Abstract
Two polycyclic aromatic hydrocarbon (PAH)-contaminated soils of pH 2 were successfully used as inoculum to enrich cultures growing on phenanthrene and pyrene at different pHs, including pH 3. Selected pyrene-utilizing cultures obtained at pH 3, pH 5, and pH 7 were further characterized. All showed rapid [14C]pyrene mineralization at pH 3 and pH 5 and grew on pyrene at pH values ranging from 2 to 6. Eubacterial and mycobacterial 16S rRNA gene denaturing gradient gel electrophoresis fingerprinting and sequencing indicated that the cultures were dominated by a single bacterium closely related to Mycobacterium montefiorense, belonging to the slow-growing Mycobacterium sp. In contrast, a culture enriched on pyrene at pH 7 from a slightly alkaline soil sampled at the same site was dominated by Pseudomonas putida and a fast-growing Mycobacterium sp. The M. montefiorense-related species dominating the pyrene-utilizing cultures enriched from the acidic soils was also the dominant Mycobacterium species in the acidic soils. Our data indicate that a slow-growing Mycobacterium species is involved in PAH degradation in that culture and show that bacteria able to degrade high-molecular-weight PAHs at low pH are present in acidic PAH-contaminated soil.
Polycyclic aromatic hydrocarbons (PAHs) are widespread contaminants that are of environmental concern due to their carcinogenic and mutagenic properties (1, 4). At coal pyrolysis sites, such as former manufactured gas plant (MGP) sites, PAHs are present as components of coal tar that was often deposited in tar pits (16, 23). Due to the presence of inorganic sulfur compounds in coal tar (23), MGP sites often contain acidic PAH-contaminated soils. Another PAH-contaminated environment with low pH is found around outdoor coal storage piles. Exposure to rain and air leads to PAH leaching from the coal and to pyrite oxidation, resulting in acidic PAH-containing runoff that contaminates the surrounding soil (10).
Most studies on PAH biodegradation have focused on microorganisms that can grow on common laboratory media at neutral pH (24). On the other hand, data on the existence of bacteria able to degrade PAHs under acidic conditions are scarce. Stapleton et al. (33) reported on the biodegradation of naphthalene by the soil microbial community of a long-term coal pile storage basin. Dore et al. (10) isolated naphthalene-utilizing bacteria at pH 3 and pH 5 from acidic soil samples (pH 2.5 to pH 3.5) surrounding an outdoor coal storage pile.
In this study, we report on cultures able to utilize pyrene, a recalcitrant four-ring PAH compound with minimal water solubility and low bioavailability in soil, as a sole source of carbon and energy up to pH 2. The cultures were enriched from acidic PAH-contaminated soils from a former MGP site. The obtained communities were compared with communities enriched for growth on pyrene from a PAH-contaminated soil sampled at the same site but with a slightly alkaline pH.
MATERIALS AND METHODS
Soil samples and physicochemical characteristics.
Soil samples TM, B7-1, and B7-2 were taken from different locations at a former MGP site in Belgium. Soil sample B3 was taken from a noncontaminated location at the MGP site. The soils were sampled from the top 0 to 30 cm, except soil B7-2, which was sampled at a 70- to 130-cm depth from the same location as that of soil B7-1. The soils were sieved at 2 mm and stored at 4°C in the dark until use. PAH concentrations, determined as described by Leys et al. (22), were around 10 mg kg−1, 500 mg kg−1, 600 mg kg−1, and 2,200 mg kg−1 for soil B3, soil TM, soil B7-1, and soil B7-2, respectively. The pHs of soil B7-1 and soil B7-2 were 2.6 and 2.4, respectively. The pHs of soil B3 and soil TM were 8.2 and 8.0, respectively. The sulfate concentrations were determined after soil leaching with water (soil/water ratio of 1:10) by using standard method DIN 38414-S4 (9) and were around 4,000 mg kg−1 and 8,400 mg kg−1 for soil B7-1 and soil B7-2, respectively. The measured sulfate concentrations in soils B3 and TM were near the detection limit of 3 mg kg−1.
Medium and enrichment of PAH-degrading bacteria.
The minimal medium (MM) used was prepared as described by Harrison (17). Phenanthrene or pyrene crystals were added as the sole carbon source. The pH of the MM was adjusted to pH 2, 3, 5, or 7 by the addition of 98% H2SO4 or 1 N H2SO4. PAHs were added to the medium after sterilization and inoculation. Solid agar plates with MM at pH 5 or pH 7 contained 25 g/liter agar (Select agar; Invitrogen, Merelbeke, Belgium), while plates with MM at pH 3 contained 35 g/liter agar. All solid media contained 0.1 g/liter cycloheximide (Sigma-Aldrich, Steinheim, Germany) to suppress fungal growth.
Enrichment of PAH-degrading bacteria was performed in 15-ml glass tubes or in 100-ml Erlenmeyer flasks containing 5 ml MM or 20 ml MM (at pH 2, 3, 5, or 7), respectively, and phenanthrene or pyrene as the sole source of carbon and energy. The recipients were inoculated with 100 μl or 400 μl of an aqueous soil extract, respectively, and incubated at 25°C on a rotary shaker at 125 rpm. A total of 0.5 ml of the suspension was transferred to 4.5 ml or 20 ml fresh MM with the same pH and the same PAH when the optical density at 600 nm (OD600) was 0.2. In all cases, control tubes were inoculated containing the same medium without added PAHs. At regular times, the pH of the enrichment cultures was checked using pH indicator strips (Merck, Darmstadt, Germany).
[14C]pyrene mineralization by enrichment cultures.
The mineralization of [4,5,9,10-14C]pyrene (58.7 mCi mmol−1 dissolved in methanol, 99.3% radiochemical purity; Sigma, Munich, Germany) by the cultures was examined in triplicate in MM at pH 3 or pH 5 in 15-ml Pyrex tubes. After the evaporation of methanol (80 μl) in 250-ml Erlenmeyer flasks, [14C]pyrene was dissolved in MM at pH 3 or MM at pH 5 to achieve a final radioactivity of 0.008 μCi/ml. The [14C]pyrene concentration in the MM was below water solubility, i.e., 0.135 mg/liter at 25°C (25). Unlabeled PAH was not added. Soil-free enrichment cultures (0.5 ml) were added to the Pyrex tubes supplied with 4.5 ml of the [14C]pyrene-containing MM. The tubes were closed with Teflon-lined stoppers equipped with alkali traps (1 ml of 0.5 M NaOH) to measure the 14CO2 produced from added [14C]pyrene and incubated at 20°C on a rotary shaker at 150 rpm. Periodically, the NaOH solution was removed from the trap and replaced with fresh alkali. The removed NaOH solution was mixed with 5 ml liquid scintillation cocktail (Ultima Gold; PerkinElmer, Boston, MA), and the mixture was kept in darkness for 8 h for the dissipation of chemiluminescence. Radioactivity was measured with a liquid scintillation counter (Packard Tri-Carb 1600CA; PerkinElmer, Boston, MA).
Molecular analysis.
DNA was extracted from soil as described by Uyttebroek et al. (36). The extraction of DNA from enrichment cultures was done in a similar fashion. Prior to extraction, 20 ml of the culture was washed twice with 0.1 M Na3PO4 (pH 8). Eubacterial 16S rRNA gene fragments were amplified by PCR using the eubacterial primers GC40-63f and 518r (11), as described previously (26). Mycobacterium 16S rRNA gene fragments were amplified by PCR using the Mycobacterium-specific primers MYCO66f and GC40-MYCO600r, as described previously (22). Denaturing gradient gel electrophoresis (DGGE) was performed on an INGENYphorU-2 system (Ingeny International, Goes, The Netherlands). DGGE analysis of the PCR products from amplification with the eubacterial primers GC40-63f and 518r was done as reported previously (26) but for 15 h at 120 V. DGGE analysis of the Mycobacterium PCR products was done as described by Leys et al. (22). Eubacterial and Mycobacterium 16S rRNA PCR products were cloned into the plasmid vector pCR2.1-TOPO, using a TOPO TA cloning kit (Invitrogen, Merelbeke, Belgium) as described earlier (22). The DGGE patterns of cloned fragments were compared with the appropriate fingerprints of the soil eubacterial or Mycobacterium community. The PCR products obtained from the appropriate clones were purified with a PCR purification kit (QIAGEN, Venlo, The Netherlands) according to the manufacturer's instructions and subjected to sequencing reactions. Sequencing reactions were performed with a Quick Start DNA sequencing kit (Beckman, Mijdrecht, The Netherlands) using the eubacterial 16S rRNA gene primer 530r (20) and analyzed on an automatic sequencer (CEQ8000; Beckman-Coulter, Fullerton, CA). The Mycobacterium 16S rRNA gene PCR product obtained from soil B7-2 was directly sequenced after purification without cloning. Partial 16S rRNA gene sequences (about 420 bp) were compared to sequences deposited in GenBank by performing a BLASTN search (2).
Nucleotide sequence accession numbers.
The reported partial 16S rRNA gene sequences were submitted to the EMBL database under accession numbers AM085768 to AM085775 and AM236045.
RESULTS
DGGE analysis of indigenous eubacteria in soil.
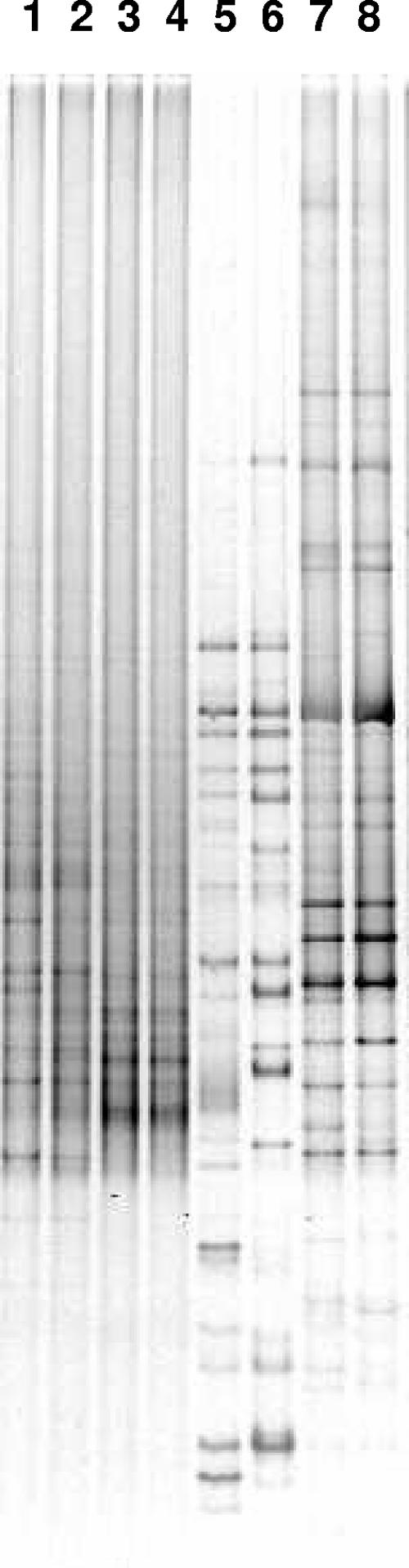
Eubacterial 16S rRNA gene DGGE fingerprints showed that the indigenous eubacterial communities were clearly different for all soil samples (Fig. 1). Both acidic contaminated soils B7-1 and B7-2 showed different dominant bands, while the alkaline contaminated soil TM and the uncontaminated soil B3 demonstrated a high number of bands without clear dominance, indicating a lower eubacterial diversity in soils B7-1 and B7-2 than in soils TM and B3. Fingerprints of replicates of soil sample B7-2 showed related but different community structures. This can be explained by the heterogeneous nature of this soil sample.
FIG. 1.
Eubacterial 16S rRNA gene DGGE analysis of the indigenous microbial community in soil samples. Lanes: 1 to 2, soil TM; 3 to 4, soil B3; 5 to 6, soil B7-2; 7 to 8, soil B7-1.
Enrichment of PAH-degrading bacteria.
From soils TM, B7-1, and B7-2, enrichment cultures that utilized phenanthrene or pyrene as the sole source of carbon and energy were obtained at different pHs (data not shown). The enrichments were set up at two different times with the same soils and gave similar results. Phenanthrene- and pyrene-utilizing cultures from soil TM were obtained at pH 5 and pH 7 but not at pH 3. In contrast, enrichment cultures growing on phenanthrene or pyrene were obtained at pH 3, 5, or 7 from the acidic soils B7-1 and B7-2. In addition, cultures utilizing pyrene at pH 2 were obtained from soils B7-1 and B7-2. The pH of the bacterial cultures was regularly checked and did not change during enrichment. Unfortunately, the enrichment cultures often failed to grow on phenanthrene or pyrene after being transferred to fresh medium. Since pyrene has a higher recalcitrance than phenanthrene (4), we emphasized further research on enrichment cultures that remained growing on pyrene after several passages into fresh medium. Those pyrene-utilizing cultures included a culture enriched at pH 7 from soil TM, a culture enriched at pH 7 from soil B7-1, and two cultures enriched for growth at either pH 3 or pH 5 from soil B7-2. CFU could never be recovered from the cultures on solid agar plates with MM and pyrene as the sole source of carbon and energy, added either as crystals or as an opaque PAH layer, and on LB solid agar plates (data not shown).
[14C]pyrene mineralization by enrichment cultures and growth at different pHs.
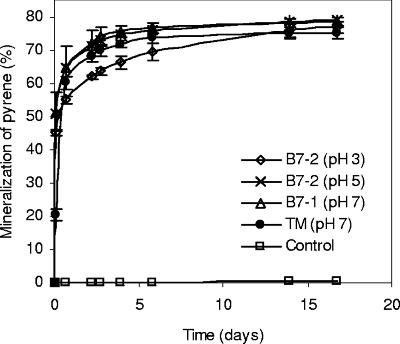
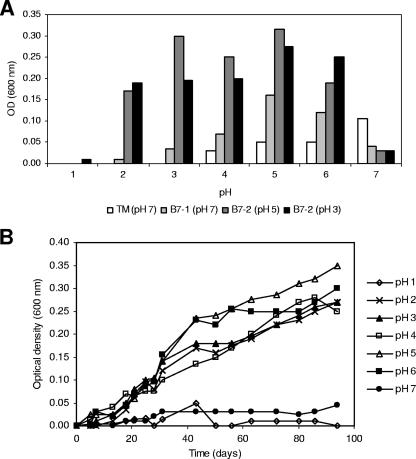
The utilization of pyrene by the four cultures enriched on pyrene was confirmed by the mineralization of [14C]pyrene in MM (Fig. 2). All cultures, including the culture obtained from soil TM enriched at pH 7, showed similar pyrene mineralization rates and extents at pH 3 and pH 5. The four enrichment cultures were tested for growth in MM with pyrene as the sole source of carbon and energy at a pH range from 1 to 7, starting from an OD of 0.001. The growth curves recorded for the culture enriched at pH 3 from soil B7-2 growing at different pHs are shown in Fig. 3B, while the ODs of all enrichment cultures after 2 months of growth are shown in Fig. 3A. If growth is considered positive at an OD of 0.05 or larger, culture TM grew from pH 5 to pH 7. In contrast, the two cultures enriched at pH 3 and pH 5 from acidic soil B7-2 grew at a pH range from 2 to 6 while the culture enriched at pH 7 from acidic soil B7-1 grew from pH 2 to pH 7. However, the culture enriched at pH 7 grew less efficiently at a lower pH than those enriched at pH 3 and pH 5. Cell numbers were at least 20- to 30-fold higher in cultures with an OD of 0.2 than in cultures with an OD of 0.001 (data not shown). The rapid mineralization of [14C]pyrene is not in contrast to the apparent slow growth of the cultures on crystalline pyrene, as the [14C]pyrene concentration in the mineralization experiment was below that for water solubility, allowing rapid uptake and conversion by the bacterial cells.
FIG. 2.
Cumulative [14C]pyrene mineralization curves at pH 3 of the pyrene-utilizing cultures enriched from soil B7-2 at pH 3 and pH 5, enriched from soil B7-1 at pH 7, and enriched from soil TM at pH 7.
FIG. 3.
(A) OD600 of the enrichment cultures after 94 days of growth in MM with pyrene as the sole source of carbon and energy at different pHs. The initial OD was <0.01. (B) Growth curves (OD600 versus time) at different pHs of the culture enriched at pH 3 from soil B7-2.
Community analysis of the enrichment cultures.
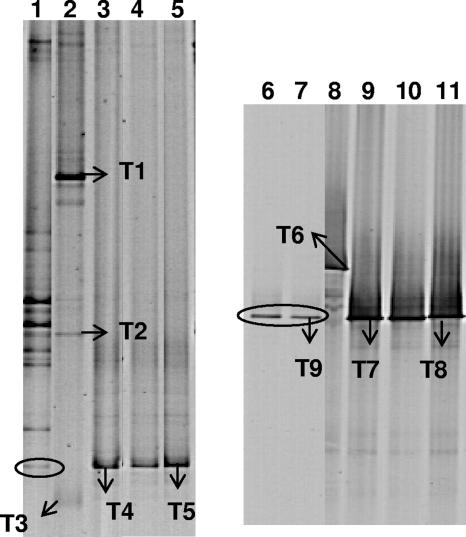
Eubacterial 16S rRNA gene DGGE fingerprinting showed that the communities of the cultures enriched from acidic soil were different from the community of the culture enriched from soil TM. The culture from soil TM showed several bands. In contrast, the cultures from soils B7-1 and B7-2 obtained at pH 3, pH 5, and pH 7 showed only one band and all at the same height (Fig. 4). Interestingly, the dominant band in the DGGE profiles of the three enrichment cultures from the acidic soils comigrated with one band present in the community fingerprint of soil B7-2. The sequences of the 16S rRNA gene bands recovered from the culture enriched at pH 3 from soil B7-2 (band T4) and the culture enriched at pH 7 from soil B7-1 (band T5) were in both cases most closely related to the 16S rRNA gene sequence of Mycobacterium montefiorense (Table 1). The DGGE profile of the culture enriched from soil TM showed several dominant bands, of which bands T1 and T2 had sequences most closely related with the 16S rRNA gene sequence of Pseudomonas putida, while the sequence of band T3 was most related with the 16S rRNA gene sequence of Mycobacterium aurum.
FIG. 4.
DGGE analysis of 16S rRNA gene fragments obtained by PCR with eubacterial primers GC40-63f and 518r (lanes 1 to 5) or with Mycobacterium-specific primers MYCO66f and GC40-MYCO600r (lanes 6 to 11) from the enrichment cultures and selected soil samples. Lanes: 1, soil B7-2; 2, culture TM (pH 7); 3, culture B7-2 (pH 3); 4, culture B7-2 (pH 5); 5, culture B7-1 (pH 7); 6, soil B7-1; 7, soil B7-2; 8, culture TM (pH 7); 9, culture B7-2 (pH 3); 10, culture B7-2 (pH 5); 11, culture B7-1 (pH 7). For more details on bands T1 to T9, see Table 1. An oval indicates the position of a band in the soil fingerprints, comigrating with bands T4 and T5 or with bands T7 and T8.
TABLE 1.
Closest GenBank matches and sequence similarities of selected 16S rRNA gene-based DGGE bands
| Banda | EMBL accession no. | Closest GenBank match (accession no.) | Similarity (%) |
|---|---|---|---|
| T1 | AM085768 | Pseudomonas putida ATCC 17494 (AF094740) | 99.8 |
| T2 | AM085769 | Pseudomonas putida ATCC 17494 (AF094740) | 97.4 |
| T3 | AM085770 | Mycobacterium aurum ATCC 23366 (X55595) | 99.0 |
| T4 | AM085771 | Mycobacterium montefiorense ATCC BAA-256 (AF330038) | 98.1 |
| T5 | AM085772 | Mycobacterium montefiorense ATCC BAA-256 (AF330038) | 98.1 |
| T6 | AM085773 | Mycobacterium aurum ATCC 23366 (X55595) | 99.0 |
| T7 | AM085774 | Mycobacterium montefiorense ATCC BAA-256 (AF330038) | 98.1 |
| T8 | AM085775 | Mycobacterium montefiorense ATCC BAA-256 (AF330038) | 97.8 |
| T9 | AM236045 | Mycobacterium montefiorense ATCC BAA-256 (AF330038) | 98.5 |
Bands T1 to T5 were detected in a PCR of the 16S rRNA gene with eubacterial primers GC40-63f and 518r, and bands T6 to T9 were detected in a PCR of the 16S rRNA gene with Mycobacterium-specific primers MYCO66f and GC40-MYCO600r. For the positions of bands, see Fig. 4.
The presence of Mycobacterium in the enrichment cultures was further investigated by Mycobacterium-specific PCR-DGGE. The Mycobacterium community fingerprints showed a single band for the cultures enriched from soils B7-1 and B7-2 (Fig. 4). Sequencing of band T7 recovered from the culture enriched at pH 3 from soil B7-2 and band T8 recovered from the culture enriched at pH 7 from soil B7-1 confirmed the presence of a species related to M. montefiorense in those cultures (Table 1). The presence of M. aurum in the TM culture was confirmed by Mycobacterium-specific fingerprinting and sequence analysis of band T6. Interestingly, soils B7-1 and B7-2 showed only one band in the Mycobacterium 16S rRNA gene DGGE fingerprints that comigrated with the dominant band in the DGGE profiles of the B7-1 and B7-2 enrichment cultures. The sequence of band T9 was highly identical to the sequence of the band in the Mycobacterium DGGE fingerprint of the pyrene-degrading cultures enriched from the acidic soils.
DISCUSSION
This study shows for the first time the enrichment of a pyrene-utilizing bacterial culture at pH 3 from an extremely acidic PAH-contaminated soil (pH 2.4). This enrichment culture grows on pyrene at pH 2 and also mineralizes 77% of the supplied [14C]pyrene at pH 3 after 5 days of incubation, probably converting the other 23% into biomass (12). PAH-utilizing cultures growing at pH 2 to pH 3 have been enriched from extremely acidic environments but only by using naphthalene as the sole carbon source (10, 33) and not a high-molecular-weight (HMW) and more recalcitrant four-ring PAH such as pyrene.
The cultures enriched at pH 3, pH 5, and pH 7 from the acidic soils B7-1 and B7-2 showed a single dominant bacterium in the eubacterial and Mycobacterium community fingerprints. Although PCR-based analysis might result in biases (34), our data suggest that the three cultures enriched from the acidic soils were dominated by highly similar Mycobacterium species. Although it cannot be excluded that the detected Mycobacterium strains grow on secondary products produced by bacteria present in the culture but not recognized by the PCR-DGGE approach, our data suggest that the strains are involved in pyrene degradation in the cultures. This seems to be confirmed by microscopic analysis of the cultures which showed only one cell type typical for Mycobacterium (data not shown). Moreover, the cultures have been transferred to fresh medium up to seven times and always gave the same fingerprint after growth (data not shown). Mycobacterium is a bacterial genus that is often isolated from PAH-contaminated soil for its ability to grow on pyrene as the sole source of carbon and energy (5, 6, 18, 22). 16S rRNA gene sequence analysis showed that the Mycobacterium isolate was most closely related to M. montefiorense, a pathogenic Mycobacterium sp. isolated from moray eels. M. montefiorense is related to Mycobacterium triplex and belongs to the so-called branch of slow-growing mycobacteria (21), which phylogenetically form a coherent line of descent, distinct from the so-called fast-growing mycobacteria (32, 35). So far, all PAH-biodegrading Mycobacterium isolates belong to the phylogenetic branch of the fast-growing mycobacteria. With a few exceptions (13, 15), all of those strains were enriched from near-neutral PAH-contaminated soils in enrichments at a pH of around 7. Also, the culture enriched at pH 7 from soil TM included Mycobacterium aurum, a species closely related to other PAH-degrading fast-growing mycobacteria, such as M. pyrenivorans, M. austroafricanum, and M. vanbaalenii (8). In addition to the Mycobacterium band, this culture showed other dominant bands in DGGE. The DNA sequences of two of them matched with the 16S rRNA gene sequence of the same P. putida strain. P. putida contains seven 16S rRNA gene copies (27) with slightly different sequences, and therefore, the bands might eventually correspond to the same P. putida strain. P. putida has been reported to use low-molecular-weight PAHs, such as naphthalene and phenanthrene, as the sole source of carbon and energy but not pyrene (14, 31).
The culture enriched from acidic soil B7-1 at pH 7 grew on pyrene from pH 2 to pH 7 but clearly did not grow as well at a lower pH as the cultures enriched from acidic soil B7-2 at pH 3 and pH 5. This indicates a higher acid tolerance of the cultures enriched at a low pH than that of the culture enriched at a neutral pH. Since the community analysis suggests that in both cultures the same strain is involved in pyrene degradation, these results might indicate that the tolerance to a low pH can be lost upon selection at a higher pH or that the cultures at pH 3 and pH 5 were obtained by genetic adaptation of the strain to a low pH during enrichment. However, the latter seems unlikely as the pyrene-degrading primary cultures enriched from the acidic soils at pH 3 developed as rapidly as and often more rapidly than those at pH 7. On the other hand, it cannot be excluded that both cultures harbor different but related organisms, as the DNA sequences of the corresponding bands in the 16S rRNA gene DGGE fingerprints showed some minor differences (Table 1), which were nonvisible on the DGGE fingerprint (22).
Our study shows the existence of acid-tolerant mycobacteria. Studies of the effect of a low pH on mycobacteria have concentrated mainly on pathogenic species such as M. tuberculosis, M. avium, and M. avium subsp. paratuberculosis (7, 28). Growth was mostly restricted at a pH lower than 6, but various mycobacteria, such as M. avium, resist exposure to pH 2 for periods of up to 24 h (3). Such a resistance to a low pH for short periods can explain the fast mineralization of [14C]pyrene (60% in 17 h) at pH 3 by the culture enriched at pH 7 from the TM soil despite its inability to grow on pyrene at pH 3. Recently, Kim et al. (19) showed that the degradation rates of phenanthrene and pyrene increased fourfold at pH 6.5 compared to those at pH 7.5. Not much is known about the mechanism of acid tolerance in mycobacteria. Proposed mechanisms include proton extrusion in M. smegmatis (30) and proton removal by glutamate decarboxylase activity in M. leprae (29).
Interestingly, the Mycobacterium community of the acidic soils was dominated by only one Mycobacterium species, which was proven to be highly related to the M. montefiorense-related populations dominating the pyrene-degrading enrichment cultures. In contrast, the Mycobacterium community of the TM soil was dominated by several Mycobacterium species (22) and even enrichment at pH 7 from the acidic soils resulted in an M. montefiorense-related population. Together, these data indicate that the slow-growing Mycobacterium species had survived over other Mycobacterium species in the B7 soils.
In conclusion, our data show that bacteria able to degrade HMW PAHs, such as pyrene, can be enriched from acidic PAH-contaminated soil and hence that bacteria able to degrade pyrene at an acidic pH are present in such soils. These data open interesting perspectives for studies on the degradation of HMW PAHs in acidic PAH-contaminated soils. The fact that in those cultures, a slow-growing Mycobacterium species, rather than a fast-growing Mycobacterium species, seems to be involved in pyrene degradation and that this species is also the major Mycobacterium species in the soil of origin is itself an interesting observation.
Acknowledgments
This study was supported by a grant from the Research Fund of K.U. Leuven to M.U. and by the EC project QLRT-1999-00326.
We thank M. Van Hessche for assistance in sequence analysis.
Footnotes
Published ahead of print on 16 March 2007.
REFERENCES
- 1.Alexander, M. 1999. Biodegradation and bioremediation. Academic Press, San Diego, CA.
- 2.Altschul, S. F., T. L. Madden, A. A. Schäffer, J. Zhang, Z. Zhang, W. Miller, and D. J. Lipman. 1997. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 25:3389-3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bodmer, T., E. Miltner, and L. E. Bermudez. 2000. Mycobacterium avium resists exposure to the acidic conditions of the stomach. FEMS Microbiol. Lett. 182:45-49. [DOI] [PubMed] [Google Scholar]
- 4.Cerniglia, C. E. 1992. Biodegradation of polycyclic aromatic hydrocarbons. Biodegradation 3:351-368. [Google Scholar]
- 5.Cheung, P. Y., and B. K. Kinkle. 2001. Mycobacterium diversity and pyrene mineralization in petroleum-contaminated soils. Appl. Environ. Microbiol. 67:2222-2229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cheung, P. Y., and B. K. Kinkle. 2005. Changes in Mycobacterium spp. population structure and pyrene mineralization in polycyclic aromatic hydrocarbon-amended soils. Soil Biol. Biochem. 37:1929-1937. [Google Scholar]
- 7.Cotter, P. D., and D. Hill. 2003. Surviving the acid test: responses of gram-positive bacteria to low pH. Microbiol. Mol. Biol. Rev. 67:429-453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Derz, K., U. Klinner, I. Schuphan, E. Stackebrandt, and R. M. Kroppenstedt. 2004. Mycobacterium pyrenivorans sp. nov., a novel polycyclic-aromatic-hydrocarbon-degrading species. Int. J. Syst. Evol. Microbiol. 54:2313-2317. [DOI] [PubMed] [Google Scholar]
- 9.Deutsches Institut für Normung. 1984. Deutsche Einheitsverfahren zur Wasser-, Abwasser- und Schlammuntersuchung-Schlamm und Sedimente-Bestimmung der Eluierbarkeit mit Wasser. DIN 38414-S4. Deutsches Institut für Normung e.V., Berlin, Germany.
- 10.Dore, S. Y., Q. E. Clancy, S. M. Rylee, and C. F. Kulpa. 2003. Naphthalene-utilizing and mercury-resistant bacteria isolated from an acidic environment. Appl. Microbiol. Biotechnol. 63:194-199. [DOI] [PubMed] [Google Scholar]
- 11.el Fantroussi, S., L. Verschuere, W. Verstraete, and E. M. Top. 1999. Effect of phenylurea herbicides on soil microbial communities estimated by analysis of 16S rRNA gene fingerprints and community-level physiological profiles. Appl. Environ. Microbiol. 65:982-988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Garcia-Junco, M., C. Gomez-Lahoz, J. L. Niqui-Arroyo, and J. J. Ortega-Calvo. 2003. Biosurfactant- and biodegradation-enhanced partitioning of polycyclic aromatic hydrocarbons from nonaqueous-phase liquids. Environ. Sci. Technol. 37:2988-2996. [DOI] [PubMed] [Google Scholar]
- 13.Grosser, R. J., D. Warshawsky, and R. J. Vestal. 1991. Indigenous and enhanced mineralization of pyrene, benzo[a]pyrene, and carbazole in soils. Appl. Environ. Microbiol. 57:3462-3469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Guerin, W. F., and S. A. Boyd. 1992. Differential bioavailability of soil-sorbed naphthalene to two bacterial species. Appl. Environ. Microbiol. 58:1142-1152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Habe, H., M. Kanemitsu, M. Nomura, T. Takemura, K. Iwata, H. Nojiri, H. Yamane, and T. Omori. 2004. Isolation and characterization of an alkaliphilic bacterium utilizing pyrene as a carbon source. J. Biosci. Bioeng. 98:306-308. [DOI] [PubMed] [Google Scholar]
- 16.Haeseler, F., D. Blanchet, V. Druelle, P. Werner, and J. P. Vandecasteele. 1999. Ecotoxicological assessment of soils of former manufactured gas plant sites: bioremediation potential and pollutant mobility. Environ. Sci. Technol. 33:4379-4384. [Google Scholar]
- 17.Harrison, A. P. 1981. Acidiphilium cryptum gen. nov., sp. nov., heterotrophic bacterium from acidic mineral environments. Int. J. Syst. Bacteriol. 31:327-332. [Google Scholar]
- 18.Kanaly, R. A., and S. Harayama. 2000. Biodegradation of high-molecular-weight polycyclic aromatic hydrocarbons by bacteria. J. Bacteriol. 182:2059-2067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kim, Y. H., J. P. Freeman, J. D. Moody, K. H. Engesser, and E. Cerniglia. 2005. Effects of pH on the degradation of phenanthrene and pyrene by Mycobacterium vanbaalenii PYR-1. Appl. Microbiol. Biotechnol. 67:275-285. [DOI] [PubMed] [Google Scholar]
- 20.Lane, D. J. 1991. 16S/23S rRNA sequencing, p. 115-175. In E. Stackebrandt and M. Goodfellow (ed.), Nucleic acid techniques in bacterial systematics. John Wiley & Sons, Chichester, United Kingdom.
- 21.Levi, M. H., J. Bartell, L. Gandolfo, S. C. Smole, S. F. Costa, L. M. Weiss, L. K. Johnson, G. Osterhout, and L. H. Herbst. 2003. Characterization of Mycobacterium montefiorense sp. nov., a novel pathogenic Mycobacterium from moray eels that is related to Mycobacterium triplex. J. Clin. Microbiol. 41:2147-2152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Leys, N. M., A. Ryngaert, L. Bastiaens, P. Wattiau, E. M. Top, W. Verstraete, and D. Springael. 2005. Occurrence and community composition of fast growing Mycobacterium in soils contaminated with polycyclic aromatic hydrocarbons. FEMS Microbiol. Ecol. 51:375-388. [DOI] [PubMed] [Google Scholar]
- 23.Luthy, R. G., D. A. Dzombak, C. A. Peters, S. B. Roy, A. Ramaswami, D. V. Nakles, and B. R. Nott. 1994. Remediating tar-contaminated soils at manufactured gas plant sites. Environ. Sci. Technol. 28:266A-276A. [DOI] [PubMed] [Google Scholar]
- 24.Margesin, R., and F. Schinner. 2001. Biodegradation and bioremediation of hydrocarbons in extreme environments. Appl. Microbiol. Biotechnol. 56:650-663. [DOI] [PubMed] [Google Scholar]
- 25.Miller, M. M., S. P. Wasik, G. L. Huang, W. Y. Shiu, and D. Mackay. 1985. Relationships between octanol-water partition coefficient and aqueous solubility. Environ. Sci. Technol. 19:522-529. [DOI] [PubMed] [Google Scholar]
- 26.Moreels, D., L. Bastiaens, F. Ollevier, R. Merckx, L. Diels, and D. Springael. 2004. Evaluation of the intrinsic methyl tert-butyl ether (MTBE) biodegradation potential of hydrocarbon contaminated subsurface soils in batch microcosm systems. FEMS Microbiol. Ecol. 49:121-128. [DOI] [PubMed] [Google Scholar]
- 27.Nelson, K. E., C. Weinel, I. T. Paulsen, R. J. Dodson, H. Hilbert, V. A. P. Martins dos Santos, D. E. Fouts, S. R. Gill, M. Pop, M. Holmes, L. Brinkac, M. Beanan, R. T. DeBoy, S. Daugherty, J. Kolonay, R. Madupu, W. Nelson, O. White, J. Peterson, H. Khouri, I. Hance, P. Chris Lee, E. Holtzapple, D. Scanlan, K. Tran, A. Moazzez, T. Utterback, M. Rizzo, K. Lee, D. Kosack, D. Moestl, H. Wedler, J. Lauber, D. Stjepandic, J. Hoheisel, M. Straetz, S. Heim, C. Kiewitz, J. Eisen, K. N. Timmis, A. Düsterhöft, B. Tümmler, and C. M. Fraser. 2002. Complete genome sequence and comparative analysis of the metabolically versatile Pseudomonas putida KT2440. Environ. Microbiol. 4:799-808. [DOI] [PubMed] [Google Scholar]
- 28.Piddington, D. L., A. Kashkouli, and N. A. Buchmeier. 2000. Growth of Mycobacterium tuberculosis in a defined medium is very restricted by acid pH and Mg2+ levels. Infect. Immun. 68:4518-4522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Prabhakaran, K., E. B. Harris, and W. F. Kirchheimer. 1983. Glutamic acid decarboxylase in Mycobacterium leprae. Arch. Microbiol. 134:320-323. [DOI] [PubMed] [Google Scholar]
- 30.Rao, M., T. L. Streur, F. E. Aldwell, and G. M. Cook. 2001. Intracellular pH regulation by Mycobacterium smegmatis and Mycobacterium bovis BCG. Microbiology 147:1017-1024. [DOI] [PubMed] [Google Scholar]
- 31.Rodrigues, A. C., S. Wuertz, A. G. Brito, and L. F. Melo. 2005. Fluorene and phenanthrene uptake by Pseudomonas putida ATCC 17514: kinetics and physiological aspects. Biotechnol. Bioeng. 90:281-289. [DOI] [PubMed] [Google Scholar]
- 32.Stahl, D. A., and J. W. Urbance. 1990. The division between fast- and slow-growing species corresponds to natural relationships among the mycobacteria. J. Bacteriol. 172:116-124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Stapleton, R. D., D. C. Savage, G. S. Sayler, and G. Stacey. 1998. Biodegradation of aromatic hydrocarbons in an extremely acidic environment. Appl. Environ. Microbiol. 64:4180-4184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Suzuki, M. T., and S. J. Giovannoni. 1996. Bias caused by template annealing in the amplification of mixtures of 16S rRNA genes by PCR. Appl. Environ. Microbiol. 62:625-630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Tortoli, E. 2003. Impact of genotypic studies on mycobacterial taxonomy: the new mycobacteria of the 1990s. Clin. Microbiol. Rev. 16:319-354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Uyttebroek, M., P. Breugelmans, M. Janssen, P. Wattiau, B. Joffe, U. Karlson, J. J. Ortega-Calvo, L. Bastiaens, A. Ryngaert, M. Hausner, and D. Springael. 2006. Distribution of the Mycobacterium community and polycyclic aromatic hydrocarbons (PAHs) among different size fractions of a long-term PAH-contaminated soil. Environ. Microbiol. 8:838-847. [DOI] [PubMed] [Google Scholar]