Abstract
Biological nitrogen fixation is a fundamental component of the nitrogen cycle and is the dominant natural process through which fixed nitrogen is made available to the biosphere. While the process of nitrogen fixation has been studied extensively with a limited set of cultivated isolates, examinations of nifH gene diversity in natural systems reveal the existence of a wide range of noncultivated diazotrophs. These noncultivated diazotrophs remain uncharacterized, as do their contributions to nitrogen fixation in natural systems. We have employed a novel 15N2-DNA stable isotope probing (5N2-DNA-SIP) method to identify free-living diazotrophs in soil that are responsible for nitrogen fixation in situ. Analyses of 16S rRNA genes from 15N-labeled DNA provide evidence for nitrogen fixation by three microbial groups, one of which belongs to the Rhizobiales while the other two represent deeply divergent lineages of noncultivated bacteria within the Betaproteobacteria and Actinobacteria, respectively. Analysis of nifH genes from 15N-labeled DNA also revealed three microbial groups, one of which was associated with Alphaproteobacteria while the others were associated with two noncultivated groups that are deeply divergent within nifH cluster I. These results reveal that noncultivated free-living diazotrophs can mediate nitrogen fixation in soils and that 15N2-DNA-SIP can be used to gain access to DNA from these organisms. In addition, this research provides the first evidence for nitrogen fixation by Actinobacteria outside of the order Actinomycetales.
Nitrogen fixation is an ancient microbial process which evolved early in the history of our planet and is of central importance to the biosphere. All known forms of life require fixed N for biosynthesis, and microbial N fixation provides the largest natural source of fixed N in the biosphere, accounting for the production of 100 to 290 Tg N yr−1 in terrestrial systems alone (7). Free-living diazotrophs in soils provide the dominant natural source of fixed N in many of these terrestrial systems (7), and yet we still have much to learn about the ecology and evolution of these organisms. Nitrogenase nifH sequences can be divided into 49 different subgroups (49). Twenty-two of these subgroups do not contain any cultivated representatives, and many of the remaining subgroups contain only one or a few members that have been cultivated successfully (49). Surveys of nitrogenase diversity in soil commonly reveal sequence types that correspond to diverse unidentified diazotrophs (6, 32, 33, 40, 45, 47), and these noncultivated diazotrophs, rather than their cultivated cousins, may be the dominant N-fixing organisms in soil systems (13, 33, 38, 44, 47). Currently, noncultivated diazotrophs can be identified only through detection of their nifH gene sequences, and since the phylogeny of the nifH gene does not consistently correspond with organismal phylogeny (36), nifH gene sequences on their own provide limited information with which to identify and characterize novel diazotrophs. 15N2-DNA stable isotope probing (15N2-DNA-SIP) can be used to link particular 16S rRNA genes to the process of nitrogen fixation as it occurs in the soil and should provide a valuable technique for characterizing noncultivated diazotrophs in a range of environments. Through 15N2-DNA-SIP, it may be possible to link 16S rRNA genes from noncultivated diazotrophs to their corresponding nifH genes, and this method may also provide a source of genome fragments that can be used to help to characterize noncultivated diazotrophs, their gene systems, and their ecological significance.
While nucleic acid SIP provides a useful tool for characterizing microbial activity under in situ conditions the method has notable limitations (10, 25, 34, 35). One limitation is the need to add labeled substrates at concentrations that are substantially higher than those typically experienced by cells in situ. Elevated substrate addition is required because cells will assimilate substrates from both native and labeled sources, resulting in the dilution of an isotopic label in the receiving community (34). Another problem encountered when performing SIP experiments is that low in situ growth rates may require prolonged incubations to permit sufficient labeling of nucleic acids. During prolonged incubations, cross-feeding and trophic cascades can result in the movement of an isotopic label into nucleic acids from nontarget functional groups (10, 17, 22, 23, 27, 28, 51). As a result, DNA from isotopically enriched environmental samples can contain a range of isotopic signatures, from 0 to 100% label incorporation. Several strategies have been developed to deal with these issues, and each requires the collection and analysis of gradient fractions and DNA fingerprinting in order to determine the degree of isotope incorporation into DNA from particular microbial groups (21, 24, 25). As a natural consequence of label dilution, the isotopic signature of nucleic acids from organisms involved in cross-feeding or secondary consumption should be less than that of primary consumers unless and until the isotopic label saturates the community (27, 28, 51). Thus, by following the incorporation of the isotopic label into the community over time and in comparison to control treatments that receive unlabeled substrates, it is possible to track the movement of the label from a substrate into particular functional groups and then into other components of the soil food web (10, 21).
There are several reasons why 15N2-DNA-SIP represents an appealing method for examining nitrogen-fixing organisms. First, incubations can be carried out at realistic substrate concentrations, since atmospheric N2 can be completely replaced with simulated air containing 15N2. Second, since nitrogen fixation is inhibited in the presence of mineral forms of nitrogen (5, 9), isotope dilution is likely to be less of a problem with 15N2 than with 13C-labeled substrates. Third, cross-feeding should be less of a problem, since the majority of N fixed by free-living diazotrophs is immobilized in microbial biomass (42), though turnover of microbial biomass may still result in subsequent secondary utilization of 15N-labeled compounds. The challenge associated with 15N-DNA-SIP is that DNA contains less than half as much N as C and thus the change in density associated with 15N-labeled DNA (0.016 g ml−1) (1) is smaller than can be achieved for 13C-labeled DNA. In addition, natural variation in genome G+C content can affect the native buoyant density of DNA and obscure the effects of isotope incorporation (21, 24, 35). The native buoyant density of DNA in CsCl gradients varies by as much as 0.05 g ml−1 over the range of genome G+C contents that occurs in complex communities (15, 30, 37). We have developed a method that makes it possible to disentangle the effects of isotope incorporation and genome G+C content on DNA buoyant density in DNA-SIP experiments, and this method makes it possible to perform 15N-DNA-SIP with complex microbial communities (2). The objective in this study was to apply this 15N-DNA-SIP method to nitrogen-fixing communities in soil by using 15N2 as a labeled substrate, to demonstrate that 15N-DNA-SIP can be used to study natural communities that possess low rates of nitrogen fixation, and to identify and characterize novel diazotrophs from soil that are engaged in nitrogen fixation in situ.
MATERIALS AND METHODS
Soil microcosm experiment.
Soil was collected from a plot on Caldwell Field (Ithaca, NY) that has been maintained as a fallow for more than 30 years. A transect was established across the plot, and three samples were taken at 15-m intervals. Each soil sample consisted of five soil cores (2.5 cm diameter and 5 cm deep) taken within a 1-m2 area and pooled. Within several hours of sampling, soils were sieved to 4 mm and then a 10-g portion of each sample was placed into 25-ml Balch tubes (Bellco Glass). Tubes were sealed and evacuated, and then the atmosphere was replaced with synthetic air containing 20% O2 and 80% N2. All samples were run in duplicate, with half receiving 15N2 containing 99.8 atom% 15N (Isotec) and half receiving unlabeled N2. Tubes were incubated horizontally in the dark at 30°C for 28 days.
Net nitrogen fixation was determined by relating the 15N enrichment of bulk soil for samples receiving 15N2 relative that for to parallel controls which received unlabeled N2. This approach controls for the effects of isotopic fractionation which may occur during incubation as the result of gaseous N loss due to nitrification and denitrification. Soil 15N enrichment was determined by using a Finnigan MAT Delta Plus mass spectrometer (Thermo Electron Corporation) plumbed to a Carlo Erba NC2500 elemental analyzer (CE Instruments) through a Conflo II open split interface for elemental and isotopic composition of solid samples (Thermo Electron Corporation).
DNA was extracted from the soil sample that showed the greatest fixation of 15N2 and from its corresponding control. DNA was extracted from four samples of 0.25 g using the UltraClean Soil DNA extraction kit (MoBio, Inc.) as per the manufacturer's instructions, and these DNA extracts were subsequently pooled. DNA was further purified by electrophoresis through a 1% agarose gel to remove DNA fragments smaller than 4 kbp. DNA of greater than 4 kbp was excised from the gel, agarose removed by digestion with agarase (New England Biolabs) as per the manufacturer's instructions, and DNA obtained by ethanol precipitation as described previously (39). A total of 1.8 μg g−1 of DNA was obtained for the 15N2-enriched soil, and 1.8 μg g−1 of DNA was also obtained for the control soil, as determined by analysis of subsamples with the Quant-iT PICO Green dsDNA assay (Invitrogen) as per the manufacturer's instructions.
CsCl density fractionation.
CsCl gradient fractionation was carried out as described previously (2). Briefly, primary CsCl gradients were formed by filling 4.7-ml polyallomer Optiseal tubes (Beckman) with 4.3 ml of gradient buffer (15 mM Tris-HCl, 15 mM KCl, 15 mM EDTA, pH 8.0) and 0.45 ml of DNA (1.8 μg) in TE buffer (50 mM Tris-HCl, 15 mM EDTA, pH 8.0) to obtain a homogeneous CsCl density of 1.69 g ml−1. Centrifugation was carried out for 66 h at 55,000 rpm (164,000 × g maximum) and 20°C in an Optima Max-E tabletop ultracentrifuge (Beckman-Coulter) equipped with a TLA110 rotor. A fraction recovery system (Beckman) was used to collect 45 fractions of 100 μl from each CsCl gradient, and the density of each fraction was determined by measurement of refractive index using an AR200 digital refractometer (Reichert). DNA of two fractions with buoyant density of 1.727 to 1.733 g ml−1 from primary gradients was resolved in secondary CsCl gradients containing bis-benzimide to disentangle the effects of isotope incorporation from genome G+C content as described elsewhere (2). These secondary CsCl gradients were prepared as described above, with the exception that 8 μl of 10-mg ml−1 bis-benzimide (Hoechst no. 33258; Sigma-Aldrich) was added to the DNA samples during the preparation of gradient media. Bis-benzimide intercalates into DNA at A-T base pairs, altering the hydration state of DNA and causing a decrease in buoyant density that is inversely proportional to DNA G+C content (15). Thus, secondary gradients containing bis-benzimide cause the separation of unlabeled DNA with high G+C content from isotopically labeled DNA of the same buoyant density (2).
Analysis of CsCl gradient fractions.
CsCl was removed from DNA by ethanol precipitation, and DNA was resuspended in 25 μl of 50 mM Tris-HCl, pH 8.0, and stored at −20°C. The distribution of DNA in CsCl gradients was determined by using either the Quant-iT PICO Green dsDNA assay (Invitrogen) or quantitative PCR as described previously (2). Briefly, quantitative PCR was conducted with primers Bact519F (5′-CAG CMG CCG CGG TAA NWC-3′) and Bact907R (5′-CCG TCA ATT CMT TTR AGT T-3′), which target bacterial 16S rRNA genes as described previously (2, 43).
DNA from primary gradient fractions was also characterized by terminal restriction fragment length polymorphism (T-RFLP) analysis of 16S rRNA genes. Bacterial 16S rRNA genes were amplified by PCR using the primer Bact8F (5′-AGA GTT TGA TCM TGG CTC AG-3′), labeled at the 5′ end with the dye 6-carboxy-fluorescein, and the primer Univ1390R (5′-GAC GGG CGG TGT GTA CAA-3′). Reactions were carried out as described previously (2), PCR products were resuspended in 50 mM Tris-HCl (pH 8.0), 250 to 400 ng of this DNA was digested with MspI (New England Biolabs) in 30-μl reaction volumes as per the manufacturer's instructions, and the enzyme was subsequently inactivated by incubation at 65°C for 20 min. The digested PCR products were desalted and concentrated again and then resolved on an Applied Biosystems Automated 3730 DNA analyzer.
Analysis of 16S rRNA and nifH genes.
Clone libraries of 16S rRNA and nifH genes were constructed with DNA obtained from targeted secondary gradient fractions for both enriched and control samples. PCR of 16S rRNA genes was carried out with primers Bact8F and Univ1390R in 50-μl volumes containing 5 μl of template DNA with each primer at a concentration of 0.3 μM, each deoxynucleoside triphosphate at a concentration of 50 μM, 0.05% Tween 20, 2.5 mM MgCl2, 5 U of AmpliTaq Gold DNA polymerase (Applied Biosystems, Foster City, CA), and 1× PCR buffer (supplied with Taq enzyme). Each PCR consisted of a 95°C hold for 5 min, followed by 35 cycles of 45 s at 95°C, 30 s at 55°C, and 30 s at 72°C, and a final extension for 15 min at 72°C. PCR of nifH genes was conducted with primers nifH-b1 (5′-GGC TGC GAT CCC AAG GCT GA-3′) (4) and CDHPnif723R (5′-GAT GTT CGC GCG GCA CGA ADT RNA TSA-3′) (41), and the conditions were as described above except that the annealing temperature used was 60°C. PCR products were cloned into pCR4.0-TOPO using a TOPO-TA cloning kit for sequencing (Invitrogen Corp., Carlsbad, CA). Clones were screened by PCR with primers flanking the cloning site as per the manufacturer's instructions (Invitrogen) to identify inserts of the expected size. Initial sequencing of 16S rRNA genes was carried out with the primer Bact8F, and additional sequencing was carried out with primers for the M13F and M13R priming sites which flank the insertion site on pCR4.0 as per the manufacturer's instructions (Invitrogen). These 16S rRNA gene sequences were screened for the presence of chimeras with the Chimera Check algorithm (8) and by examining base-pair complementarily in 16S rRNA secondary structure.
Phylogenetic analyses were performed by using the programs ARB (2.5 ed.; O. Strunk and W. Ludwig, Department of Microbiology, Technical University of Munich, Munich, Germany, 1997) and PHYLIP 3.64 (J. Felsenstein, Department of Genome Sciences, University of Washington, Seattle, 2005). The 16S rRNA gene sequences were initially aligned by using the ARB automatic aligner and then were verified and corrected manually. The nifH sequences were translated and aligned to the Pfam Fer4_nifH amino acid seed alignment (12). All nifH gene sequences available in GenBank were downloaded and likewise aligned to facilitate accurate estimation of nifH phylogeny relative to existing sequence groups. Regions of ambiguous alignment were identified and excluded from subsequent phylogenetic analyses. A total of 1,334 aligned 16S rRNA gene positions and 105 nifH amino acid positions were used in the construction of phylogenetic trees. Phylogenetic trees were generated by performing parsimony (D. L. Swofford, PAUP, 3.0 ed., 1991; Illinois Natural History Survey, Champaign, IL) and maximum-likelihood analyses (31). During tree construction, the sequence composition of trees and outgroups was varied. Bootstrapping was performed using Phylip with 100 randomizations.
Nucleotide sequence accession numbers.
The nucleotide sequences of the 95 16S rRNA gene clones described in this study and the 34 nifH sequences have been deposited in GenBank under accession numbers EF520944 to EF521038 and EF521039 to EF521072, respectively.
RESULTS
Soil microcosms labeled with 15N2.
Soil microcosms were incubated in synthetic air with atmospheric N2 replaced by 99.8% atom-enriched 15N2. Nitrogen fixation was assessed after 28 days by determining the change in soil 15N enrichment relative to that for controls that were incubated in parallel. The soil had a δ15N content of 2.1‰ ± 0.8‰ (mean ± standard deviation [SD]; n = 3) prior to incubation, and following incubation the δ15N content of soil that received 15N2 was 9.3‰ ± 8.9‰ (mean ± SD; n = 3) and that of control soils was −3.3‰ ± 1.5‰ (mean ± SD; n = 3). Thus, incubation conditions resulted in depletion of soil 15N, but soils receiving 15N2 were significantly enriched relative to controls (P < 0.05, Mann-Whitney U test; 12.6‰ mean difference), indicating significant nitrogen fixation in these samples. The total N content of the soil did not change significantly during the course of incubation and was 2.3 ± 0.6 mg N g−1 dry weight (mean ± SD; n = 9) across all samples. The sample with the greatest amount of nitrogen fixation had a δ15N enrichment of 21.2‰ relative to controls, and this soil sample was selected for analysis by SIP. A nitrogen fixation rate of 0.48 nmol N g−1 day−1 would be required to generate a net δ15N enrichment of 21.2‰ in this experiment, and when integrated over the top 10 cm of soil, this would represent approximately 0.2 kg N ha−1 month−1. In addition, based on the total N content and δ15N content of soil, it is possible to estimate that there was 203 ng 15N g−1 soil, which corresponds to approximately 60 ng 15N-DNA g−1 soil (based on the simplifying assumption that all fixed N is present in cell biomass and that a cell on average is composed of 12% N, has a total dry weight of 2.84 × 10−13 g, and contains 10 × 10−15 g DNA (29).
Effect of 15N2 on buoyant density of DNA from soil.
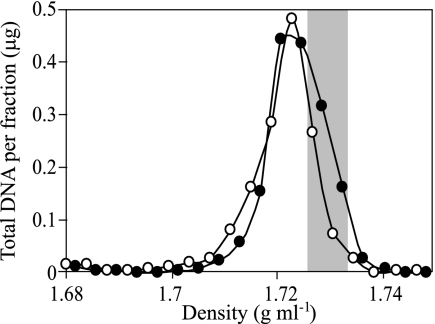
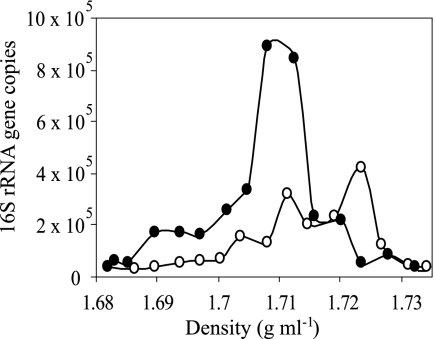
DNA was extracted from 1 g of the control or enriched microcosm soil, yielding 1.8 μg DNA from each sample. Primary CsCl gradient fractionation of these samples revealed a slight increase in the buoyant density of DNA in the 15N2-enriched sample relative to the control sample (Fig. 1). Based on the observed buoyant density in primary gradients, DNA with a buoyant density of 1.727 to 1.733 g ml−1 (Fig. 1) was selected for further analysis. DNA in this density range was expected to have a G+C content of 66% to 75% if unlabeled and 51% to 58% if completely 15N labeled (based on the established relationship between DNA G+C content and buoyant density in CsCl gradients and the expectation that the buoyant density of completely 15N-labeled DNA will increase by 0.016 g ml−1 [1]). These primary fractions were pooled for each sample, and equal proportions were equilibrated in secondary gradients containing bis-benzimide (representing 190 ng DNA from the 15N2-enriched gradient and 135 ng DNA from the control gradient). Following secondary gradient fractionation, the number of 16S rRNA genes present in each gradient fraction was determined by quantitative PCR (Fig. 2). In the presence of bis-benzimide, DNA with a G+C content of 66% to 75% was expected to decrease in buoyant density by 0.013 to 0.007 g ml−1 and DNA with a G+C content of 51% to 58% to decrease by 0.024 to 0.018 g ml−1 (14, 15). The dominant peak for the control sample occurred at a density of 1.723 g ml−1, corresponding to a reduction of buoyant density by 0.010 to 0.004 g ml−1 in response to bis-benzimide (Fig. 2), consistent with unenriched DNA of high G+C content. The majority of DNA in the enriched sample was observed to have a buoyant density of 1.708 to 1.713 g ml−1, which corresponds to a buoyant density decrease of 0.025 to 0.014 g ml−1 in response to bis-benzimide (Fig. 2). Thus, the DNA in secondary fractions with densities of 1.708 and 1.713 g ml−1 from the enriched sample must be dominated by DNA that is completely labeled with 15N. Based on the total amount of DNA added to secondary gradients and the total number of 16S rRNA genes detected in these gradients, it was possible to estimate 1.9 fg genomic DNA per 16S rRNA gene (which roughly corresponds to an average rrn copy number in the range of three to five per Escherichia coli-size genome). Using this value, it was possible to estimate that secondary fractions with densities of 1.708 and 1.713 g ml−1 contained 82 ng and 25 ng of DNA for 15N2-treated and control samples, respectively. The net difference of these values, 57 ng, corresponds to the amount of 15N-labeled DNA present in the secondary gradient.
FIG. 1.
DNA buoyant density was resolved in primary CsCl gradients, and total DNA was quantified in each gradient fraction. DNA was extracted from soil incubated in the presence of either artificial air (○) or artificial air containing 99.8 atom% 15N2 (•). The shaded region corresponds to fractions of buoyant density of 1.727 to 1.733 g ml−1 which were selected for further analysis in secondary gradients containing bis-benzimide.
FIG. 2.
Secondary CsCl gradients containing bis-benzimide were used to disentangle the effects of 15N incorporation and G+C content on the buoyant density of DNA from targeted primary gradient fractions. The total number of 16S rRNA genes in each gradient fraction was determined by quantitative PCR. The secondary gradients were loaded with DNA that had a buoyant density of 1.727 to 1.733 g ml−1 in primary gradients. The symbols correspond to DNA from soil incubated in the presence either of artificial air (○) or of artificial air containing 99.8 atom% 15N2 (•).
Analysis of 16S rRNA genes from 15N-labeled fractions.
PCR amplification of 16S rRNA genes from the 1.708- to 1.713-g ml−1 fractions of secondary gradients resulted in an amplified product of the expected size from the 15N-enriched DNA, but the quantity of the amplified product generated from the control DNA was insufficient to be visualized as a band by gel electrophoresis (data not shown). While a band was not visible in the control, quantitative PCR indicated that 16S rRNA genes were present in this sample (Fig. 2), and it was possible to generate 16S rRNA gene clone libraries from both the 15N-enriched and control DNA (Table 1). The 15N-enriched library contained 51 16S rRNA sequences, while the control library contained 44 sequences. A total of 33 different microbial groups were detected in both libraries (Table 1). Only three groups, the Rhodoplanes, the unclassified Betaproteobacteria, and the unclassified Actinobacteria, were obtained in sufficient numbers that their abundance in the enriched library was unlikely to be due to chance (Fisher's exact test; P < 0.05). The significant overrepresentation of these groups in the enriched library would be expected if these organisms were involved in nitrogen fixation in soil.
TABLE 1.
Phylogenetic classification of 16S rRNA genes in clone libraries generated from DNA-SIP of soil incubated with or without 15N2
| Taxonomic designation | No. of 16S rRNA sequences for soil witha:
|
|
|---|---|---|
| 14N2 | 15N2 | |
| Alphaproteobacteria | ||
| Rhodospirillales | ||
| Unclassified Rhodospirillales | 1 | 1 |
| Rhizobiales | ||
| Methylocystis | 0 | 1 |
| Bradyrhizobium | 3 | 4 |
| Rhodoplanes | 2 | 11ψ |
| Labrys | 1 | 0 |
| Unclassified Alphaproteobacteria | 1 | 2 |
| Betaproteobacteria | ||
| Burkholderiales | ||
| Massilia | 1 | 0 |
| Ralstonia | 2 | 1 |
| Delftia | 2 | 0 |
| Insertae sedis 5 | 2 | 0 |
| Unclassified Betaproteobacteria | 0 | 5ψ |
| Gammaproteobacteria | ||
| Xanthomonadales | 1 | 1 |
| Unclassified Gammaproteobacteria | 0 | 1 |
| Deltaproteobacteria | ||
| Myxococcales | ||
| Nannocystaceae | 2 | 1 |
| Desulfuromonales | ||
| Geobacter | 2 | 0 |
| Unclassified Desulfuromonales | 1 | 0 |
| Unclassified Deltaproteobacteria | 2 | 1 |
| WS3 | ||
| Unclassified WS3 | 0 | 2 |
| Actinobacteria | ||
| Acidimicrobiales | ||
| Acidimicrobium | 2 | 2 |
| Actinomycetales | ||
| Mycobacterium | 0 | 1 |
| Propionibacterium | 1 | 1 |
| Unclassified Actinobacteria | 0 | 5ψ |
| Firmicutes | ||
| Clostridiales | ||
| Unclassified Acidaminococcaceae | 0 | 1 |
| Acidobacteria | ||
| Acidobacteriales | ||
| Acidobacterium | 2 | 2 |
| Unclassified Acidobacteriaceae | 5 | 5 |
| Planctomycetes | ||
| Planctomycetales | ||
| Isosphaera | 1 | 0 |
| Pirellula | 1 | 0 |
| Planctomyces | 3 | 0 |
| Unclassified Planctomycetaceae | 1 | 0 |
| Verrucomicrobia | ||
| Unclassified Verrucomicrobia | 2 | 1 |
| Gemmatimonadetes | ||
| Gemmatimonadales | ||
| Gemmatimonas | 1 | 0 |
| Cyanobacteria | ||
| Subsection 4 | ||
| Unclassified family 4.1 | 0 | 1 |
| Unclassified Bacteria | 2 | 1 |
ψ, phylogenetic groups for which the number of 16S rRNA sequences recovered in the 15N2 library is significantly greater than that observed in the control library (Fisher's exact test; P < 0.05).
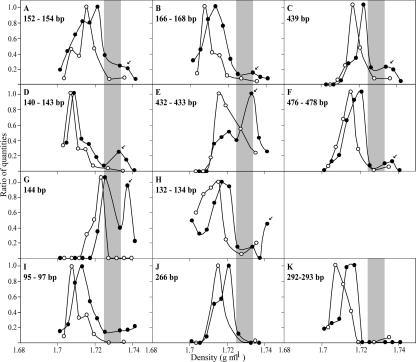
The cloned 16S rRNA sequences were evaluated to determine the sizes of the terminal restriction fragments (TRFs) they would generate after digestion with the enzyme MspI. The distribution of these TRFs was then evaluated in the primary gradient as a function of gradient density (Fig. 3). The 11 Rhodoplanes sequences possessed a total of four TRFs: 134 bp (one sequence), 152 to 154 bp (eight sequences), 166 bp (one sequence), and 439 bp (one sequence). The TRFs of 152 to 154 bp, 166 bp, and 439 bp show secondary peaks in fluorescent intensity at 1.733 g ml−1, which are shifted 0.018 to 0.026 g ml−1 relative to the dominant peaks in the unlabeled controls (Fig. 3A, B, and C). It should be noted that since the analytical resolution of gradient fractionation is 0.0025 g ml−1 (2), the buoyant density differences observed between the dominant unlabeled TRFs of the control and enriched experiments (i.e., TRF of 439 bp in Fig. 3C has peaks at 1.716 g ml−1 and 1.721 g ml−1 for the control and enriched samples, respectively) are not significant. The five unclassified betaproteobacterium sequences possessed a total of three TRFs: 142 to 143 bp (two sequences), 432 bp (one sequence), and 477 to 478 bp (two sequences). These TRFs all show secondary peaks in fluorescent intensity that are shifted 0.018 to 0.026 g ml−1 relative to the dominant peak in the unlabeled controls (Fig. 3D, E, and F). The five unclassified Actinobacteria sequences possessed two TRFs: 132 to 134 bp (four sequences) and 144 bp (one sequence). The 132- to 134-bp TRF has a secondary peak in fluorescent intensity that is shifted 0.026 g ml−1 relative to the dominant peak in the unlabeled controls (Fig. 3H), while the shift for the 144-bp TRF is 0.014 g ml−1 (Fig. 3G). For comparison, the TRFs of Acidobacteria sequences were also determined. Sequences from Acidobacteria were found in equal numbers in enriched and control libraries, and thus, their presence in target fractions in not likely due to 15N labeling of DNA. The Acidobacteria 16S rRNA sequences possessed a total of six TRFs: 96 bp (two sequences), 148 bp (one sequence), 152 bp (six sequences), 266 bp (two sequences), 293 bp (two sequences), and 402 bp (one sequence). Four of these TRFs were detected in the T-RFLP analysis of 16S rRNA genes in the primary gradient; three of these did not evince a secondary peak in enriched samples, and this is consistent with the absence of N fixation by these organisms (Fig. 3, panels I to K). The final TRF from Acidobacteria (152 bp) could not be definitively resolved from the 152- to 154-bp TRF predicted for the Rhodoplanes.
FIG. 3.
T-RFLP analysis of 16S rRNA genes was used to examine the buoyant densities of particular TRFs in primary gradient fractions. The TRFs shown are those predicted for cloned 16S rRNA gene sequences described in Table 1 and correspond to Rhodoplanes (A, B, and C), Betaproteobacteria (D, E, and F), unclassified Actinobacteria (G and H), and unclassified Acidobacteriacea (I, J, and K). Symbols correspond to DNA from soil with artificial air (○) or artificial air containing 99.8 atom% 15N2 (•). TRF peak height was normalized as a function of maximum peak height in each gradient. Arrows are used to indicate putative 15N-labeled DNA, and shading is used to represent DNA from fractions that were added to secondary gradients as described in the legend to Fig. 1.
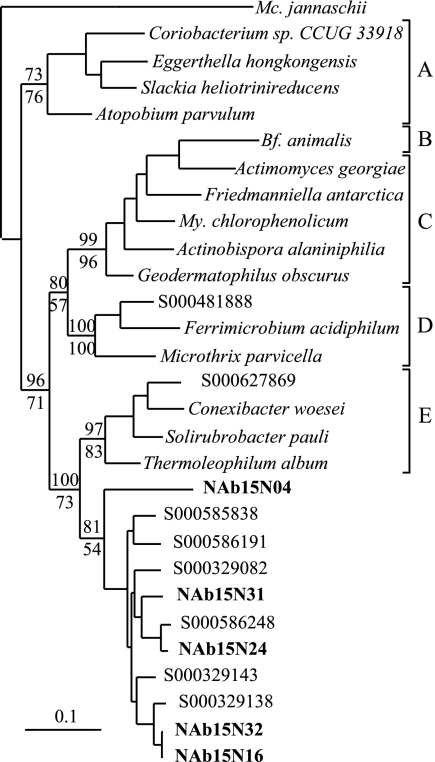
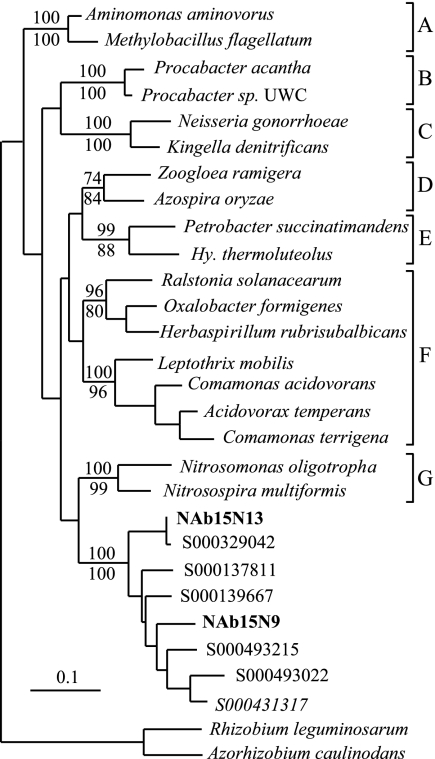
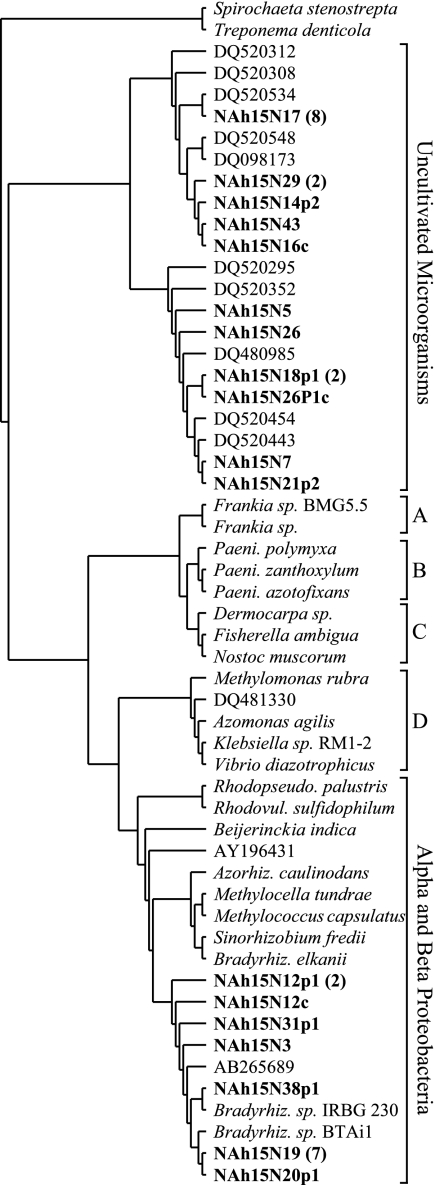
Phylogenetic analyses of 16S rRNA genes from the unclassified Betaproteobacteria and unclassified Antinobacteria revealed that these groups represent previously uncharacterized and deeply divergent lineages within their respective phyla (Fig. 4 and 5). A search of GenBank revealed 88 16S rRNA gene sequences corresponding to the unclassified Betaproteobacteria group and 95 16S rRNA gene sequences from the unclassified Actinobacteria group, and these sequences were included in subsequent phylogenetic analyses. The unclassified Actinobacteria group is most closely related to the order Rubrobacteriales, but phylogenetic analyses consistently indicate that it is a monophyletic group and suggest that it is sufficiently divergent from the Rubrobacteriales that it may represent a new order within the Actinobacteria (Fig. 4). Likewise, the unclassified Betaproteobacteria group represents a distinct lineage that may represent a new order within the Betaproteobacteria (Fig. 5). Phylogenetic analyses consistently indicated that the unclassified Betaproteobacteria group is monophyletic but were unable to demonstrate a consistent affiliation between this group and any of the characterized orders within the Betaproteobacteria.
FIG. 4.
Phylogenetic tree showing the unclassified Actinobacteria group that has been implicated in N fixation in relation to the main orders within Actinobacteria. The tree was constructed from 1,343 16S rRNA positions by using maximum-likelihood analysis (PHYLIP 3.64; J. Felsenstein, Department of Genome Sciences, University of Washington, Seattle, 2005). The numbers listed above and below branches indicate bootstrap values based on 100 replications made using either parsimony (DNApars) or maximum-likelihood (DNAml) analysis, respectively (PHYLIP 3.64). Sequences from this study are in bold type. Designations which begin with S000 are Ribosomal Database Project identifiers representing sequences that are currently unclassified. The scale bar represents a sequence difference of 0.1 nucleotide per position. The letters are used to denote the different orders within the Actinobacteria as follows: A, Coriobacteriales; B, Bifidobacteriales; C, Actinomycetales; D, Acidimicrobiales; E, Rubrobacteriales. The abbreviation Mc. is used for Methanococcus, Bf. for Bifidobacterium, and My. for Mycobacterium.
FIG. 5.
Phylogenetic tree showing the relationship between the unclassified Betaproteobacteria group and the other described orders within the Betaproteobacteria. The tree was constructed from 1,334 16S rRNA positions by using maximum-likelihood analysis (PHYLIP 3.64). The numbers listed above and below branches indicate bootstrap values based on 100 replications made using either parsimony (DNApars) or maximum-likelihood (DNAml) analysis, respectively (PHYLIP 3.64). Sequences from this study are in bold type. Designations which begin with S000 are Ribosomal Database Project identifiers representing sequences that are currently unclassified. The scale bar represents a sequence difference of 0.1 nucleotide per position. The letters are used to denote the different orders within the Betaproteobacteria as follows: A, Methylophilales; B, Procabacteriales; C, Neisseriales; D, Rhodocyclales; E, Hydrogenophilales; F, Burkholderiales; G, Nitrosomonadales. The abbreviation Hy. is used for Hydrogenophilus.
Analysis of nifH genes from 15N-labeled fractions.
PCR amplification of nifH genes from the 1.708- to 1.713-g ml−1 fractions of secondary gradients resulted in a product of the expected size from the 15N-enriched sample but no visible band from the control (data not shown). A total of 34 nifH clones were identified in the library obtained from the 1.708- to 1.713-g ml−1 fraction of the enriched sample, but attempts to recover nifH clones from the corresponding fraction of the control were unsuccessful, suggesting that nifH genes were either absent or very rare in the control fraction. The 34 nifH clones represented 18 different amino acid sequences, and phylogenetic analysis determined that these sequences fell into three different groups within nifH cluster I (Fig. 6). One of these nifH groups associated with nifH sequences from the Alpha- and Betaproteobacteria and was most closely related to sequences from within the Rhizobiales. The other two nifH groups represented distinct monophyletic groups within a larger group of uncultivated nifH sequences that is deeply divergent within nifH cluster I. This uncultivated nifH group has been documented previously in a range of soils (3, 4, 11, 18, 48, 50) and is represented by more than 900 nifH sequences currently in GenBank. The group appears to contain numerous coherent subgroups, the majority of which are dominated by nifH sequences obtained from soil samples (data not shown).
FIG. 6.
This phylogenetic tree shows the relationship between the nifH sequences identified through 15N2-DNA-SIP of soil and other representative sequences from nifH cluster I. The tree was constructed from 105 amino acid positions by using maximum-likelihood analysis (PHYLIP 3.64). Unique sequences from this study are in bold type, and the number in parentheses represents the number of times that each sequence was recovered in the clone library. The letters are used to denote the different nifH groups as follows: A, Frankia; B, Paenibacillus; C, Cyanobacteria; D, Gammaproteobacteria. The abbreviation Paeni. is used for Paenibacillus, Rhodopseudo. for Rhodopseudomonas, Rhodovul. for Rhodovulum, Azorhiz. for Azorhizobium, and Bradyrhiz. for Bradyrhizobium.
DISCUSSION
The potential for cross-feeding or trophic cascades is a serious concern that must be considered when interpreting results from nucleic acid SIP experiments. The amount of 15N-labeled N mineralized by N-fixing organisms is likely to be small (42) and will be diluted considerably by unlabeled N in the soil. Thus, with certain exceptions, only those organisms that are actively engaged in N fixation should become highly labeled with 15N in response to incubation with 15N2. Exceptions are when high N fixation rates are maintained over long incubation periods (i.e., when 15N2 fixation makes significant contributions to total N pools in the local environment) or when there is direct transfer of N between organisms (such as during symbiosis). Cross-feeding and trophic effects are unlikely to have resulted in significant 15N labeling of DNA in this experiment, since 15N represented only 0.009 atom% of total soil N at the end of the incubation period. Due to the low proportion of 15N in the total soil N pool, organisms obtaining 15N through cross-feeding or trophic cascades would likely have very low amounts of 15N incorporation due to dilution by unlabeled sources of N. This partially 15N-labeled DNA should easily be discriminated from DNA that is completely 15N labeled by its response to bis-benzimide in secondary gradients. Thus, DNA recovered from secondary gradients in this experiment, due to its response to bis-benzimide (Fig. 2), is likely to be heavily 15N labeled, and this degree of labeling should occur only for organisms involved in N fixation. Another problem associated with nucleic acid SIP experiments involves the potential for isotopic dilution (34). Isotopic dilution is not a significant concern with 15N2-DNA-SIP, since unlabeled 14N2 can be removed completely by evacuation and since significant N fixation occurs only when fixed (and unlabeled) forms of nitrogen are unavailable to the cell (5, 9).
Analysis of 16S rRNA genes from target fractions provided evidence for three groups of organisms involved in N fixation in soil: a group associated with the Rhodoplanes, an unclassified group of noncultivated Betaproteobacteria, and an unclassified group of noncultivated Actinobacteria (Table 1). While N-fixing organisms are widespread within the Alphaproteobacteria and within the Rhizobiales in particular, N fixation has not previously been attributed to members of the Rhodoplanes. One of the nifH groups from the target fraction was observed to closely associate with members of the Rhizobiales (Fig. 6), which suggests that these nifH genes may originate from the Rhodoplanes group. Unfortunately, due to the mingling of nifH genes from Alpha- and Betaproteobacteria in the nifH phylogeny (possibly as a result of horizontal gene transfer) (36) and the fact that nifH genes have yet to be identified in members of the Rhodoplanes, it is not yet possible to conclusively link these groups of genes at this time. It is also compelling to note that we observed in the target fraction two groups of unclassified noncultivated 16S rRNA sequences and two groups of unclassified noncultivated nifH sequences (Fig. 4, 5, and 6). While it is possible that the two noncultivated nifH groups that we identified (Fig. 6) correspond to the novel Betaproteobacteria and Actinobacteria that we observed (Fig. 4 and 5), it is not possible to provide a conclusive link between these genes at this time.
While nitrogen fixation is widespread in the Betaproteobacteria, the 16S rRNA genes that we recovered represent a deeply divergent lineage that may constitute a new order within this group (Fig. 5). More than 88 16S rRNA sequences from GenBank currently fall within this group, none are from cultivated isolates, and more than 75% of these sequences were obtained from soils with samples representing five continents (data not shown). Of even greater interest is our evidence for nitrogen fixation within a deeply divergent lineage of the Actinobacteria. Nitrogen fixation is relatively uncommon in the Actinobacteria, being found only in Frankia and two recently isolated strains of Actinomycetales (46). In addition, within the Actinobacteria, nitrogen-fixing bacteria have not previously been observed outside of the order Actinomycetales. The group that we have identified, while most closely related to the Rubrobacteriales, likely represents a new order of Actinobacteria (Fig. 4) for which no cultivated isolate is currently available. This group currently encompasses more than 95 16S rRNA sequences in GenBank, 86% of which were obtained from soils originating from six continents (data not shown).
Since this research represents the first attempt to use 15N2 in SIP experiments and since few attempts have been made to use 15N isotopes in nucleic acid SIP, it is important to consider the evidence that supports our assertion that the groups identified are involved in nitrogen fixation. While an obvious change in bulk DNA density was not observed in response to 15N2 labeling in primary gradients (Fig. 1), such a response would in fact be unlikely in this experiment. The rate of nitrogen fixation in this experiment was low, resulting in a small amount of 15N-labeled DNA relative to unlabeled DNA (an estimated 60 ng 15N target DNA relative to 1,800 ng nontarget DNA). In addition, completely 15N-labeled DNA is expected to change density by only 0.016 g ml−1, while variation of DNA G+C content in soil will cause wide variation in the buoyant density of DNA from soil (1). Thus, secondary gradients containing bis-benzimide were used to resolve 15N-labeled DNA. DNA samples from target fractions in primary gradients were observed to respond to bis-benzimide incorporation in secondary gradients in a manner which is consistent with 100% 15N incorporation into DNA (Fig. 2). Clearly, however, while target fractions from the secondary gradient showed a significant response to 15N2 enrichment, some contaminating unlabeled DNA was still present (Fig. 2 and Table 1). The recovery of unlabeled DNA in target fractions has been observed in previous nucleic acid SIP experiments (20, 22, 25) and could result from failure to completely equilibrate nucleic acids in CsCl gradients, DNA fragmentation, reagent contamination, or carryover of DNA between fractions during fractionation. Thus, it is essential to use proper controls to distinguish isotopically labeled DNA from unlabeled DNA. The 16S rRNA gene libraries constructed from the 1.708- to 1.713-g ml−1 fractions of secondary gradients indicate that certain groups occur more frequently in the enriched library than in the control library and that this difference is unlikely to be due to chance (Table 1). This observation is consistent with the conclusion that these groups were involved in 15N2 fixation (considering that any PCR bias that may affect the abundance of PCR products should be consistent between enriched and control samples (e.g., see reference 16). Finally, the recovery of nifH genes from target secondary fractions (1.708 to 1.713 g ml−1) and failure to obtain nifH genes from corresponding unenriched control fractions suggest that the difference in the amount of DNA observed between these fractions (Fig. 2) is likely attributable to N-fixing organisms. This observation is also supported by the fact that the difference in the amount of DNA present in secondary gradient target fractions from 15N2-treated and control samples was 57 ng, a value which corresponds with the 60 ng of 15N-DNA estimated per g soil based on measurements of soil 15N enrichment resulting from 15N2 fixation.
Independent confirmation that fixation of 15N2 caused an increase in the DNA density of these groups is provided by examining the distribution of their 16S rRNA TRFs in primary gradients. When interpreting this evidence, it is important to consider that multiple organisms can possess TRFs of the same size, and this problem can complicate interpretation of T-RFLP patterns (19, 26). This problem is ameliorated to some degree in the current experiment by the fact that organisms that share a TRF but have different genome G+C content will occur in different positions in a CsCl gradient as a function of their DNA buoyant density (2). As a result, however, when conducting nucleic acid SIP experiments, careful comparison is needed between DNA fingerprint profiles of isotopically enriched and control samples in density gradients. For example, TRFs of 476 to 478 bp (Fig. 3F) and 132 to 134 bp (Fig. 3H) have indications of a small TRF peak in the control sample at the same density where 15N-labeled DNA would be expected to occur. This result could occur if organisms with a high G+C content share a TRF with N-fixing organisms. Thus, while the data from these TRFs is consistent with 15N2 fixation, this result could also occur if a change in the abundance of certain high-G+C organisms between treatment and control samples took place.
In other cases, examining the distribution of 16S rRNA TRFs in primary gradients provided unambiguous evidence of 15N incorporation into DNA. It was possible to observe TRFs from the Rhodoplanes (439 bp) (Fig. 3C), the noncultivated Betaproteobacteria group (140 to 143 bp and 432 to 433 bp) (Fig. 3D and E), and the noncultivated Actinobacteria group (144 bp) (Fig. 3G) that clearly show an increase in DNA density in enriched samples relative to that for controls. The presence of these heavy peaks in the enriched treatment but not in the control is consistent with 15N incorporation into DNA. The change in the DNA buoyant density of these TRFs in enriched treatments relative to that for controls was greater than 0.014 g ml−1, consistent with 100% 15N incorporation into DNA. In contrast, TRFs from the Acidobacteria (Fig. 3I to K), a group detected in control and enriched samples in equal numbers and thus not expected to be involved in N fixation (Table 1), did not demonstrate heavy TRFs, a result that is inconsistent with 15N incorporation into DNA. It is interesting to note that all TRFs from organisms involved in 15N2 fixation retained a distinct unlabeled DNA peak (Fig. 3). This observation suggests that nitrogen fixation occurred within certain microsites, while in other microsites, either fixed forms of nitrogen were available or growth did not occur. Such microheterogeneity in soil would result in the simultaneous presence of both completely labeled DNA and unlabeled DNA, as was observed in this experiment. This observation is also consistent with the expectation that organisms that are fixing nitrogen will not simultaneously incorporate unlabeled mineral forms of nitrogen from soil.
This research represents the first application of 15N2-DNA-SIP and the first application of nucleic acid 15N-SIP that has been able to identify microorganisms associated with 15N incorporation in the environment. We have identified three microbial groups that we suggest are able to carry out nitrogen fixation in soil. None of these groups has previously been implicated in this process, and two of them, the unclassified Betaproteobacteria and the unclassified Actinobacteria, remain completely uncharacterized despite being widespread in soils. Our data also suggest the hypothesis that these two 16S rRNA groups may correspond to the two noncultivated groups of nifH genes that we observed. Thus, the application of 15N2-DNA-SIP may make it possible to identify previously uncharacterized groups of nitrogenase genes. The two groups of nifH genes that we identified belong to a larger family of more than 900 nifH sequences that are deeply divergent within the nitrogenase cluster I and which currently remains completely uncharacterized due to the lack of any cultivated representatives within this group. Work is currently under way to either isolate these organisms by using the nifH or 16S rRNA genes that we have identified as a marker or use DNA from SIP experiments to isolate genome fragments that would allow more-thorough characterization of these novel diazotrophs.
Acknowledgments
This research was supported by the National Science Foundation under award number MCB-0447586. This research was also supported in part by the National Research Initiative of the USDA Cooperative State Research, Education and Extension Service, grant number 2005-35107-15266.
Footnotes
Published ahead of print on 16 March 2007.
REFERENCES
- 1.Birnie, G. D., and D. Rickwood (ed.). 1978. Centrifugal separations in molecular and cell biology. Butterworths, Boston, MA.
- 2.Buckley, D. H., V. Huangyutitham, S.-F. Hsu, and T. A. Nelson. 2007. Stable isotope probing with 15N achieved by disentangling the effects of genome G+C content and isotope enrichment on DNA density. Appl. Environ. Microbiol. 73:3189-3195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bürgmann, H., S. Meier, M. Bunge, F. Widmer, and J. Zeyer. 2005. Effects of model root exudates on structure and activity of a soil diazotroph community. Environ. Microbiol. 7:1711-1724. [DOI] [PubMed] [Google Scholar]
- 4.Bürgmann, H., F. Widmer, W. Von Sigler, and J. Zeyer. 2004. New molecular screening tools for analysis of free-living diazotrophs in soil. Appl. Environ. Microbiol. 70:240-247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Burris, R. H., and P. W. Wilson. 1946. Ammonia as an intermediate in nitrogen fixation by Azotobacter. J. Bacteriol. 52:505-512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chelius, M. K., and J. E. Lepo. 1999. Restriction fragment length polymorphism analysis of PCR-amplified nifH sequences from wetland plant rhizosphere communities. Environ. Technol. 20:883-889. [Google Scholar]
- 7.Cleveland, C. C., A. R. Townsend, D. S. Schimel, H. Fisher, R. W. Howarth, L. O. Hedin, S. S. Perakis, E. F. Latty, J. C. Von Fischer, A. Elseroad, and M. F. Wasson. 1999. Global patterns of terrestrial biological nitrogen (N-2) fixation in natural ecosystems. Global Biogeochem. Cycles 13:623-645. [Google Scholar]
- 8.Cole, J. R., B. Chai, R. J. Farris, Q. Wang, S. A. Kulam, D. M. McGarrell, G. M. Garrity, and J. M. Tiedje. 2005. The Ribosomal Database Project (RDP-II): sequences and tools for high-throughput rRNA analysis. Nucleic Acids Res. 33:D294-D296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Daesch, G., and L. E. Mortenson. 1972. Effect of ammonia on the synthesis and function of the N2-fixing enzyme system in Clostridium pasteurianum. J. Bacteriol. 110:103-109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.DeRito, C. M., G. M. Pumphrey, and E. L. Madsen. 2005. Use of field-based stable isotope probing to identify adapted populations and track carbon flow through a phenol-degrading soil microbial community. Appl. Environ. Microbiol. 71:7858-7865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Deslippe, J. R., and K. N. Egger. 2006. Molecular diversity of nifH genes from bacteria associated with high arctic dwarf shrubs. Microb. Ecol. 51:516-525. [DOI] [PubMed] [Google Scholar]
- 12.Finn, R. D., J. Mistry, B. Schuster-Böckler, S. Griffiths-Jones, V. Hollich, T. Lassmann, S. Moxon, M. Marshall, A. Khanna, R. Durbin, S. R. Eddy, E. L. L. Sonnhammer, and A. Bateman. 2006. Pfam: clans, web tools and services. Nucleic Acids Res. D247-D251. [DOI] [PMC free article] [PubMed]
- 13.Hamelin, J., N. Fromin, S. Tarnawski, S. Teyssier-Cuvelle, and M. Aragno. 2002. nifH gene diversity in the bacterial community associated with the rhizosphere of Molinia coerulea, an oligonitrophilic perennial grass. Environ. Microbiol. 4:477-481. [DOI] [PubMed] [Google Scholar]
- 14.Holben, W. E., K. P. Feris, A. Kettunen, and J. H. A. Apajalahti. 2004. GC fractionation enhances microbial community diversity assessment and detection of minority populations of bacteria by denaturing gradient gel electrophoresis. Appl. Environ. Microbiol. 70:2263-2270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Holben, W. E., and D. Harris. 1995. DNA-based monitoring of total bacterial community structure in environmental samples. Mol. Ecol. 4:627-631. [DOI] [PubMed] [Google Scholar]
- 16.Hughes, J. B., J. J. Hellman, T. H. Ricketts, and B. J. M. Bohannan. 2001. Counting the uncountable: statistical approaches to estimating microbial diversity. Appl. Environ. Microbiol. 67:4399-4406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hutchens, E., S. Radajewski, M. G. Dumont, I. R. McDonald, and J. C. Murrell. 2004. Analysis of methanotrophic bacteria in Movile Cave by stable isotope probing. Environ. Microbiol. 6:111-120. [DOI] [PubMed] [Google Scholar]
- 18.Izquierdo, J. A., and K. Nusslein. 2006. Distribution of extensive nifH gene diversity across physical soil microenvironments. Microb. Ecol. 51:441-452. [DOI] [PubMed] [Google Scholar]
- 19.Liu, W. T., T. L. Marsh, H. Cheng, and L. J. Forney. 1997. Characterization of microbial diversity by determining terminal restriction fragment length polymorphisms of genes encoding 16S rRNA. Appl. Environ. Microbiol. 63:4516-4522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lu, Y. H., T. Lueders, M. W. Friedrich, and R. Conrad. 2005. Detecting active methanogenic populations on rice roots using stable isotope probing. Environ. Microbiol. 7:326-336. [DOI] [PubMed] [Google Scholar]
- 21.Lueders, T., M. Manefield, and M. W. Friedrich. 2004. Enhanced sensitivity of DNA- and rRNA-based stable isotope probing by fractionation and quantitative analysis of isopycnic centrifugation gradients. Environ. Microbiol. 6:73-78. [DOI] [PubMed] [Google Scholar]
- 22.Lueders, T., B. Wagner, P. Claus, and M. W. Friedrich. 2004. Stable isotope probing of rRNA and DNA reveals a dynamic methylotroph community and trophic interactions with fungi and protozoa in oxic rice field soil. Environ. Microbiol. 6:60-72. [DOI] [PubMed] [Google Scholar]
- 23.Mahmood, S., G. I. Paton, and J. I. Prosser. 2005. Cultivation-independent in situ molecular analysis of bacteria involved in degradation of pentachlorophenol in soil. Environ. Microbiol. 7:1349-1360. [DOI] [PubMed] [Google Scholar]
- 24.Manefield, M., A. S. Whiteley, R. I. Griffiths, and M. J. Bailey. 2002. RNA stable isotope probing, a novel means of linking microbial community function to phylogeny. Appl. Environ. Microbiol. 68:5367-5373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Manefield, M., A. S. Whiteley, N. Ostle, P. Ineson, and M. J. Bailey. 2002. Technical considerations for RNA-based stable isotope probing: an approach to associating microbial diversity with microbial community function. Rapid Commun. Mass Spectrom. 16:2179-2183. [DOI] [PubMed] [Google Scholar]
- 26.Marsh, T. L., P. Saxman, J. Cole, and J. Tiedje. 2000. Terminal restriction fragment length polymorphism analysis program, a web-based research tool for microbial community analysis. Appl. Environ. Microbiol. 66:3616-3620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.McDonald, I. R., S. Radajewski, and J. C. Murrell. 2005. Stable isotope probing of nucleic acids in methanotrophs and methylotrophs: a review. Org. Geochem. 36:779-787. [Google Scholar]
- 28.Morris, S. A., S. Radajewski, T. W. Willison, and J. C. Murrell. 2002. Identification of the functionally active methanotroph population in a peat soil microcosm by stable-isotope probing. Appl. Environ. Microbiol. 68:1446-1453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Neidhardt, F. C., and J. L. Ingraham (ed.). 1987. Escherichia coli and Salmonella typhimurium: cellular and molecular biology. ASM Press, Washington, DC.
- 30.Nüsslein, K., and J. M. Tiedje. 1998. Characterization of the dominant and rare members of a young Hawaiian soil bacterial community with small-subunit ribosomal DNA amplified from DNA fractionated on the basis of its guanine and cytosine composition. Appl. Environ. Microbiol. 64:1283-1289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Olsen, G. J., H. Matsuda, R. Hagstrom, and R. Overbeek. 1994. FasDNAml: a tool for construction of phylogenetic trees of DNA sequences using maximum likelihood. Comput. Appl. Biosci. 10:41-48. [DOI] [PubMed] [Google Scholar]
- 32.Piceno, Y. M., and C. R. Lovell. 2000. Stability in natural bacterial communities. I. Nutrient addition effects on rhizosphere diazotroph assemblage composition. Microb. Ecol. 39:32-40. [DOI] [PubMed] [Google Scholar]
- 33.Poly, F., L. Ranjard, S. Nazaret, F. Gourbiere, and L. J. Monrozier. 2001. Comparison of nifH gene pools in soils and soil microenvironments with contrasting properties. Appl. Environ. Microbiol. 67:2255-2262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Radajewski, S., P. Ineson, N. R. Parekh, and J. C. Murrell. 2000. Stable-isotope probing as a tool in microbial ecology. Nature 403:646-649. [DOI] [PubMed] [Google Scholar]
- 35.Radajewski, S., I. R. McDonald, and J. C. Murrell. 2003. Stable-isotope probing of nucleic acids: a window to the function of uncultured microorganisms. Curr. Opin. Biotechnol. 14:296-302. [DOI] [PubMed] [Google Scholar]
- 36.Raymond, J., J. L. Siefert, C. R. Staples, and R. E. Blankenship. 2004. The natural history of nitrogen fixation. Mol. Biol. Evol. 21:541-554. [DOI] [PubMed] [Google Scholar]
- 37.Rolfe, R., and M. S. Meselson. 1959. The relative homogeneity of microbial DNA. Proc. Natl. Acad. Sci. USA 45:1039-1043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Rösch, C., and H. Bothe. 2005. Improved assessment of denitrifying, N2-fixing, and total-community bacteria by terminal restriction fragment length polymorphism analysis using multiple restriction enzymes. Appl. Environ. Microbiol. 71:2026-2035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Sambrook, J., and D. W. Russell (ed.). 2001. Molecular cloning: a laboratory manual. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- 40.Shaffer, B. T., F. Widmer, L. A. Porteous, and R. J. Seidler. 2000. Temporal and spatial distribution of the nifH gene of N2 fixing bacteria in forests and clearcuts in western Oregon. Microb. Ecol. 39:12-21. [DOI] [PubMed] [Google Scholar]
- 41.Steward, G. F., B. D. Jenkins, B. B. Ward, and J. P. Zehr. 2004. Development and testing of a DNA microarray to assess nitrogenase (nifH) gene diversity. Appl. Environ. Microbiol. 70:1455-1465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Stewart, W. D. P. 1982. Nitrogen-fixation—its current relevance and future potential. Isr. J. Bot. 31:5-44. [Google Scholar]
- 43.Stubner, S. 2002. Enumeration of 16S rDNA of Desulfotomaculum lineage 1 in rice field soil by real-time PCR with SybrGreen detection. J. Microbiol. Methods 50:155-164. [DOI] [PubMed] [Google Scholar]
- 44.Tan, X. Y., T. Hurek, and B. Reinhold-Hurek. 2003. Effect of N-fertilization, plant genotype and environmental conditions on nifH gene pools in roots of rice. Environ. Microbiol. 5:1009-1015. [DOI] [PubMed] [Google Scholar]
- 45.Ueda, T., Y. Suga, N. Yahiro, and T. Matsuguchi. 1995. Remarkable N2-fixing bacterial diversity detected in rice roots by molecular evolutionary analysis of nifH gene sequences. J. Bacteriol. 177:1414-1417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Valdés, M. A., N.-O. Pérez, P. Estrada-de los Santos, J. Caballero-Mellado, J. J. Peña-Cabriales, P. Normand, and A. M. Hirsch. 2005. Non-Frankia actinomycetes isolated from surface-sterilized roots of Casuarina equisetifolia fix nitrogen. Appl. Environ. Microbiol. 71:460-466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Widmer, F., B. T. Shaffer, L. A. Porteous, and R. J. Seidler. 1999. Analysis of nifH gene pool complexity in soil and litter at a Douglas fir forest site in the Oregon Cascade Mountain Range. Appl. Environ. Microbiol. 65:374-380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Yeager, C. M., D. E. Northup, C. C. Grow, S. M. Barns, and C. R. Kuske. 2005. Changes in nitrogen-fixing and ammonia-oxidizing bacterial communities in soil of a mixed conifer forest after wildfire. Appl. Environ. Microbiol. 71:2713-2722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Zehr, J. P., B. D. Jenkins, S. M. Short, and G. F. Steward. 2003. Nitrogenase gene diversity and microbial community structure: a cross-system comparison. Environ. Microbiol. 5:539-554. [DOI] [PubMed] [Google Scholar]
- 50.Zhang, Y. G., D. Q. Li, H. M. Wang, Q. M. Xiao, and X. D. Liu. 2006. Molecular diversity of nitrogen-fixing bacteria from the Tibetan Plateau, China. FEMS Microbiol. Lett. 260:134-142. [DOI] [PubMed] [Google Scholar]
- 51.Ziegler, S. E., P. M. White, D. C. Wolf, and G. J. Thoma. 2005. Tracking the fate and recycling of C-13-labeled glucose in soil. Soil Sci. 170:767-778. [Google Scholar]