Abstract
Despite the economic and sanitary problems caused by harmful biofilms, biofilms are nonetheless used empirically in industrial environmental and bioremediation processes and may be of potential use in medical settings for interfering with pathogen development. Escherichia coli is one of the bacteria with which biofilm formation has been studied in great detail, and it is especially appreciated for biotechnology applications because of its genetic amenability. Here we describe the development of two new genetic tools enabling the constitutive and inducible expression of any gene or operon of interest at its native locus. In addition to providing valuable tools for complementation and overexpression experiments, these two compact genetic cassettes were used to modulate the biofilm formation capacities of E. coli by taking control of two biofilm-promoting factors, autotransported antigen 43 adhesin and the bscABZC cellulose operon. The modulation of the biofilm formation capacities of E. coli or those of other bacteria capable of being genetically manipulated may be of use both for reducing and for improving the impact of biofilms in a number of industrial and medical applications.
Most natural and artificial surfaces available in the environment are prone to bacterial colonization. Following the initial adhesion event, cell-to-cell adhesion and the secretion of an extracellular matrix rapidly lead to the formation of a surface-attached multicellular structure known as a biofilm (3, 43, 75, 84). Besides being resistant to environmental shear forces, biofilm communities are also phenotypically more resistant to antibiotic and biocide treatments, a trait that poses important sanitary and economic problems. Indeed, biofilms formed by bacterial pathogens on medically relevant surfaces are difficult to eradicate and are thus often involved in the development of infections (12, 13, 48). Moreover, industrial biofouling resulting from bacterial biofilm formation is a major cause of pipe biocorrosion and reduces the efficiency of pharmaceutical or food bioprocesses (2, 10, 70, 71).
While recent studies have focused mainly on the negative impact of biofilms, bacterial biofilms can also have valuable applications, including those involving bioremediation and wastewater treatment bioreactor processes and the improvement of biomineralization or plant-bacteria symbiosis (1, 14, 40, 42, 52, 62, 73, 74, 80). Beneficial biofilms may also have medical applications, and the use of protective innocuous bacterial biofilms that interfere with the development of bacterial pathogens is considered a promising approach (19).
Escherichia coli is a gram-negative enterobacterium that has been used extensively as a model to study biofilm development due to its relevance to the human biotic environment and its genetic amenability. In E. coli, various cell surface appendages were shown to be necessary to achieve mature biofilm development (78). Flagella, type I fimbriae, and curli are implicated in early adhesion steps, while the production of a polysaccharide-rich matrix (cellulose, colanic acid, and poly-β-1,6-N-acetylglucosamine) and of short adhesins such as antigen 43 (Ag43) and conjugative plasmid pili contributes to biofilm maturation (11, 16-18, 30, 32, 59, 60, 63, 67, 79, 83, 85).
We previously showed that the ability of E. coli K-12 to form a biofilm could be enhanced by increasing the expression of specific adhesin genes (68). Here we describe the genetic engineering of different E. coli strains whose capacity to develop as biofilms was tightly controlled by the modulation of the expression levels of different biofilm-promoting factors. We developed new genetic tools designed to enable the cloning-free, site-directed insertion of either inducible or constitutive promoters in front of genes of interest. We chose genes coding for either adhesin Ag43 or cellulose production to show that this new expression strategy can be used to create plasmid-free E. coli strains with defined and tightly controlled biofilm-forming abilities. This approach may be used to improve the biofilm potential of laboratory and natural bacterial strains used in industrial and medical bioprocesses.
MATERIALS AND METHODS
Bacterial strains and plasmids.
Bacterial strains and plasmids used in this study are listed in Table 1. Strains were constructed by transformation and the λ red linear DNA gene inactivation method (see below), followed by P1vir transduction into a fresh E. coli background when possible.
TABLE 1.
Strains and plasmids used in this study
| Strain or plasmid | Relevant characteristicsa | Source or reference |
|---|---|---|
| Strains | ||
| MG1655 | F− lambda−ilvG rfb-50 rph-1 | Laboratory collection |
| MG1655λatt-kmgfp | Insertion, at the λatt site, of the gfp gene under the control of the constitutive λpR promoter; source of the kmPcL and kmPcLrbs cassettes; Kmr | This study |
| MG1655λatt-ampgfp | Insertion, at the λatt site, of the gfp gene under the control of the constitutive λpR promoter; source of the ampPcL and ampPcLrbs cassettes; Ampr | This study |
| MG1655Δflu | Δflu::cat Cmr | 68 |
| MG1655ΔoxyR | ΔoxyR::aph Kmr | 68 |
| MG1655ΔoxyRΔflu | ΔoxyR::aph Δflu::cat Kmr Cmr | 68 |
| MG1655kmRExTETrbs-flu | flu placed under the control of the kmRExTETrbs cassette PLtetO-1 promoter; Kmr | This study |
| MG1655kmPcLflu | flu with its own RBS sequence placed under the control of the kmPcL cassette λpR promoter; Kmr | This study |
| MG1655kmRExTETlacZ | lacZ with its own RBS sequence placed under the control of the kmRExTET cassette PLtetO-1 promoter; Kmr | This study |
| MG1655kmRExTETrbs-lacZ | lacZ placed under the control of the kmRExTETrbs cassette PLtetO-1 promoter; Kmr | This study |
| MG1655kmPcLlacZ | lacZ with its own RBS sequence placed under the control of the kmPcL cassette λpR promoter; Kmr | This study |
| MG1655kmPcLrbs-lacZ | lacZ placed under the control of the kmPcLrbs cassette λpR promoter; Kmr | This study |
| MG1655ampPcLlacZ | lacZ with its own RBS sequence placed under the control of the ampPcL cassette λpR promoter; Ampr | This study |
| MG1655ampPcLrbs-lacZ | lacZ placed under the control of the ampPcLrbs cassette λpR promoter; Ampr | This study |
| 1094 | E. coli commensal strain | 18 |
| 1094ΔbcsABZC | ΔbcsABZC::aph Kmr | 18 |
| 1094kmRExTETrbs-bcsA | bcsABZC operon placed under the control of the kmRExTETrbs cassette PLtetO-1 promoter; Kmr | This study |
| DH5αZ1 | DH5α with integrated transcription units encoding LacI and TetR at the λatt site | 49 |
| Plasmids | ||
| pZE21-gfp | gfp under the control of the synthetic PLtetO-1 promoter; Kmr | 49 |
| pZEtetR21-gfp | Same as pZE21-gfp with an insertion of PN25-tetR-T1 between the nptII gene and the terminator t0; Kmr | This study |
| pZE2R-gfp | gfp under the control of the constitutive λpR promoter; Kmr | Gift from C. C. Guet |
| pZE1R-gfp | gfp under the control of the constitutive λpR promoter; Ampr | Gift from C. C. Guet |
| pAg43 | Pflu::lacZ transcriptional fusion in pQF50; Ampr | 81 |
Kmr, kanamycin resistance; Ampr, ampicillin resistance; Cmr, chloramphenicol resistance.
Growth conditions.
All experiments were performed in M63B1 0.4% glucose minimal medium (M63B1glu) or in lysogeny broth (LB) medium at 37°C. The following antibiotics at the indicated concentrations were added when required: kanamycin (50 μg/ml), chloramphenicol (25 μg/ml), and ampicillin (100 μg/ml). Repression-expression TetR-controlled (RExTET) cassette constructs were induced with anhydrotetracycline (aTc) at the concentrations indicated in the figures. As previously shown, we observed that aTc did not have any effect on bacterial growth at concentrations below 500 ng/ml (55).
Three-step PCR.
In order to place chromosomal target genes (lacZ, flu, and the bcsABZC operon) under the control of the RExTET cassette and the constitutive lambda promoter (PcL) cassettes, we used a three-step PCR procedure as described in references 9, 20, 21, and 47 and detailed at http://www.pasteur.fr/recherche/unites/Ggb/3SPCRprotocol.html. The primers used to insert the cassettes upstream of the target genes are listed in Table 2.
TABLE 2.
Primers used in this study
| Primer name | Sequence | Target gene |
|---|---|---|
| Primers used to generate kmRExTET and PcL cassette insertions | ||
| lacZACTIV.A1.500-5 | CTCAGGTCAAATTCAGACGGC | lacZ |
| lacZACTIV.B1.500-3 | CTCAGGTCAAATTCAGACGGC | lacZ |
| lacZ.CATptetO.A2.L-3 | GAGAATCCAAGCACTAGTAACCACAATTCCACACAACATA | lacZ |
| lacZ.CATptetO.B2.L-5 | GCACATCAGCAGGACGCACTGACAGCGGATAACAATTTCACAC | lacZ |
| lacZ.CATptetOB2LrbstetO5 | CATTAAAGAGGAGAAAGGTACCATGACCATGATTACGGATTC | lacZ |
| lacZ.ext-5 | CATTGGGTCACCAGCAAATC | lacZ |
| lacZ.CATBAD.ext-3 | CCAGATAACTGCCGTCACTC | lacZ |
| flu.KmRExTET.500-5 | CCCGAATTCTGCGGTGGACCGGATATTTG | flu |
| flu.KmRExTET.500-3 | ATGACGGTTCCTGTGGCTATC | flu |
| flu.KmRExTET.ext-3 | GCCCGGTATCACCGTTTTCTG | flu |
| flu.KmRExTET.ext-5 | ATACGCTGGTCAGTGCGCTC | flu |
| flu.KmRExTET.Lbrs-5 | CATTAAAGAGGAGAAAGGTACCATGAAACGACATCTGAATAC | flu |
| flu.KmRExTET.L-3 | GAGAATCCAAGCACTAGTAACCACATTGAGGGTGAATAAAAAAG | flu |
| flu.PcL.A2.L-3 | GTGAGAATTACTAACTTGAGCGAATTGAGGGTGAATAAAAAAG | flu |
| flu.PcL.B2.L-5 | CGGTGATAATGGTTGCATGTACTATCTAAGGAAAAGCTGATGAAACGA | flu |
| bcsA-ext-5 | CGCATTAGCCTGGTCATTAC | bcsA |
| yhjQ.KmRExTET.bcsA.ext-3 | AGAAATCAGCGAGAAGGTGAC | bcsA |
| KmRExTET.bcsA.L-3 | GAGAATCCAAGCACTAGTAACCACTTATGATGCACTCCCGACTGGCGTTTTC | bcsA |
| yhjQ.KmRExTET.bcsA.Lbrs-5 | CATTAAAGAGGAGAAAGGTACCatgaGTATCCTGACCCGGTGG | bcsA |
| yhjQ.KmRExTET.bcsA.ext-5 | TTGTCTGATTATCAGTTTAC | bcsA |
| Primers used to verify cassette insertions | ||
| KmRExTET.verif-5 | GCGAAACGATCCTCATCCTG | |
| KmRExTET.verif-3 | CATTGCTTATCAATTTGTTGC | |
| PcL-km-verif-5 | CAGAGCAGCCGATTGTCTGTTG | |
| PcL-km-verif-3 | CTTCCTCGTGCTTTACGGTATCG | |
| PcL-amp-verif-5 | CGAAAACTCTCAAGGATCTTAC | |
| PcL-amp-verif-3 | TGGTTTATTGCTGATAAATCTG | |
| Primers used to sequence the junction between cassettes and the target gene ATG codon | ||
| lacZATG + 100-3 | GGGGGATGTGCTGCAAGGCGATTAAG | lacZ |
| fluATG + 100-3 | GACGTGACTGCGGCAAGAGACAGTG | flu |
| KmRExTET.bcsA.500-3 | GAGGATCAACCGCCGCGCCCCG | bcsA |
| Primers used to amplify kmRExTET and PcL cassettes | ||
| tetR.ptetOgfp.ampl-5 | GAAGATCCTTTGATCTTTTC | |
| tetR.ptetOgfp.ampl-3 | TGCCCATTAACATCACCATC | |
| km.Rex.Tet.ampli-5 | CACTTTATGCTTCCGGCTCGTATG | |
| km.Rex.Tet.ampli-3 | CGCCAGGGTTTTCCCAGTCACGAC | |
| PcL-ampli-5 | CTCTGGCAAGCGCCTCGATTACTG | |
| PcL-ampli-3 | CATCACCTTCACCCTCTCCACTGAC | |
| Primers used for pZEtet21-gfp plasmid construction | ||
| tetR-T1-sacI-5 | CGCGGGGAGCTCGCGCAACGCAATTAATGTAAG | |
| tetR-T1.sacI-3bis | CAGAACGAGCTCGATTTGTCCTACTCAGGAGAG |
Construct verification.
All constructs were checked by PCR with specific primers (Table 2). The integrity of the cassettes was verified by sequencing the junction between the RExTET cassette or the PcL cassette and the target gene by using primers described in Table 2.
β-Galactosidase activity assay.
To determine the level of β-galactosidase enzyme activity, the different cultures were grown in LB for 8 h. Cultures were then diluted 1:100 in LB or M63B1glu medium containing 1 mM IPTG (isopropyl-β-d-thiogalactopyranoside) or various amounts of aTc when needed and grown overnight (16 to 18 h) at 37°C. The enzyme activity of each strain was assayed in triplicate as described in reference 50 and expressed in arbitrary Miller units.
Immunodetection of Ag43 and immunofluorescence microscopy.
Amounts of overnight cultures equivalent to an optical density at 600 nm (OD600) of 0.2 were analyzed on sodium dodecyl phosphate-10% polyacrylamide gel electrophoresis gel, followed by the immunodetection of Ag43. Equivalent loads of proteins in the lanes were verified by staining the nitrocellulose membranes with Ponceau S. Immunodetection was performed using a 1:10,000 dilution of polyclonal rabbit antiserum raised against the α domain of Ag43, a kind gift of P. Owen.
Immunofluorescence microscopy analysis was performed as follows. Overnight cultures of the different strains were grown at 37°C in LB medium without aTc or, for strain MG1655kmRExTETrbs-flu, with 50 ng of aTc/ml. Cells were diluted to an OD600 of 1 in LB medium, and an aliquot was loaded onto 0.1% poly-l-lysine-treated immunofluorescence microscope slides. Slides were washed three times with phosphate-buffered saline (PBS) between each step of this protocol. Cells were fixed with 3% paraformaldehyde for 10 min before quenching with 50 mM NH4Cl in PBS for 3 min. Slides were then saturated for 15 min with 0.5% bovine serum albumin in PBS before being incubated first for 45 min with a 1:1,000 dilution of primary polyclonal rabbit antiserum raised against the α domain of Ag43 and next with a 1:300 dilution of a secondary polyclonal goat anti-rabbit serum coupled to Alexa488 (Molecular Probes-Invitrogen) along with 10 μg of 4′,6-diamidino-2-phenylindole (DAPI)/ml. Finally, the slides were mounted with Mowiol 4088 (Calbiochem) and observed under an epifluorescence microscope with green fluorescent protein and DAPI filters.
Ag43 switching frequency.
The Ag43 switching frequency was calculated as previously described (58, 81). Five white (phase-“off”) and five blue (phase-“on”) LB-grown colonies of the MG1655/pAg43 strain were serially diluted for examination. Dilutions were plated onto LB agar supplemented with 100 μg of 5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside (X-Gal)/ml, and plates were incubated at 37°C. Both total counts of viable cells (N) and the number of colonies that switched from the phenotype of the original inoculum (M) were determined. Based on the assumption that predominantly phase-on and phase-off colonies are derived from phase-on and phase-off cells, respectively, the following equation was used to calculate the frequency of phase switching: switching frequency (per cell per generation) = [1 − gth root of (1 − M/N)], where g is the number of growth generations, calculated as g = (log N/log 2).
Aggregation assay.
Aggregation assays were performed as described in reference 68. Briefly, overnight cultures were adjusted to an OD600 of 2.5 by dilution with LB medium. Three-milliliter aliquots of cultures were incubated in 5-ml standing tubes at room temperature, and the OD600 of the upper parts of the cultures were measured every hour for 6 h and after 24 h before image capture. MG1655kmRExTETrbs-flu cultures were grown in the presence of various concentrations of aTc.
Calcofluor phenotype assays.
Two-microliter aliquots of overnight cultures grown at 37°C in LB medium (with added antibiotics and aTc when needed) were spotted onto LB plates containing 0.02% calcofluor (Sigma; reference no. F-3543) and 1 mM HEPES with or without 200 ng of aTc/ml. The spotted drops were allowed to dry, and the plates were incubated for 24 h at 30 or 37°C. The fluorescence of a spot under UV light revealed the binding of calcofluor, indicating cellulose production.
Biofilm formation assay in microfermentors.
All biofilm formation experiments were performed in triplicate with M63B1glu minimal medium supplemented or not with 20 ng of aTc/ml at 37°C. Sixty-milliliter microfermentors containing a removable glass slide were configured as continuous-flow culture bioreactors with a 40-ml h−1 flow rate as described in references 5 and 30 and at http://www.pasteur.fr/recherche/unites/Ggb/biofilmfermenter.html. Because of the different natures of the biofilms, for the study of flu expression or cellulose production, two distinct protocols were used to determine biofilm biomasses. In the case of the flu gene, overnight cultures grown in M63B1glu supplemented with appropriate antibiotics and 20 ng of aTc/ml when required were diluted to an OD600 of 2 in M63B1 medium. Microfermentors were inoculated by dipping the removable glass slides into 15 ml of the diluted cultures for 1 min, followed by a brief rinsing in M63B1 medium before insertion into the microfermentor. Biofilms were grown for 30 h under nonbubbling conditions, and pictures were taken before the resuspension of the biofilms in the microfermentors for OD600 measurements. In the case of the bcsABZC operon, microfermentors were inoculated by direct injection with an amount equivalent to an OD600 of 1 of overnight cultures grown in M63B1glu supplemented with appropriate antibiotics and 20 ng of aTc/ml when required. Biofilms were grown for 26 h under bubbling conditions before the resuspension of the biofilms in the microfermentors for OD600 measurements.
RESULTS
Construction of chromosomal inducible-promoter insertion cassettes.
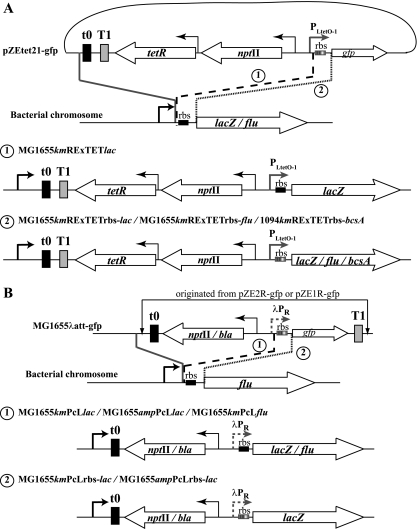
We recently described a selectable repression-expression cassette that places target genes under the control of the inducible PBAD promoter directly at their native chromosomal loci (the RExBAD cassette) (68). While this cassette proved to be an appropriate tool in many applications, the use of a potential carbon source (here, the sugar arabinose) as an inducer may introduce a phenotypic bias. This issue is of particular relevance in the study of biofilms, in which the production of a polysaccharide-based matrix is strongly dependent on bacterial sugar metabolism (see below). As an alternative to the arabinose-inducible RExBAD cassette, we chose to create a new repression-expression cassette based on a TetR-controlled system. TetR binds very tightly to the tet operators (tetO) of the tetA promoter of the Tn10 tetracycline resistance operon (38). TetR-controlled expression systems have been used efficiently in different bacteria (25, 29, 41, 49, 61). This system is induced either by tetracycline, which displays a high affinity for the TetR repressor, or by the tetracycline derivative aTc, a gratuitous nonmetabolizable inducer. To generate the RExTET cassette, we first amplified the sequence encompassing the tetR gene controlled by the constitutive promoter PN25 up to transcription terminator T1 of the rrnB operon from strain DH5αZ1 (49). The PCR fragment was then cloned between the kanamycin resistance gene and the t0 terminator from phage lambda into the pZE21-gfp plasmid, where the gfp gene is under the control of the PLtetO-1 promoter (49), to create pZEtetR21-gfp (see Fig. 1A). pZEtetR21-gfp carries the resulting RExTET cassette, a compact genetic element composed of (i) the tandem and constitutively expressed nptII selectable marker gene (Kmr) and the TetR repressor-encoding gene and, (ii) in the opposite direction, the synthetic TetR-regulated and aTc-inducible PLtetO-1 promoter. Terminators t0 and T1 were placed downstream of the tetR gene to prevent read-through transcription from potential external promoters. The presence of the tetR gene in the RExTET cassette overcomes the need for a specific bacterial background that already expresses this gene. Moreover, the direct insertion of the RExTET cassette-controlled PLtetO-1 promoter by using λ red-mediated homologous recombination can be done either with the original ribosome binding site (RBS) sequence of the chromosomal target gene (kmRExTET cassette) (Fig. 1A, step 1) or with the RBS included in the cassette (kmRExTETrbs) (Fig. 1A, step 2).
FIG. 1.
Construction of the RExTET and PcL transcriptional fusions. (A) RExTET fusions were constructed in three steps, as follows: (i) cloning of the tetR gene with its own constitutive promoter and terminator T1 downstream of the nptII gene encoding resistance to kanamycin and upstream of the t0 terminator, on the pZE21-gfp plasmid; (ii) amplification of the 2,537-bp kmRExTET or the 2,555-bp kmRExTETrbs cassette by using primers annealing to the 3′ end of the t0 terminator and just upstream from the 5′ end of the RBS sequence (1; kmRExTET; no RBS) or just upstream from the 5′ end of the gfp gene (2; kmRExTETrbs; RBS present in the cassette); (iii) insertion of the kmRExTET or kmRExTETrbs cassette by three-step PCR upstream of the RBS sequence or at the start codon of the target gene (lacZ, flu, or bcsA, as indicated), respectively. (B) Construction of constitutive PcL fusions by (i) amplification of the 1,222-bp kmPcL, 1,281-bp ampPcL, 1,254 kmPcLrbs, or 1,313-bp ampPcLrbs cassette by using primers annealing to the 3′ end of the t0 terminator and just upstream from the 5′ end of the RBS sequence (1; kmPcL and ampPcL; no RBS) or just upstream from the 5′ end of the gfp gene (2; kmPcLrbs and ampPcLrbs; RBS present in the cassette) and (ii) insertion of the PcL or PcLrbs cassette by three-step PCR upstream of the RBS sequence or at the start codon of the target gene (lacZ or flu, as indicated), respectively.
To test the functionality of the cassette in the pZEtetR21-gfp plasmid, E. coli strain MG1655 was transformed with the plasmid and the expression of the gfp gene was evaluated via the monitoring of fluorescence. In the absence of the aTc inducer, none of the cells were green, whereas in the presence of 1 μg of aTc/ml, all the cells were fluorescent (data not shown). This result indicated that a large amount of the TetR repressor was produced from the multicopy plasmid and that it could, in the absence of aTc, repress the PLtetO-1 promoter, blocking gfp transcription. Plasmid pZEtetR21-gfp thus served as a template to amplify the 2,527-bp kmRExTET and 2,555-bp kmRExTETrbs cassettes.
Construction of chromosomal constitutive-promoter insertion cassettes.
In order to enable the constitutive expression of chromosomal target genes at their original loci, we took advantage of the pZE2R-gfp and pZE1R-gfp plasmids, which carry gfp under the control of the constitutive λpR promoter (Table 1).
To create the chromosomal kmPcL and ampPcL cassettes, fragments containing the plasmid region from terminator t0 to terminator T1 were amplified and inserted using λ red-mediated homologous recombination at the λatt site in strain MG1655. The resulting strains, MG1655λatt-kmgfp and MG1655λatt-ampgfp, constitutively expressed the green fluorescent protein and fluoresced (data not shown). They served as templates for the amplification of the 1,222-bp kmPcL, 1,254-bp kmPcLrbs, 1,281-bp ampPcL, and 1,313-bp ampPcLrbs cassettes (Fig. 1B). These cassettes comprise (i) a constitutively expressed selectable marker (kanamycin or ampicillin resistance) terminating with the t0 terminator sequence and (ii) the constitutive λpR promoter oriented in the opposite direction (Fig. 1B).
Inducible or constitutive expression of chromosomal target genes.
To test the functionality of both the inducible RExTET and the constitutive PcL cassettes, we inserted the different cassettes upstream of the lacZ gene, between the native lacZ promoter and the lacZ RBS or start codon, in the E. coli K-12 MG1655 chromosome (Fig. 1). These events replaced the native lacZ promoter with the aTc-inducible promoter PLtetO-1 or with the constitutive λpR promoter, creating strains MG1655kmRExTETlacZ (with the native lacZ RBS sequence) or MG1655kmRExTETrbs-lacZ (with the cassette's RBS sequence) and MG1655kmPcLlacZ and MG1655ampPcLlacZ (with the native lacZ RBS sequence) or MG1655kmPcLrbs-lacZ and MG1655ampPcLrbs-lacZ (with the cassette's RBS sequence), respectively.
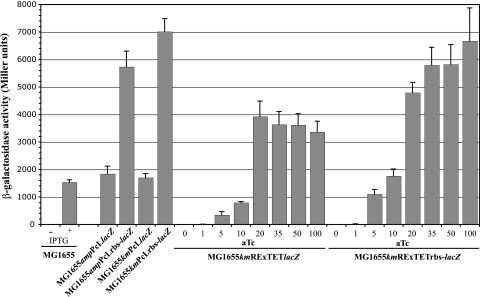
All site-directed promoter replacements resulted in functional reporter transcriptional lacZ fusions, as shown in Fig. 2. PcL cassette-lacZ fusions containing the lacZ RBS (PcLlacZ fusions), associated with either ampicillin or kanamycin resistance, displayed a level of constitutive activation of lacZ comparable to that of the naturally occurring induction of lacZ expression by IPTG in a wild-type background. Interestingly, the transcriptional activation of PcL cassette-lacZ fusions containing the cassette's RBS (PcLrbs-lacZ fusions) resulted in β-galactosidase activities that were at least threefold higher than those resulting from the transcriptional activation of PcLlacZ fusions (Fig. 2). This finding suggests that the RBS sequence of the PcL cassette (present in PcLrbs-lacZ fusions) promotes more efficient translation than the native lacZ RBS.
FIG. 2.
Gene expression can be modulated by RExTET cassettes and can be taken over by PcL cassettes. β-Galactosidase activity measurements of RExTET and PcLlacZ fusions. Strains were grown up until stationary phase in M63B1glu medium at 37°C. Concentrations (nanograms per milliliter) of aTc, when added, are indicated below the bars. MG1655 grown in the presence of 1 mM IPTG was considered to demonstrate the wild-type expression of the lac operon. The experiments were performed in triplicate; error bars represent standard deviations of the means. All the results were qualitatively the same with cultures grown in LB medium (data not shown), with slight changes in the levels of enzymatic activity and aTc concentrations needed to reach the plateau. +, present; −, absent.
To evaluate the extent of the modulation of target gene expression by the kmRExTET cassette, we assessed the range of expression of the lacZ gene in cultures supplemented with various concentrations of aTc (Fig. 2). Both kmRExTET and kmRExTETrbs cassettes displayed a strongly repressed state in the absence of an inducer, with no detectable β-galactosidase activity. Progressive induction was achieved with increasing aTc concentrations of up to 10 ng/ml; higher concentrations led to a steep induction that quickly reached a plateau at around 20 ng of aTc/ml for the lacZ RBS-containing cassette (kmRExTET) and around 35 ng of aTc/ml for the cassette containing the cassette's RBS (kmRExTETrbs). A concentration as low as 10 ng of aTc/ml was sufficient to induce a wild-type level of transcriptional activation (with IPTG) of the kmRExTETrbs-lacZ fusion.
As observed for the constitutive λpR promoter of the PcL cassettes, the origin of the RBS had an influence on the final level of induction of the inducible PLtetO-1 promoter of the RExTET cassette. The same activity was exhibited by the fully induced kmRExTETrbs-lacZ and kmPcLrbs-lacZ and ampPcLrbs-lacZ fusions, which was approximately 1.5-fold the maximal activity of the kmRExTET-lacZ fusion (Fig. 2). In the case of lacZ, the RBS from the cassettes seemed better suited to achieving high levels of activity.
These results show that the RExTET cassette is tightly repressed in the absence of aTc and can be used to modulate target gene expression levels, from no expression to a level approximately fourfold higher than the wild-type level of activation.
Modulation of autotransported Ag43 adhesin production.
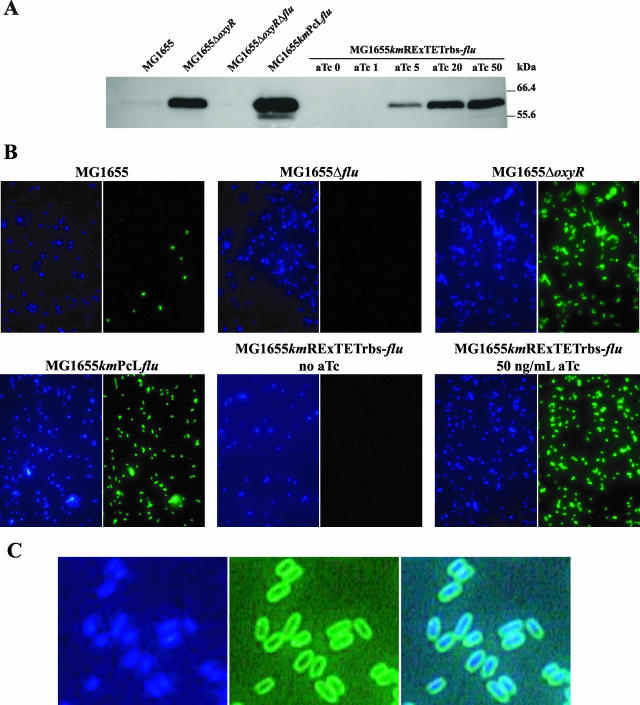
To demonstrate that RExTET and PcL expression systems can be used to control biofilm-promoting factors in E. coli, we introduced into the E. coli K-12 strain MG1655 the kmRExTETrbs and kmPcL cassettes upstream of flu (3.120 kb), the gene encoding the self-recognizing autotransported adhesin Ag43 (23, 57). Autotransporter proteins possess a modular structure with a C-terminal β domain allowing the insertion of the protein into the outer membrane and an N-terminal α passenger domain, exposed at the cell surface, which carries the activity of the protein (35, 36, 56). Ag43 is a major surface protein of E. coli that promotes biofilm formation through cell-to-cell interaction and microcolony formation (16, 44). The expression of Ag43 is phase variable, and the shift from the Ag43+ to the Ag43− phenotype is governed by a mechanism involving the concerted action of both Dam, the GATC site DNA-methylating enzyme deoxyadenosine methylase (activation), and the transcriptional regulator OxyR (repression) (34, 58, 82). In a wild-type situation, each bacterium is either in an Ag43-off situation (if, after DNA replication, the OxyR protein manages to bind to its consensus site before DNA methylation, thus stopping RNA polymerase progression) or in an Ag43-on situation (if DNA methylation occurs before OxyR can bind to DNA). To check that kmRExTETrbs-flu and kmPcLflu constructions could take full control of Ag43 production while maintaining the localization properties of the protein, we monitored the quantities of the α domain of Ag43 in these strains by immunodetection (Fig. 3A) and performed immunofluorescence experiments to locate the protein at the single-cell level (Fig. 3B and C). As shown in Fig. 3B, the on or off state of each wild-type MG1655 bacterium was reflected, respectively, by the presence or absence of Ag43 at the cell surface, and wild-type cells were predominantly in the Ag43-off state. We calculated that in LB medium, the rate of switching from on to off in strain MG1655 bearing pAg43, a plasmid containing the lacZ gene under the control of the flu promoter (81), is ca. 7 × 10−3 switches per cell per generation and the rate of switching from off to on is ca. 10−3 switches per cell per generation. These data are in good agreement with switching frequencies observed previously for chromosomal Ag43 expression in other E. coli strains (58, 66). This finding explains why most MG1655 wild-type cells are in the Ag43-off state (Fig. 3B) and is consistent with the detection of low levels of Ag43 in wild-type MG1655 cells (Fig. 3A). As expected, the deletion of flu or of oxyR resulted, respectively, in the absence of Ag43 or in the detection of high levels of Ag43, which localized at the cell surface (Fig. 3A and B).
FIG. 3.
Modulation of Ag43 production by using PcL and RExTET cassettes. (A) RExTET and PcL cassettes can modulate Ag43 production. An amount of each culture equivalent to an OD600 of 0.2 was loaded onto a sodium dodecyl sulfate-10% polyacrylamide gel electrophoresis gel. Immunodetection was performed using a polyclonal rabbit antiserum raised against the α domain of Ag43. Concentrations (in nanograms per milliliter) of aTc, when added, are indicated above the lanes. (B) Overnight cultures of the different strains were grown at 37°C in LB medium without aTc or, for strain MG1655kmRExTETrbs-flu, with 50 ng of aTc/ml. After fixation and successive incubations with a 1:1,000 dilution of a primary polyclonal rabbit antiserum raised against the α domain of Ag43 and with a 1:300 dilution of a secondary polyclonal goat anti-rabbit serum coupled to Alexa488 along with DAPI, cells were observed with a 600× objective under oil immersion. A DAPI filter was used to reveal the bacterial nucleoid (in blue), and a green fluorescent protein filter was use to reveal the presence of Ag43 (in green) at the cell surface. (C) Closeup of a culture of MG1655kmRExTETrbs-flu in which the Ag43 production had been induced with 50 ng of aTc/ml. The cell surface localization of Ag43 is clearly demonstrated, as is the integrity of the bacterial membrane. The right panel shows the superimposition of the two preceding images.
Consistent with the previously observed kmPcL-mediated activation of lacZ, large amounts of Ag43 were produced when the PcL cassette was introduced in front of the flu gene (Fig. 3A), and all bacteria in the culture exposed Ag43 at the cell surface (Fig. 3B). In the absence of aTc, no Ag43 could be detected in cell extracts from strain MG1655kmRExTETrbs-flu (Fig. 3A) and no bacterium displayed detectable Ag43 at the cell surface (Fig. 3B). These results confirmed that expression from the PLtetO-1 promoter in the RExTET cassette was tightly repressed in the absence of an inducer, therefore mimicking the phenotype of a strain carrying a deletion of flu. They also demonstrated that, in this situation, the natural phase variation of the intact flu promoter located upstream of RExTET does not interfere with the repression process, probably because of the presence of transcriptional terminators in the RExTET cassette. Upon the addition of aTc, a strong correlation between the aTc concentration and the level of Ag43 production from the RExTET cassette was observed (Fig. 3A). Maximal Ag43 production was achieved with 50 ng of aTc/ml, with all bacteria expressing Ag43 at the cell surface (Fig. 3B).
These results demonstrate that PcL and RExTET cassettes can be used to tightly regulate the expression of a naturally phase-variable adhesin-encoding gene such as that for autotransporter Ag43 without affecting the surface localization of the adhesin or the cell membrane integrity, as shown by the regular shapes of the bacteria revealed by the detection of Ag43 (Fig. 3C).
Application of RExTET and PcL cassettes to the study of biofilm phenotypes of E. coli K-12.
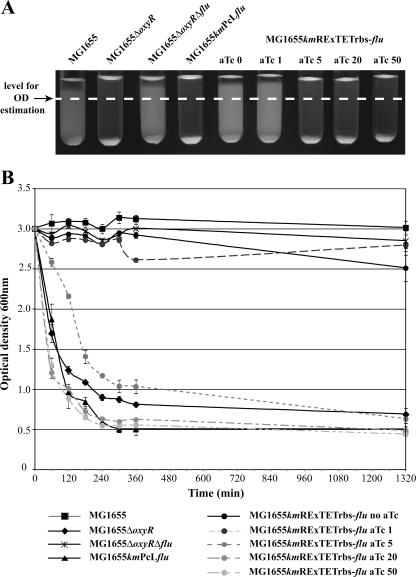
Immunodetection and immunofluorescence experiments (Fig. 3) showed that the expression of Ag43 could be finely regulated under the control of the RExTET and PcL cassettes. To further investigate the phenotype associated with flu expression, we studied the capacities of strains MG1655kmPcL-flu and MG1655kmRExTETrbs-flu to aggregate. As shown in Fig. 4, the constitutive expression of flu led to rapid cell aggregation in standing tubes, and increasing amounts of aTc correlated with increasing bacterial aggregation in tubes. Although the amount of Ag43 in MG1655kmRExTETrbs-flu cells induced with 20 ng of aTc/ml was clearly smaller than that in MG1655kmPcL-flu cells (Fig. 3A), the cells of the two strains aggregated at the same rate (Fig. 4). This suggests that a maximal aggregation pattern is reached at a certain Ag43 concentration, after which an increase in the protein amount does not significantly influence the aggregative phenotype.
FIG. 4.
Modulation of bacterial autoaggregation via controlled production of the Ag43 protein. The different strains were grown overnight in LB with increasing concentrations of aTc ranging from 1 to 50 ng/ml when indicated. After growth, cells were diluted to an OD600 of 2.5 in a 3-ml volume and the autoaggregation of each strain over 24 h at room temperature was assessed by capturing images of the tubes at 24 h (A) and determining the OD600 of the upper part of the standing culture tubes (white dashed line) at the indicated time (B). Aggregation tests were performed in triplicate; error bars represent the standard deviations of the means.
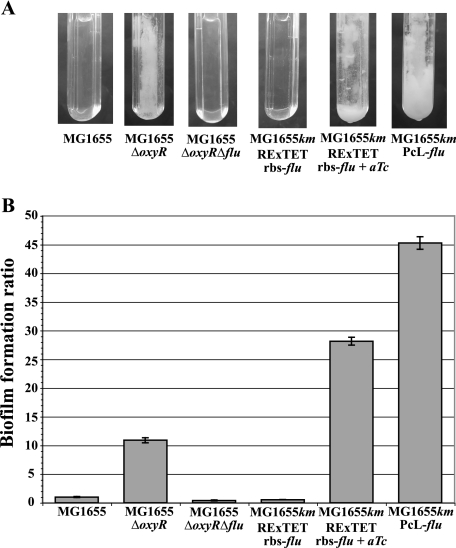
Ag43 has been shown to play a role in biofilm formation and structure (16, 44). Indeed, the derepression of flu in strain MG1655ΔoxyR resulted in a 10-fold increase in the level of biofilm formation compared to that by the wild-type strain in a continuous-flow culture system (Fig. 5) (4). We thus tested the effects of the modulation of the expression of the flu adhesin-encoding gene on strain biofilm ability. As shown in Fig. 5, strain MG1655kmPcL-flu displayed the greatest biofilm capacity of the tested strains, with a 45-fold increase in biofilm biomass compared to that of MG1655. In the absence of an inducer, strain MG1655kmRExTETrbs-flu formed a small biofilm comparable to that formed by strain MG1655ΔoxyRΔflu (Fig. 5). However, upon aTc induction, the MG1655kmRExTETrbs-flu biofilm biomass increased approximately 30-fold compared to that of wild-type MG1655 and 3-fold compared to that of MG1655ΔoxyR.
FIG. 5.
Modulation of biofilm formation via RExTET- and PcL-mediated control of Ag43 production. The ability of the different strains to form a biofilm in a microfermentor was tested in M63B1glu medium. aTc at 20 ng/ml was added to the medium where indicated. Images of the 30-h-old biofilms in microfermentors (A) were captured before the biofilms were resuspended and OD600 measurements were taken (B). The histogram shows the ratios of the biofilm biomasses of the different strains to that of wild-type strain MG1655. Data are the averages of results from three independent experiments; error bars represent standard deviations of the means.
Consequently, the controlled expression of Ag43 through the RExTET and PcL cassettes enabled the modulation of E. coli K-12 biofilm formation abilities.
Modulation of biofilm formation in a natural E. coli isolate by using the RExTET cassette.
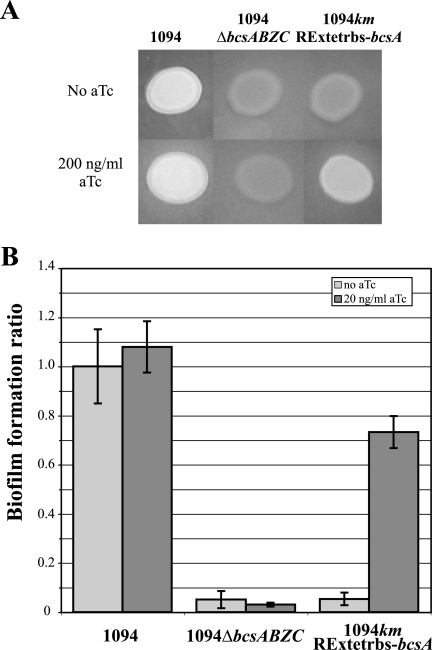
One of the rationales behind creating a new repression-expression cassette lies in the impossibility of using arabinose as an inducer to study biofilm matrix-encoding genes (see above). The biofilm matrix is a complex hydrated milieu that contains proteins, DNA, RNA, ions, and polysaccharide polymers (7). In E. coli, three types of polysaccharides in the biofilm matrix have been detected and demonstrated to be important for biofilm formation. We recently showed that the nonsequenced commensal E. coli 1094 strain possesses genes involved in cellulose production (bcsABZC and bcsEFG operons) and that, in this strain, cellulose production is required for biofilm formation (18). Cellulose has the property of binding calcofluor, which then fluoresces under UV light, thereby facilitating the monitoring of cellulose production in bacterial colonies. We had previously observed that the presence of arabinose on LB plates containing calcofluor modifies the fluorescence phenotype of the E. coli cellulose-producing strain 1094 (S. Da Re, unpublished results). In order to test whether the promoter cassettes described in this study could be used to modulate the expression of biofilm matrix-encoding genes, we introduced the RExTETrbs cassette in front of the bcsABZC cellulose operon. First, we showed that the presence of aTc influenced neither 1094's cellulose production on LB-calcofluor plates nor its capacity for biofilm formation in a microfermentor (Fig. 6). In the absence of aTc, the 1094kmRExTETrbs-bcsA strain did not fluoresce nor form a biofilm and it exhibited the same phenotype as a bcsABZC deletion mutant (Fig. 6). In contrast, aTc could induce the expression of the bcsABZC operon in the 1094kmRExTETrbs-bcsA strain, as demonstrated by fluorescence on the plate (Fig. 6A), and the induction with 20 ng of aTc/ml was sufficient to restore the strain's capacity for biofilm formation to a level close to that of the wild type (Fig. 6B).
FIG. 6.
Modulation by the RExTETrbs cassette of cellulose production and biofilm formation by an E. coli natural isolate. The ability of the different 1094-derivative strains to produce cellulose was tested. (A) Calcofluor binding and fluorescence under UV light. Two-microliter volumes of overnight cultures were spotted onto LB-calcofluor (upper row) and LB-calcofluor-aTc (lower row) plates and incubated for 24 h at 37°C; the aTc concentration on the plate was 200 ng/ml. (B) Capacities for the formation of biofilms in microfermentors. Biofilms were grown for 26 h at 37°C in M63B1glu supplemented or not with 20 ng of aTc/ml. The levels of biofilm formation by the different strains are expressed as the ratios of the OD readings for the resuspended biofilms of the different strains to that for the 1094 biofilm in the absence of aTc. Data are averages of results from three independent experiments; error bars represent standard deviations of the means.
These results show that the use of the gratuitous inducer aTc is an appropriate alternative for matrix polysaccharide gene induction under conditions in which arabinose cannot be used, and they also highlight the possibility provided by our promoter cassettes to modulate the biofilm formation capacities of natural isolates.
DISCUSSION
In the present study, we chose two previously described biofilm-promoting factors, autotransporter adhesin Ag43 and extracellular matrix cellulose, to demonstrate that precise genetic control of the corresponding genes enables the fine modulation of E. coli biofilm formation. Ag43 belongs to a family of self-associating autotransporters comprising other adhesins like AidA-I and TibA that can interact with one another and have been shown to play a role in interspecies contacts (44, 45). Cellulose production is associated with biofilm formation, root colonization, and the persistence of many gram-negative bacteria in natural environments (6, 46).
In order to design strains with high or controlled biofilm formation capacities, we developed two compact promoter insertion cassettes containing either an inducible (RExTET cassette) or a constitutive (PcL cassette) expression system that can be easily introduced upstream of any genes on the E. coli chromosome.
The RExTET cassette was first validated by using transcriptional fusions with the lacZ and flu genes, and in the absence of the inducer aTc, strong repression of the expression of both genes was observed: neither Ag43 production nor β-galactosidase activity associated with the RExTETrbs-flu and RExTETrbs-lacZ or RExTETlacZ fusions could be detected. Under noninducing conditions, the RExTET cassettes therefore mimic the phenotype of a bona fide deletion. Compared to the RExBAD approach that we developed previously, the RExTET construction effectively led to better repression in the absence of an inducer (data not shown) (68). On the other hand, in the presence of increasing concentrations of the aTc inducer, both flu expression and β-galactosidase activity could be induced to increase from levels below those in the wild type to even higher levels. The RExTET cassettes can thus be used to study the functions of the genes of E. coli and any related eubacteria at the genes' native loci. These genetic tools allow for both deletion-like (medium without inducer) and complementation (addition of the gratuitous aTc inducer) conditions and therefore constitute a valuable alternative to conditional-mutant construction. Our approach also provides the advantage of using a gratuitous, nonmetabolizable inducer, thus alleviating the need for the creation of a nonmetabolizing mutant (i.e., one carrying the Δara mutation when the inducer is arabinose), necessary when long-term and homogeneous regulation is required (51, 68). As shown in Fig. 3B, all cells presented Ag43 at the surface when RExTET constructs and the aTc inducer were used, thus demonstrating that the heterogeneity due to lac and PBAD promoter autocatalytic induction phenomena was not observed with this expression system (51).
Recent works demonstrated that the use of antibiotics in biofilm experiments can affect or even induce bacterial biofilm formation (39, 69). The RExTET and PcL expression systems do not require the use of antibiotic selective pressure for stable insertion into the chromosome (data not shown), and this feature is well adapted to the study of multicellular complex communities such as biofilms.
The design of both RExTET and PcL cassettes enables the construction of transcriptional fusions carrying either the RBS of the target gene or the RBS of the cassette. In the case of lacZ, we showed that the choice between the RExTET or PcL and the RExTETrbs or PcLrbs expression systems provides an additional possibility for modulating the production of the protein(s) of interest through translation efficiency.
Our data also showed that, although the strong promoter λpR included in the PcL cassette drove the constitutive expression of the outer membrane protein Ag43, this constitutive expression did not perturb cell growth or cell membrane shape. Nevertheless, in certain cases, it is possible that too-strong expression will perturb cell physiology and morphology. The construction of a set of cassettes with known promoters of different strengths could then be considered for constitutively expressing target genes at different levels.
In addition to the functionality of the RExTET cassette, PcL cassettes enable constitutive gene expression, thus alleviating the need for an external inducer and its potentially associated pleiotropic metabolic effects. The use of such a constitutive cassette is therefore particularly indicated in performing in vivo assays in which plasmids are difficult to maintain over a long period of time and the inducer cannot be regularly and homogeneously provided over the course of the in vivo test. The PcL cassettes described here were consistently and successfully used to constitutively express different adhesin genes in a uropathogenic E. coli strain and to analyze the effects of the overexpression of these genes on early and long-term colonization of the bladder in a mouse model of urinary-tract infection (77a). Such strategies could thus be envisaged for in vivo studies with different pathogens that are genetically amenable.
Beyond the creation of new tools to facilitate the functional study of genes with unknown expression conditions, we propose that the PcL and RExTET expression strategies, or those of other promoter insertion cassettes optimized for gram-positive bacteria, could also be used to engineer a wide range of bacterial strains with high or controlled biofilm formation capacities. Indeed, although many efforts are being directed toward fighting biofilm formation or engineering strains with reduced biofilm formation abilities for biotechnological utilization (76), applications which exploit the biofilm formation abilities of harmless bacteria in a medical environment are presently being considered (19). A recent study demonstrating that, compared to planktonic cells, E. coli biofilms display enhanced high-copy-number-plasmid maintenance and heterologous protein production is also encouraging the use of biofilms in industrial applications (54).
We showed in an in vitro experiment that a commensal MG1655 strain that constitutively expresses the Ag43 autotransporter adhesin (MG1655kmPcLflu) was more efficient than the isogenic MG1655Δflu strain at competing for biofilm formation with a pathogenic enteroaggregative E. coli strain (Fig. S1 in the supplemental material). In vivo approaches using the controlled biofilm capacities of innocuous bacteria could thus be envisaged (i) to challenge deleterious biofilms found in both industrial and medical settings through bacterial interference or competitive adhesion to the surface (77) or through the production of toxic or matrix-dissolving compounds such as cellulase and dispersins, (ii) to optimize the persistence of probiotic strains (15, 19, 24, 27, 28, 31, 33, 37, 64), and (iii) to improve—via stronger heterologous bacterial interactions—the establishment, in nonsterile soil microcosms or in mixed bioreactors, of strains with desirable or genetically engineered features and their development as biosensors or agents in bioremediation processes (8, 22, 26, 53, 65, 72).
In conclusion, our study demonstrates that improved knowledge of bacterial biofilm formation at the molecular level can be used to control cell surface adhesion in bacterial strains of interest. This fine-tuning of cell-to-surface and cell-to-cell interactions of all sorts of bacteria may contribute to the opening up of new perspectives in situations of industrial and medical relevance.
Supplementary Material
Acknowledgments
We are grateful to Peter Owen for his gift of Ag43 antiserum and to Shaynoor Dramsi and Marie-Elise Caliot for their help with immunofluorescence experiments. We thank C. C. Guet for providing different plasmids.
S.D.R. is supported by Sanofi-Pasteur. B.L.Q. is supported by a MENESR (Ministère Français de l'Éducation Nationale, de l'Enseignement Supérieur et de la Recherche) fellowship. This work was supported by grants from the Institut Pasteur, CNRS URA 2172, the Network of Excellence EuroPathoGenomics, the European Community (LSHB-CT-2005-512061), and the Fondation BNP PARIBAS.
Footnotes
Published ahead of print on 23 March 2007.
Supplemental material for this article may be found at http://aem.asm.org/.
REFERENCES
- 1.Aouad, G., J. L. Crovisier, V. A. Geoffroy, J. M. Meyer, and P. Stille. 2006. Microbially-mediated glass dissolution and sorption of metals by Pseudomonas aeruginosa cells and biofilm. J. Hazard. Mater. 136:889-895. [DOI] [PubMed] [Google Scholar]
- 2.Beech, I. B., J. A. Sunner, and K. Hiraoka. 2005. Microbe-surface interactions in biofouling and biocorrosion processes. Int. Microbiol. 8:157-168. [PubMed] [Google Scholar]
- 3.Beloin, C., S. Da Re, and J.-M. Ghigo. August 2005, posting date. Chapter 8.3.1.3, Colonization of abiotic surfaces. In R. Curtiss III, A. Böck, J. L. Ingraham, J. B. Kaper, F. C. Neidhardt, M. Riley, and C. L. Squires (ed.), EcoSal—Escherichia coli and Salmonella: cellular and molecular biology. ASM Press, Washington, DC. http://www.ecosal.org.
- 4.Beloin, C., K. Michaelis, K. Lindner, P. Landini, J. Hacker, J.-M. Ghigo, and U. Dobrindt. 2006. The transcriptional antiterminator RfaH represses biofilm formation in Escherichia coli. J. Bacteriol. 188:1316-1331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Beloin, C., J. Valle, P. Latour-Lambert, P. Faure, M. Kzreminski, D. Balestrino, J. A. Haagensen, S. Molin, G. Prensier, B. Arbeille, and J.-M. Ghigo. 2004. Global impact of mature biofilm lifestyle on Escherichia coli K-12 gene expression. Mol. Microbiol. 51:659-674. [DOI] [PubMed] [Google Scholar]
- 6.Bokranz, W., X. Wang, H. Tschape, and U. Romling. 2005. Expression of cellulose and curli fimbriae by Escherichia coli isolated from the gastrointestinal tract. J. Med. Microbiol. 54:1171-1182. [DOI] [PubMed] [Google Scholar]
- 7.Branda, S. S., S. Vik, L. Friedman, and R. Kolter. 2005. Biofilms: the matrix revisited. Trends Microbiol. 13:20-26. [DOI] [PubMed] [Google Scholar]
- 8.Chalova, V., C. L. Woodward, and S. C. Ricke. 2006. Application of an Escherichia coli green fluorescent protein-based lysine biosensor under nonsterile conditions and autofluorescence background. Lett. Appl. Microbiol. 42:265-270. [DOI] [PubMed] [Google Scholar]
- 9.Chaveroche, M. K., J.-M. Ghigo, and C. d'Enfert. 2000. A rapid method for efficient gene replacement in the filamentous fungus Aspergillus nidulans. Nucleic Acids Res. 28:E97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Coetser, S. E., and T. E. Cloete. 2005. Biofouling and biocorrosion in industrial water systems. Crit. Rev. Microbiol. 31:213-232. [DOI] [PubMed] [Google Scholar]
- 11.Cookson, A. L., W. A. Cooley, and M. J. Woodward. 2002. The role of type 1 and curli fimbriae of Shiga toxin-producing Escherichia coli in adherence to abiotic surfaces. Int. J. Med. Microbiol. 292:195-205. [DOI] [PubMed] [Google Scholar]
- 12.Costerton, J. W., L. Montanaro, and C. R. Arciola. 2005. Biofilm in implant infections: its production and regulation. Int. J. Artif. Organs 28:1062-1068. [DOI] [PubMed] [Google Scholar]
- 13.Costerton, J. W., P. S. Stewart, and E. P. Greenberg. 1999. Bacterial biofilms: a common cause of persistent infections. Science 284:1318-1322. [DOI] [PubMed] [Google Scholar]
- 14.Crespo, J. G., S. Velizarov, and M. A. Reis. 2004. Membrane bioreactors for the removal of anionic micropollutants from drinking water. Curr. Opin. Biotechnol. 15:463-468. [DOI] [PubMed] [Google Scholar]
- 15.Cursino, L., D. Smajs, J. Smarda, R. M. Nardi, J. R. Nicoli, E. Chartone-Souza, and A. M. Nascimento. 2006. Exoproducts of the Escherichia coli strain H22 inhibiting some enteric pathogens both in vitro and in vivo. J. Appl. Microbiol. 100:821-829. [DOI] [PubMed] [Google Scholar]
- 16.Danese, P. N., L. A. Pratt, S. L. Dove, and R. Kolter. 2000. The outer membrane protein, antigen 43, mediates cell-to-cell interactions within Escherichia coli biofilms. Mol. Microbiol. 37:424-432. [DOI] [PubMed] [Google Scholar]
- 17.Danese, P. N., L. A. Pratt, and R. Kolter. 2000. Exopolysaccharide production is required for development of Escherichia coli K-12 biofilm architecture. J. Bacteriol. 182:3593-3596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Da Re, S., and J.-M. Ghigo. 2006. A CsgD-independent pathway for cellulose production and biofilm formation in Escherichia coli. J. Bacteriol. 188:3073-3087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Darouiche, R. O., J. I. Thornby, C. Cerra-Stewart, W. H. Donovan, and R. A. Hull. 2005. Bacterial interference for prevention of urinary tract infection: a prospective, randomized, placebo-controlled, double-blind pilot trial. Clin. Infect. Dis. 41:1531-1534. [DOI] [PubMed] [Google Scholar]
- 20.Datsenko, K. A., and B. L. Wanner. 2000. One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proc. Natl. Acad. Sci. USA 97:6640-6645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Derbise, A., B. Lesic, D. Dacheux, J.-M. Ghigo, and E. Carniel. 2003. A rapid and simple method for inactivating chromosomal genes in Yersinia. FEMS Immunol. Med. Microbiol. 1590:1-4. [DOI] [PubMed] [Google Scholar]
- 22.Diaz, E., A. Ferrandez, M. A. Prieto, and J. L. Garcia. 2001. Biodegradation of aromatic compounds by Escherichia coli. Microbiol. Mol. Biol Rev. 65:523-569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Diderichsen, B. 1980. flu, a metastable gene controlling surface properties of Escherichia coli. J. Bacteriol. 141:858-867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Duncker, S. C., A. Lorentz, B. Schroeder, G. Breves, and S. C. Bischoff. 2006. Effect of orally administered probiotic E. coli strain Nissle 1917 on intestinal mucosal immune cells of healthy young pigs. Vet. Immunol. Immunopathol. 111:239-250. [DOI] [PubMed] [Google Scholar]
- 25.Ehrt, S., X. V. Guo, C. M. Hickey, M. Ryou, M. Monteleone, L. W. Riley, and D. Schnappinger. 2005. Controlling gene expression in mycobacteria with anhydrotetracycline and Tet repressor. Nucleic Acids Res. 33:e21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Erable, B., I. Goubet, S. Lamare, M. D. Legoy, and T. Maugard. 2006. Bioremediation of halogenated compounds: comparison of dehalogenating bacteria and improvement of catalyst stability. Chemosphere 65:1146-1152. [DOI] [PubMed] [Google Scholar]
- 27.Focareta, A., J. C. Paton, R. Morona, J. Cook, and A. W. Paton. 2006. A recombinant probiotic for treatment and prevention of cholera. Gastroenterology 130:1688-1695. [DOI] [PubMed] [Google Scholar]
- 28.Fric, P., and M. Zavoral. 2003. The effect of non-pathogenic Escherichia coli in symptomatic uncomplicated diverticular disease of the colon. Eur. J. Gastroenterol. Hepatol. 15:313-315. [DOI] [PubMed] [Google Scholar]
- 29.Geissendorfer, M., and W. Hillen. 1990. Regulated expression of heterologous genes in Bacillus subtilis using the Tn10 encoded tet regulatory elements. Appl. Microbiol. Biotechnol. 33:657-663. [DOI] [PubMed] [Google Scholar]
- 30.Ghigo, J.-M. 2001. Natural conjugative plasmids induce bacterial biofilm development. Nature 412:442-445. [DOI] [PubMed] [Google Scholar]
- 31.Grabig, A., D. Paclik, C. Guzy, A. Dankof, D. C. Baumgart, J. Erckenbrecht, B. Raupach, U. Sonnenborn, J. Eckert, R. R. Schumann, B. Wiedenmann, A. U. Dignass, and A. Sturm. 2006. Escherichia coli strain Nissle 1917 ameliorates experimental colitis via Toll-like receptor 2- and Toll-like receptor 4-dependent pathways. Infect. Immun. 74:4075-4082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hammar, M., A. Arnqvist, Z. Bian, A. Olsen, and S. Normark. 1995. Expression of two csg operons is required for production of fibronectin- and congo red-binding curli polymers in Escherichia coli K-12. Mol. Microbiol. 18:661-670. [DOI] [PubMed] [Google Scholar]
- 33.Hejnova, J., U. Dobrindt, R. Nemcova, C. Rusniok, A. Bomba, L. Frangeul, J. Hacker, P. Glaser, P. Sebo, and C. Buchrieser. 2005. Characterization of the flexible genome complement of the commensal Escherichia coli strain A0 34/86 (O83 : K24 : H31). Microbiology 151:385-398. [DOI] [PubMed] [Google Scholar]
- 34.Henderson, I. R., M. Meehan, and P. Owen. 1997. A novel regulatory mechanism for a novel phase-variable outer membrane protein of Escherichia coli. Adv. Exp. Med. Biol. 412:349-355. [DOI] [PubMed] [Google Scholar]
- 35.Henderson, I. R., F. Navarro-Garcia, M. Desvaux, R. C. Fernandez, and D. Ala'Aldeen. 2004. Type V protein secretion pathway: the autotransporter story. Microbiol. Mol. Biol. Rev. 68:692-744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Henderson, I. R., F. Navarro-Garcia, and J. P. Nataro. 1998. The great escape: structure and function of the autotransporter proteins. Trends Microbiol. 6:370-378. [DOI] [PubMed] [Google Scholar]
- 37.Henker, J., F. Schuster, and K. Nissler. 2001. Successful treatment of gut-caused halitosis with a suspension of living non-pathogenic Escherichia coli bacteria: a case report. Eur. J. Pediatr. 160:592-594. [DOI] [PubMed] [Google Scholar]
- 38.Hillen, W., and C. Berens. 1994. Mechanisms underlying expression of Tn10 encoded tetracycline resistance. Annu. Rev. Microbiol. 48:345-369. [DOI] [PubMed] [Google Scholar]
- 39.Hoffman, L. R., D. A. D'Argenio, M. J. MacCoss, Z. Zhang, R. A. Jones, and S. I. Miller. 2005. Aminoglycoside antibiotics induce bacterial biofilm formation. Nature 436:1171-1175. [DOI] [PubMed] [Google Scholar]
- 40.Jeong, Y., B. G. Park, and J. S. Chung. 2005. High performance biofilm process for treating wastewater discharged from coal refining plants containing nitrogen, cyanide and thiocyanate. Water Sci. Technol. 52:325-334. [PubMed] [Google Scholar]
- 41.Ji, Y., B. Zhang, S. F. Van Horn, P. Warren, G. Woodnutt, M. K. Burnham, and M. Rosenberg. 2001. Identification of critical staphylococcal genes using conditional phenotypes generated by antisense RNA. Science 293:2266-2269. [DOI] [PubMed] [Google Scholar]
- 42.Karthikeyan, S., and T. J. Beveridge. 2002. Pseudomonas aeruginosa biofilms react with and precipitate toxic soluble gold. Environ. Microbiol. 4:667-675. [DOI] [PubMed] [Google Scholar]
- 43.Kierek-Pearson, K., and E. Karatan. 2005. Biofilm development in bacteria. Adv. Appl. Microbiol. 57:79-111. [DOI] [PubMed] [Google Scholar]
- 44.Kjaergaard, K., M. A. Schembri, C. Ramos, S. Molin, and P. Klemm. 2000. Antigen 43 facilitates formation of multispecies biofilms. Environ. Microbiol. 2:695-702. [DOI] [PubMed] [Google Scholar]
- 45.Klemm, P., R. M. Vejborg, and O. Sherlock. 2006. Self-associating autotransporters, SAATs: functional and structural similarities. Int. J. Med. Microbiol. 296:187-195. [DOI] [PubMed] [Google Scholar]
- 46.Lehner, A., K. Riedel, L. Eberl, P. Breeuwer, B. Diep, and R. Stephan. 2005. Biofilm formation, extracellular polysaccharide production, and cell-to-cell signaling in various Enterobacter sakazakii strains: aspects promoting environmental persistence. J. Food Prot. 68:2287-2294. [DOI] [PubMed] [Google Scholar]
- 47.Lesic, B., S. Bach, J.-M. Ghigo, U. Dobrindt, J. Hacker, and E. Carniel. 2004. Excision of the high-pathogenicity island of Yersinia pseudotuberculosis requires the combined actions of its cognate integrase and Hef, a new recombination directionality factor. Mol. Microbiol. 52:1337-1348. [DOI] [PubMed] [Google Scholar]
- 48.Lindsay, D., and A. von Holy. 13 October 2006. Bacterial biofilms within the clinical setting: what healthcare professionals should know. J. Hosp. Infect. 64:313-325. doi: 10.1016/j.jhin.2006.06.028. [Epub ahead of print.] [DOI] [PubMed] [Google Scholar]
- 49.Lutz, R., and H. Bujard. 1997. Independent and tight regulation of transcriptional units in Escherichia coli via the LacR/O, the TetR/O and AraC/I1-I2 regulatory elements. Nucleic Acids Res. 25:1203-1210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Miller, J. H. 1992. A short course in bacterial genetics: a laboratory manual and handbook for Escherichia coli and related bacteria. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- 51.Morgan-Kiss, R. M., C. Wadler, and J. E. Cronan, Jr. 2002. Long-term and homogeneous regulation of the Escherichia coli araBAD promoter by use of a lactose transporter of relaxed specificity. Proc. Natl. Acad. Sci. USA 99:7373-7377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Morikawa, M. 2006. Beneficial biofilm formation by industrial bacteria Bacillus subtilis and related species. J. Biosci. Bioeng. 101:1-8. [DOI] [PubMed] [Google Scholar]
- 53.Norman, A., L. H. Hansen, and S. J. Sorensen. 2006. A flow cytometry-optimized assay using an SOS-green fluorescent protein (SOS-GFP) whole-cell biosensor for the detection of genotoxins in complex environments. Mutat. Res. 603:164-172. [DOI] [PubMed] [Google Scholar]
- 54.O'Connell, H. A., C. Niu, and E. S. Gilbert. 20 October 2006. Enhanced high copy number plasmid maintenance and heterologous protein production in an Escherichia coli biofilm. Biotechnol. Bioeng. doi: 10.1002/bit.21240. [Epub ahead of print.] [DOI] [PubMed]
- 55.Oliva, B., G. Gordon, P. McNicholas, G. Ellestad, and I. Chopra. 1992. Evidence that tetracycline analogs whose primary target is not the bacterial ribosome cause lysis of Escherichia coli. Antimicrob. Agents Chemother. 36:913-919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Oomen, C. J., P. van Ulsen, P. van Gelder, M. Feijen, J. Tommassen, and P. Gros. 2004. Structure of the translocator domain of a bacterial autotransporter. EMBO J. 23:1257-1266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Owen, P. 1983. Antigens of the Escherichia coli cell envelope, p. 347-373. In O. J. Bjerrum (ed.), Electroimmunochemical analysis of membrane proteins. Elsevier Science Publishing, Amsterdam, The Netherlands.
- 58.Owen, P., M. Meehan, H. de Loughry-Doherty, and I. Henderson. 1996. Phase-variable outer membrane proteins in Escherichia coli. FEMS Immunol. Med. Microbiol. 16:63-76. [DOI] [PubMed] [Google Scholar]
- 59.Pratt, L. A., and R. Kolter. 1998. Genetic analysis of Escherichia coli biofilm formation: roles of flagella, motility, chemotaxis and type I pili. Mol. Microbiol. 30:285-293. [DOI] [PubMed] [Google Scholar]
- 60.Prigent-Combaret, C., G. Prensier, T. T. Le Thi, O. Vidal, P. Lejeune, and C. Dorel. 2000. Developmental pathway for biofilm formation in curli-producing Escherichia coli strains: role of flagella, curli and colanic acid. Environ. Microbiol. 2:450-464. [DOI] [PubMed] [Google Scholar]
- 61.Qian, F., and W. Pan. 2002. Construction of a tetR-integrated Salmonella enterica serovar Typhi CVD908 strain that tightly controls expression of the major merozoite surface protein of Plasmodium falciparum for applications in human vaccine production. Infect. Immun. 70:2029-2038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Ramey, B. E., M. Koutsoudis, S. B. von Bodman, and C. Fuqua. 2004. Biofilm formation in plant-microbe associations. Curr. Opin. Microbiol. 7:602-609. [DOI] [PubMed] [Google Scholar]
- 63.Reisner, A., J. A. Haagensen, M. A. Schembri, E. L. Zechner, and S. Molin. 2003. Development and maturation of Escherichia coli K-12 biofilms. Mol. Microbiol. 48:933-946. [DOI] [PubMed] [Google Scholar]
- 64.Rembacken, B. J., A. M. Snelling, P. M. Hawkey, D. M. Chalmers, and A. T. Axon. 1999. Non-pathogenic Escherichia coli versus mesalazine for the treatment of ulcerative colitis: a randomised trial. Lancet 354:635-639. [DOI] [PubMed] [Google Scholar]
- 65.Rizk, S. S., M. J. Cuneo, and H. W. Hellinga. 2006. Identification of cognate ligands for the Escherichia coli phnD protein product and engineering of a reagentless fluorescent biosensor for phosphonates. Protein Sci. 15:1745-1751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Roche, A., J. McFadden, and P. Owen. 2001. Antigen 43, the major phase-variable protein of the Escherichia coli outer membrane, can exist as a family of proteins encoded by multiple alleles. Microbiology 147:161-169. [DOI] [PubMed] [Google Scholar]
- 67.Romling, U. 2005. Characterization of the rdar morphotype, a multicellular behaviour in Enterobacteriaceae. Cell. Mol. Life Sci. 62:1234-1246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Roux, A., C. Beloin, and J.-M. Ghigo. 2005. Combined inactivation and expression strategy to study gene function under physiological conditions: application to identification of new Escherichia coli adhesins. J. Bacteriol. 187:1001-1013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Sailer, F. C., B. M. Meberg, and K. D. Young. 2003. Beta-lactam induction of colanic acid gene expression in Escherichia coli. FEMS Microbiol. Lett. 226:245-249. [DOI] [PubMed] [Google Scholar]
- 70.Schmidt, F. R. 2004. Recombinant expression systems in the pharmaceutical industry. Appl. Microbiol. Biotechnol. 65:363-372. [DOI] [PubMed] [Google Scholar]
- 71.Shiloach, J., and R. Fass. 2005. Growing E. coli to high cell density: historical perspective on method development. Biotechnol. Adv. 23:345-357. [DOI] [PubMed] [Google Scholar]
- 72.Shinkyo, R., M. Kamakura, S. Ikushiro, K. Inouye, and T. Sakaki. 2006. Biodegradation of dioxins by recombinant Escherichia coli expressing rat CYP1A1 or its mutant. Appl. Microbiol. Biotechnol. 72:584-590. [DOI] [PubMed] [Google Scholar]
- 73.Singh, R., D. Paul, and R. K. Jain. 2006. Biofilms: implications in bioremediation. Trends Microbiol. 14:389-397. [DOI] [PubMed] [Google Scholar]
- 74.Skiadas, I. V., H. N. Gavala, J. E. Schmidt, and B. K. Ahring. 2003. Anaerobic granular sludge and biofilm reactors. Adv. Biochem. Eng. Biotechnol. 82:35-67. [DOI] [PubMed] [Google Scholar]
- 75.Stoodley, P., K. Sauer, D. G. Davies, and J. W. Costerton. 2002. Biofilms as complex differentiated communities. Annu. Rev. Microbiol. 56:187-209. [DOI] [PubMed] [Google Scholar]
- 76.Sung, B. H., C. H. Lee, B. J. Yu, J. H. Lee, J. Y. Lee, M. S. Kim, F. R. Blattner, and S. C. Kim. 2006. Development of a biofilm production-deficient Escherichia coli strain as a host for biotechnological applications. Appl. Environ. Microbiol. 72:3336-3342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Trautner, B. W., R. A. Hull, and R. O. Darouiche. 2003. Escherichia coli 83972 inhibits catheter adherence by a broad spectrum of uropathogens. Urology 61:1059-1062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77a.Ulett, G. C., J. Valle, C. Beloin, O. Sherlock, J.-M. Ghigo, and M. A. Schembri. Functional analysis of antigen 43 in uropathogenic Escherichia coli reveals a role in long-term persistence in the urinary tract. Infect. Immun., in press. [DOI] [PMC free article] [PubMed]
- 78.Van Houdt, R., and C. W. Michiels. 2005. Role of bacterial cell surface structures in Escherichia coli biofilm formation. Res. Microbiol. 156:626-633. [DOI] [PubMed] [Google Scholar]
- 79.Vidal, O., R. Longin, C. Prigent-Combaret, C. Dorel, M. Hooreman, and P. Lejeune. 1998. Isolation of an Escherichia coli K-12 mutant strain able to form biofilms on inert surfaces: involvement of a new ompR allele that increases curli expression. J. Bacteriol. 180:2442-2449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Viggiani, A., G. Olivieri, L. Siani, A. Di Donato, A. Marzocchella, P. Salatino, P. Barbieri, and E. Galli. 2006. An airlift biofilm reactor for the biodegradation of phenol by Pseudomonas stutzeri OX1. J. Biotechnol. 123:464-477. [DOI] [PubMed] [Google Scholar]
- 81.Waldron, D. E., P. Owen, and C. J. Dorman. 2002. Competitive interaction of the OxyR DNA-binding protein and the Dam methylase at the antigen 43 gene regulatory region in Escherichia coli. Mol. Microbiol. 44:509-520. [DOI] [PubMed] [Google Scholar]
- 82.Wallecha, A., V. Munster, J. Correnti, T. Chan, and M. van der Woude. 2002. Dam- and OxyR-dependent phase variation of agn43: essential elements and evidence for a new role of DNA methylation. J. Bacteriol. 184:3338-3347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Wang, X., J. F. Preston III, and T. Romeo. 2004. The pgaABCD locus of Escherichia coli promotes the synthesis of a polysaccharide adhesin required for biofilm formation. J. Bacteriol. 186:2724-2734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Webb, J. S., M. Givskov, and S. Kjelleberg. 2003. Bacterial biofilms: prokaryotic adventures in multicellularity. Curr. Opin. Microbiol. 6:578-585. [DOI] [PubMed] [Google Scholar]
- 85.Zogaj, X., M. Nimtz, M. Rohde, W. Bokranz, and U. Romling. 2001. The multicellular morphotypes of Salmonella typhimurium and Escherichia coli produce cellulose as the second component of the extracellular matrix. Mol. Microbiol. 39:1452-1463. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.