fragments can vary from the mean genome G+C content. Thus, as a genome is cut into smaller and smaller fragments, the variation between the G+C contents of the individual fragments and the G+C content of the genome will tend to increase (44). The anticipated result would be that the range of densities occupied by DNA from a given genome (i.e., the width of the band in the gradient) would increase as the molecular weight of DNA fragments decreases. Thus, the G+C content of DNA fragments that are smaller than 5 kbp can vary by as much as 13% for a single genome (44). These effects may help to explain why low but detectable amounts of nucleic acids can be recovered throughout density gradients (8, 16, 19, 25, 37). Removal of DNA fragments smaller than 4 kbp (see Fig. S1 in the supplemental material) prior to ultracentrifugation is therefore a reasonable precaution that can potentially increase the resolution of DNA within density gradients.
One common concern associated with nucleic acid SIP experiments is that cross-feeding and trophic cascades can result in the movement of an isotopic label into nucleic acids from nontarget functional groups (5, 35, 36). The isotopic signature of nucleic acids from organisms involved in cross-feeding or secondary consumption should, as a natural consequence of label dilution, be less than that of primary consumers until the isotopic label saturates the community. Thus, by following the incorporation of the isotopic label into the community over time, it is possible to track the movement of the label from a substrate into particular functional groups and then into other components of the soil food web (5, 13, 18, 21). As a result, DNA from isotopically enriched environmental samples can contain a range of isotopic signatures from 0% to 100% label incorporation (28, 30, 45). It is possible to use nucleic acid fingerprinting methods to determine the degree of isotope incorporation by particular microbial groups by fingerprinting gradient fractions from isotopically labeled samples relative to controls (19, 20, 25). However, to identify the organisms that assimilated the label, it is necessary to match TRFs from particular gradient fractions with sequences obtained in clone libraries, a task which can prove difficult with complex communities (4, 7, 17). Also, the possibility that two or more organisms may share a particular TRF (7, 14, 27) can lead to difficulties in interpreting these data. The use of secondary density gradients containing bis-benzimide to purify isotopically labeled DNA from unlabeled DNA should be useful in these applications.
It is important to note that the behavior of DNA in density gradients differs considerably from that of rRNA. rRNA has greater buoyant density and lower molecular weight than DNA. It also has a more narrow range of G+C contents (more than 90% of 16S rRNAs have G+C content between 50% and 60%; personal observation calculated from 24,000 16S rRNA sequences) and extensive regions of secondary and tertiary structure (10). Variability in denaturing conditions during equilibration of rRNA molecules in density gradients can affect rRNA secondary structure, and as a result, rRNA from different species can have buoyant densities that range over 0.08 g ml−1 (19). Thus, isotopically labeled rRNA may be expected to cooccur with unlabeled rRNA under certain circumstances but, unlike the case with DNA, this phenomenon is less likely to be driven by differences in nucleic acid G+C content. Thus, additional experiments and validation would be needed to determine whether the method we describe would be applicable to RNA-SIP applications.
The centrifugation and fractionation conditions that we describe are optimized to resolve DNA molecules that have very small differences in buoyant density. The collection of 100-μl fractions from density gradients allows for increased resolution of buoyant density relative to existing methods and results in highly reproducible density profiles with respect to fraction number (data not shown). In addition, the removal of small DNA fragments prior to centrifugation is a simple step that should theoretically enhance gradient resolution (see Fig. S1 in the supplemental material), but the magnitude of this effect still needs to be confirmed experimentally. These efforts to improve gradient resolution likely contributed to our ability to resolve differences in T-RFLP profiles and the distribution of individual TRFs that result from the effect of genome G+C content on DNA buoyant density in complex communities (Fig. 3). The use of gradient fractionation and T-RFLP fingerprinting of gradient fractions make it possible to identify gradient fractions that contain isotopically labeled DNA even when it cooccurs with unlabeled DNA. Subsequent purification of those target fractions in secondary density gradients containing bis-benzimide will facilitate separation of labeled DNA from unlabeled DNA and enable the identification and characterization of isotopically labeled organisms. This approach will make it possible to effectively use 15N-labeled compounds in DNA-SIP experiments and will be beneficial in a range of 13C-DNA-SIP applications.
Supplementary Material
Acknowledgments
This research was supported by the National Science Foundation under award number MCB-0447586. This research was also supported in part by the National Research Initiative of the USDA Cooperative State Research, Education and Extension Service, grant number 2005-35107-15266.
Footnotes
Published ahead of print on 16 March 2007.
Supplemental material for this article may be found at http://aem.asm.org/.
REFERENCES
- 1.Birnie, G. D., and D. Rickwood (ed.). 1978. Centrifugal separations in molecular and cell biology. Butterworths, Boston, MA.
- 2.Cadisch, G., M. Espana, R. Causey, M. Richter, E. Shaw, J. A. W. Morgan, C. Rahn, and G. D. Bending. 2005. Technical considerations for the use of N-15-DNA stable-isotope probing for functional microbial activity in soils. Rapid Commun. Mass Spectrom. 19:1424-1428. [DOI] [PubMed] [Google Scholar]
- 3.Cupples, A. M., E. A. Shaffer, J. C. Chee-Sanford, and G. K. Sims. DNA buoyant density shifts during 15N-DNA stable isotope probing. Microbiol. Res., in press. [DOI] [PubMed]
- 4.Derakshani, M., T. Lukow, and W. Liesack. 2001. Novel bacterial lineages at the (sub)division level as detected by signature nucleotide-targeted recovery of 16S rRNA genes from bulk soil and rice roots of flooded rice microcosms. Appl. Environ. Microbiol. 67:623-631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.DeRito, C. M., G. M. Pumphrey, and E. L. Madsen. 2005. Use of field-based stable isotope probing to identify adapted populations and track carbon flow through a phenol-degrading soil microbial community. Appl. Environ. Microbiol. 71:7858-7865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Dumont, M. G., and J. C. Murrell. 2005. Stable isotope probing—linking microbial identity to function. Nat. Rev. Microbiol. 3:499-504. [DOI] [PubMed] [Google Scholar]
- 7.Dunbar, J., L. O. Ticknor, and C. R. Kuske. 2001. Phylogenetic specificity and reproducibility and new method for analysis of terminal restriction fragment profiles of 16S rRNA genes from bacterial communities. Appl. Environ. Microbiol. 67:190-197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Freitag, T. E., L. Chang, and J. I. Prosser. 2006. Changes in the community structure and activity of betaproteobacterial ammonia-oxidizing sediment bacteria along a freshwater-marine gradient. Environ. Microbiol. 8:684-696. [DOI] [PubMed] [Google Scholar]
- 9.Friedrich, M. W. 2006. Stable-isotope probing of DNA: insights into the function of uncultivated microorganisms from isotopically labeled metagenomes. Curr. Opin. Biotechnol. 17:59-66. [DOI] [PubMed] [Google Scholar]
- 10.Glotz, C., C. Zwieb, R. Brimacombe, K. Edwards, and H. Kossel. 1981. Secondary structure of the large subunit ribosomal RNA from Escherichia coli, Zea mays chloroplast, and human and mouse mitochondrial ribosomes. Nucleic Acids Res. 9:3287-3306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Holben, W. E., K. P. Feris, A. Kettunen, and J. H. A. Apajalahti. 2004. GC fractionation enhances microbial community diversity assessment and detection of minority populations of bacteria by denaturing gradient gel electrophoresis. Appl. Environ. Microbiol. 70:2263-2270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Holben, W. E., and D. Harris. 1995. DNA-based monitoring of total bacterial community structure in environmental samples. Mol. Ecol. 4:627-631. [DOI] [PubMed] [Google Scholar]
- 13.Hutchens, E., S. Radajewski, M. G. Dumont, I. R. McDonald, and J. C. Murrell. 2004. Analysis of methanotrophic bacteria in Movile Cave by stable isotope probing. Environ. Microbiol. 6:111-120. [DOI] [PubMed] [Google Scholar]
- 14.Kent, A. D., D. J. Smith, B. J. Benson, and E. W. Triplett. 2003. Web-based phylogenetic assignment tool for analysis of terminal restriction fragment length polymorphism profiles of microbial communities. Appl. Environ. Microbiol. 69:6768-6776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Liu, W. T., T. L. Marsh, H. Cheng, and L. J. Forney. 1997. Characterization of microbial diversity by determining terminal restriction fragment length polymorphisms of genes encoding 16S rRNA. Appl. Environ. Microbiol. 63:4516-4522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lu, Y. H., T. Lueders, M. W. Friedrich, and R. Conrad. 2005. Detecting active methanogenic populations on rice roots using stable isotope probing. Environ. Microbiol. 7:326-336. [DOI] [PubMed] [Google Scholar]
- 17.Ludemann, H., I. Arth, and W. Liesack. 2000. Spatial changes in the bacterial community structure along a vertical oxygen gradient in flooded paddy soil cores. Appl. Environ. Microbiol. 66:754-762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lueders, T., R. Kindler, A. Miltner, M. W. Friedrich, and M. Kaestner. 2006. Identification of bacterial micropredators distinctively active in a soil microbial food web. Appl. Environ. Microbiol. 72:5342-5348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lueders, T., M. Manefield, and M. W. Friedrich. 2004. Enhanced sensitivity of DNA- and rRNA-based stable isotope probing by fractionation and quantitative analysis of isopycnic centrifugation gradients. Environ. Microbiol. 6:73-78. [DOI] [PubMed] [Google Scholar]
- 20.Lueders, T., B. Pommerenke, and M. W. Friedrich. 2004. Stable-isotope probing of microorganisms thriving at thermodynamic limits: syntrophic propionate oxidation in flooded soil. Appl. Environ. Microbiol. 70:5778-5786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lueders, T., B. Wagner, P. Claus, and M. W. Friedrich. 2004. Stable isotope probing of rRNA and DNA reveals a dynamic methylotroph community and trophic interactions with fungi and protozoa in oxic rice field soil. Environ. Microbiol. 6:60-72. [DOI] [PubMed] [Google Scholar]
- 22.Madsen, E. L. 2006. The use of stable isotope probing techniques in bioreactor and field studies on bioremediation. Curr. Opin. Biotechnol. 17:92-97. [DOI] [PubMed] [Google Scholar]
- 23.Magurran, A. E. 1988. Ecological diversity and its measurement. Princeton University Press, Princeton, NJ.
- 24.Mahmood, S., G. I. Paton, and J. I. Prosser. 2005. Cultivation-independent in situ molecular analysis of bacteria involved in degradation of pentachlorophenol in soil. Environ. Microbiol. 7:1349-1360. [DOI] [PubMed] [Google Scholar]
- 25.Manefield, M., A. S. Whiteley, R. I. Griffiths, and M. J. Bailey. 2002. RNA stable isotope probing, a novel means of linking microbial community function to phylogeny. Appl. Environ. Microbiol. 68:5367-5373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Manefield, M., A. S. Whiteley, N. Ostle, P. Ineson, and M. J. Bailey. 2002. Technical considerations for RNA-based stable isotope probing: an approach to associating microbial diversity with microbial community function. Rapid Commun. Mass Spectrom. 16:2179-2183. [DOI] [PubMed] [Google Scholar]
- 27.Marsh, T. L., P. Saxman, J. Cole, and J. Tiedje. 2000. Terminal restriction fragment length polymorphism analysis program, a web-based research tool for microbial community analysis. Appl. Environ. Microbiol. 66:3616-3620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.McDonald, I. R., S. Radajewski, and J. C. Murrell. 2005. Stable isotope probing of nucleic acids in methanotrophs and methylotrophs: a review. Org. Geochem. 36:779-787. [Google Scholar]
- 29.Meselson, M. S., and F. W. Stahl. 1958. The replication of DNA in Escherichia coli. Proc. Natl. Acad. Sci. USA 44:671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Morris, S. A., S. Radajewski, T. W. Willison, and J. C. Murrell. 2002. Identification of the functionally active methanotroph population in a peat soil microcosm by stable-isotope probing. Appl. Environ. Microbiol. 68:1446-1453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Neidhardt, F. C., and J. L. Ingraham (ed.). 1987. Escherichia coli and Salmonella typhimurium: cellular and molecular biology. ASM Press, Washington, DC.
- 32.Nusslein, K., and J. M. Tiedje. 1998. Characterization of the dominant and rare members of a young Hawaiian soil bacterial community with small-subunit ribosomal DNA amplified from DNA fractionated on the basis of its guanine and cytosine composition. Appl. Environ. Microbiol. 64:1283-1289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Nusslein, K., and J. M. Tiedje. 1999. Soil bacterial community shift correlated with change from forest to pasture vegetation in a tropical soil. Appl. Environ. Microbiol. 65:3622-3626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ogram, A., G. S. Sayler, and T. Barkay. 1987. The extraction and purification of microbial DNA from sediments. J. Microbiol. Methods 7:57-66. [Google Scholar]
- 35.Radajewski, S., P. Ineson, N. R. Parekh, and J. C. Murrell. 2000. Stable-isotope probing as a tool in microbial ecology. Nature 403:646-649. [DOI] [PubMed] [Google Scholar]
- 36.Radajewski, S., I. R. McDonald, and J. C. Murrell. 2003. Stable-isotope probing of nucleic acids: a window to the function of uncultured microorganisms. Curr. Opin. Biotechnol. 14:296-302. [DOI] [PubMed] [Google Scholar]
- 37.Rangel-Castro, J. I., K. Killham, N. Ostle, G. W. Nicol, I. C. Anderson, C. M. Scrimgeour, P. Ineson, A. Meharg, and J. I. Prosser. 2005. Stable isotope probing analysis of the influence of liming on root exudate utilization by soil microorganisms. Environ. Microbiol. 7:828-838. [DOI] [PubMed] [Google Scholar]
- 38.Rickwood, R. (ed.). 1984. Centrifugation: a practical approach. IRL Press, Washington, DC.
- 39.Rolfe, R., and M. S. Meselson. 1959. The relative homogeneity of microbial DNA. Proc. Natl. Acad. Sci. USA 45:1039-1043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Sambrook, J., and D. W. Russell (ed.). 2001. Molecular cloning: a laboratory manual. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- 41.Stubner, S. 2002. Enumeration of 16S rDNA of Desulfotomaculum lineage 1 in rice field soil by real-time PCR with SybrGreen detection. J. Microbiol. Methods 50:155-164. [DOI] [PubMed] [Google Scholar]
- 42.Whiteley, A. S., M. Manefield, and T. Lueders. 2006. Unlocking the ‘microbial black box’ using RNA-based stable isotope probing technologies. Curr. Opin. Biotechnol. 17:67-71. [DOI] [PubMed] [Google Scholar]
- 43.Young, B. D. 1984. Measurement of sedimentation coefficients and computer simulation of rate-zonal separations. In D. Rickwood (ed.), Centrifugation: a practical approach. IRL Press, Washington, DC.
- 44.Zavala, A., H. Naya, H. Romero, V. Sabbia, R. Piovani, and H. Musto. 2005. Genomic GC content prediction in prokaryotes from a sample of genes. Gene 357:137-143. [DOI] [PubMed] [Google Scholar]
- 45.Ziegler, S. E., P. M. White, D. C. Wolf, and G. J. Thoma. 2005. Tracking the fate and recycling of C-13-labeled glucose in soil. Soil Sci. 170:767-778. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.