Abstract
Intestinal pathogenic Escherichia coli represents a global health problem for mammals, including humans. At present, diarrheagenic E. coli bacteria are grouped into seven major pathotypes that differ in their virulence factor profiles, severity of clinical manifestations, and prognosis. In this study, we developed and evaluated a one-step multiplex PCR (MPCR) for the straightforward differential identification of intestinal pathotypes of E. coli. The specificity of this novel MPCR was validated by using a subset of reference strains and further confirmed by PCR-independent pheno- and genotypic characterization. Moreover, we tested 246 clinical E. coli isolates derived from diarrhea patients from several distinct geographic regions. Interestingly, besides strains belonging to the defined and well-described pathotypes, we identified five unconventional strains expressing intermediate virulence factor profiles. These strains have been further characterized and appear to represent intermediate strains carrying genes and expressing factors associated with enteropathogenic E. coli, Shiga toxin-producing E. coli, enterotoxigenic E. coli, and enteroaggregative E. coli alike. These strains represent further examples of the extraordinary plasticity of the E. coli genome. Moreover, this implies that the important identification of specific pathotypes has to be based on a broad matrix of indicator genes. In addition, the presence of intermediate strains needs to be accounted for.
Diarrheal diseases are leading causes of morbidity and mortality, especially in Africa, Asia, and Latin America (18). Although commonly regarded as a nonpathogenic beneficial inhabitant of the gastrointestinal tract, Escherichia coli is an important bacterial pathogen. Several highly adapted clones have acquired specific virulence factors that are accountable for a variety of intestinal and extraintestinal diseases including diarrhea, acute inflammation, hemorrhagic colitis, urinary tract infections, septicemia, and neonatal meningitis (18, 25, 31, 32). Currently, diarrheagenic E. coli bacteria can be grouped into seven major categories: enteropathogenic E. coli (EPEC), atypical EPEC (ATEC), locus of enterocyte effacement (LEE)-positive and LEE-negative Shiga toxin (Stx)-producing E. coli (STEC), enterotoxigenic E. coli (ETEC), enteroinvasive E. coli (EIEC), and enteroaggregative E. coli (EAEC) (31).
EPEC is a major etiological agent of infant diarrhea predominantly in developing countries (7, 8, 18). EPEC colonizes the small intestine and causes typical attaching-and-effacing lesions characterized by the degeneration of microvilli and intimate adherence of bacteria to epithelial membranes. Virulence factors necessary for full pathogenicity are encoded on a 35-kb pathogenicity island (PAI), the LEE PAI (9, 13). ATEC lacks the EAF (EPEC adherence factor) plasmid encoding the bundle-forming pili (BFP) that in EPEC are responsible for localized adherence and the formation of microcolonies on host cells (2, 14). Interestingly, ATEC strains are identified with increasing frequency and—in industrialized areas—have become a more frequent cause of diarrhea than typical EPEC and, just like STEC, resemble emerging pathogens (15, 44).
Infection with Stx-producing E. coli (STEC) can result in a spectrum of outcomes ranging from asymptomatic carriage to uncomplicated diarrhea but also in bloody diarrhea and the hemolytic-uremic syndrome (HUS) (42). The defining features of this pathotype are the bacteriophage-encoded Stxs that instigate cytotoxic effects on endothelial cells in the kidneys, pancreas, heart, and brain. Stxs have been subdivided, on the basis of toxin neutralization assays and sequence analysis of stx genes, into two families, Stx1 and Stx2. Each of these families consists of the major Stx type and several variants.
ETEC is the leading cause of traveler's diarrhea of adults in industrialized countries and children in developing countries. ETEC bacteria are largely defined by their production of the plasmid-encoded heat-labile (LT) and heat-stable (STIa/STIb) toxins (31).
EIEC bacteria are associated with watery diarrhea and inflammation with fever resembling Shigella (23). After invading enterocytes, EIEC characteristically induces polarized actin comets, propelling the bacteria through the cytoplasm of host cells, also allowing cell-to-cell spread. All of the necessary effector proteins described thus far are carried on a 140-MDa virulence plasmid, pInv.
Although EAEC has been associated with sporadic and persistent diarrhea, thus far detailed studies of EAEC virulence mechanisms have been hindered by the high phenotypic and genotypic diversity of this pathogroup. All EAEC bacteria are characterized by their aggregative-adherence (AA) pattern, designated the stacked-brick configuration, that is mostly mediated by aggregative-adherence fimbriae (AAF) encoded on a 60-MDa plasmid (17). Most EAEC bacteria harbor additional virulence factors such as, for example, EAST1 (EAEC heat-stable enterotoxin, a homolog of ST of ETEC) and serine proteases like Pet and Pic.
Because of the pronounced differences in virulence factor profiles of E. coli pathotypes, the clinical manifestations, and the severity of intestinal diseases, therapeutic options and prognosis are dependent on the differential diagnosis of the causative agent. At present, routine detection and differentiation of diarrheagenic E. coli are usually based on a combination of biochemical tests, serotyping, phenotypic assays based on virulence characteristics, and molecular detection methods. To simplify and accelerate differential diagnosis, we designed and evaluated a novel multiplex PCR (MPCR) for the simultaneous detection and differentiation of the seven major categories of intestinal pathogenic E. coli strains. This was achieved by combining 12 specific primer pairs in a single reaction mixture. Validation with a collection of reference strains proved this novel MPCR to be highly specific. Interestingly, we could identify several strains that expressed unusual virulence factor profiles apparently representing intermediate pathotypes. This not only serves as a further example of the plasticity of the E. coli genome but, moreover, also emphasizes the need for the differential identification of specific pathotypes in order to facilitate appropriate countermeasures.
(This study is part of the Ph.D. thesis of D.M.)
MATERIALS AND METHODS
Bacterial strains.
The 246 diarrheagenic E. coli strains investigated in this study were isolated from patients with diarrhea in seven different countries (Brazil, Denmark, France, Germany, Great Britain, Mexico, and the United States). Strains were kindly provided by G. Schmidt (Borstel, Germany), K. Jann (Freiburg, Germany), H.-G. Sonntag (Heidelberg, Germany), G. Peters (Münster, Germany), A. Cravioto (Mexico City, Mexico), C. Jallat (Clermont-Ferrand, France), S. Knutton (Birmingham, United Kingdom), C. Le Bouguénec (Paris, France), L. W. Riley (New York, NY), S. Moseley (Seattle, WA), F. Ørskov and I. Ørskov (Copenhagen, Denmark), and L. Trabulsi (Saõ Paulo, Brazil), and K. Wachsmuth (Atlanta, GA). Additional strains are from our collection. A subset of these strains was analyzed for stx genotypes, bfpB, and escV in a preceding study (29) and described previously (5, 40, 41). The nine reference strains used as controls in the MPCR included EPEC strain E2348/69 (LEE positive, bfp positive), ATEC strain 9812 (LEE positive), STEC strain EDL933 (LEE positive, stx1 positive, stx2 positive), STEC strain 04-3175 (stx1 positive, stx2 positive), ETEC strain 164/82 (elt positive, estIa positive), ETEC strain 117/86 (estIb positive), EIEC strain 99-10282 (invE positive), and EAEC strain 02-1850 (astA positive, aggR positive, pic positive). E. coli strain C600 (uidA positive) served as a negative control for virulence genes in all PCRs. Serotyping and identification of rough strains were performed by the National Reference Center for Bacterial Gastroenteritis at the Robert Koch Institute by using the whole spectrum of typing sera for E. coli O and H antigens. Rough strains were detected by standard slide agglutination assay.
Detection of selected virulence determinants by PCR.
The genes selected for incorporation into the MPCR, uidA, escV, bfpB, stx1, stx2, elt, estIa, estIb, invE, astA, aggR, and pic, were amplified by colony PCR with the optimized primer pairs listed in Table 1. The MPCR was performed in 200-μl reaction tubes with a 25-μl reaction mixture consisting of 2 U of Taq DNA polymerase (Segenetic, Borken, Germany), 84 mM Tris HCl (pH 8.5), 2.1 mM MgCl2, 50 mM KCl, 14 mM 2-mercaptoethanol, 0.14% Triton X-100, 0.3 mM each deoxynucleoside triphosphate, and PCR primers at the concentrations listed in Table 1. Thermocycling conditions were as follows: 94°C for 5 min, 30 cycles of 94°C for 30 s, 63°C for 30 s, and 72°C for 1.5 min; and a final extension at 72°C for 5 min. As templates, single bacterial colonies were picked from freshly incubated standard I agar plates and resuspended for 1 min in the reaction mixture on ice. PCR-amplified fragments (10 μl) were separated on 2.0% (wt/vol) agarose gels and visualized under UV light after staining with ethidium bromide.
TABLE 1.
Primer pairs used for detection of marker genes indicative of a particular pathotype
| Pathovar and target gene | Primer | Sequence (5′ to 3′) | Product size (bp) | Concn (μM) | Annealing temp (°C) |
|---|---|---|---|---|---|
| LEE-positive strains (EPEC, ATEC, STEC) | |||||
| escV | MP3-escV-F | ATTCTGGCTCTCTTCTTCTTTATGGCTG | 544 | 0.4 | 63 |
| MP3-escV-R | CGTCCCCTTTTACAAACTTCATCGC | 0.4 | |||
| Typical EPEC | |||||
| bfpB | MP3-bfpB-F | GACACCTCATTGCTGAAGTCG | 910 | 0.1 | 63 |
| MP3-bfpB-R | CCAGAACACCTCCGTTATGC | 0.1 | |||
| STEC | |||||
| stx1 | MP4-stx1A-F | CGATGTTACGGTTTGTTACTGTGACAGC | 244 | 0.2 | 63 |
| MP4-stx1A-R | AATGCCACGCTTCCCAGAATTG | 0.2 | |||
| stx2 | MP3-stx2A-F | GTTTTGACCATCTTCGTCTGATTATTGAG | 324 | 0.4 | 63 |
| MP3-stx2A-R | AGCGTAAGGCTTCTGCTGTGAC | 0.4 | |||
| ETEC | |||||
| elt | MP2-LT-F | GAACAGGAGGTTTCTGCGTTAGGTG | 655 | 0.1 | 63 |
| MP2-LT-R | CTTTCAATGGCTTTTTTTTGGGAGTC | 0.1 | |||
| estIa | MP4-STIa-F | CCTCTTTTAGYCAGACARCTGAATCASTTG | 157 | 0.4 | 63 |
| MP4-STIa-R | CAGGCAGGATTACAACAAAGTTCACAG | 0.4 | |||
| estIb | MP2-STI-F | TGTCTTTTTCACCTTTCGCTC | 171 | 0.2 | 63 |
| MP2-STI-R | CGGTACAAGCAGGATTACAACAC | ||||
| EIEC | |||||
| invE | MP2-invE-F | CGATAGATGGCGAGAAATTATATCCCG | 766 | 0.2 | 63 |
| MP2-invE-R | CGATCAAGAATCCCTAACAGAAGAATCAC | 0.2 | |||
| EAEC | |||||
| astA | MP-astA-F | TGCCATCAACACAGTATATCCG | 102 | 0.4 | 63 |
| MP2-astA-R | ACGGCTTTGTAGTCCTTCCAT | 0.4 | |||
| aggR | MP2-aggR-F | ACGCAGAGTTGCCTGATAAAG | 400 | 0.2 | 63 |
| MP2-aggR-R | AATACAGAATCGTCAGCATCAGC | 0.2 | |||
| pic | MP2-pic-F | AGCCGTTTCCGCAGAAGCC | 1,111 | 0.2 | 63 |
| MP2-pic-R | AAATGTCAGTGAACCGACGATTGG | 0.2 | |||
| E. coli | |||||
| uidA | MP2-uidA-F | ATGCCAGTCCAGCGTTTTTGC | 1,487 | 0.2 | 63 |
| MP2-uidA-R | AAAGTGTGGGTCAATAATCAGGAAGTG | 0.2 | |||
| ShET2 homologue | |||||
| ent | ent-F | TGGGCTAAAAGAAGACACACTG | 629 | 0.4 | 58 |
| ent-R | CAAGCATCCTGATTATCTCACC | 0.4 |
Sequence analysis of PCR products.
The identities of representative PCR products were confirmed by DNA sequencing. PCR products obtained with the reference strains were purified (PCR Purification Kit; QIAGEN, Hilden, Germany) and sequenced by SEQLAB (Göttingen, Germany).
Identification of clinical isolates to the species level.
To further confirm that all of the pathogens detected were indeed E. coli, the isolates were analyzed for E. coli-specific metabolic reactions with selective indicator media such as MacConkey broth, Simmons citrate agar, and SIM medium (Merck, Darmstadt, Germany). A subset of strains was tested with the Bactident E. coli kit (Merck, Darmstadt, Germany). All isolates detected by the novel MPCR exhibited E. coli-specific reactions. The strains isolated were serotyped by a micromethod as previously described (34).
Cytotoxicity and toxin immunoreactivity.
Cytotoxicity assays of bacterial cell lysates and purified toxin preparations were performed with Vero cells by the method of Gentry and Dalrymple (14). The 50% Vero cell cytotoxic dose is expressed per milliliter of culture supernatant and is the reciprocal of the highest dilution of toxin that caused 50% Vero cell death. Confluent Vero cells were infected with sterile bacterial supernatant and incubated for 48 h. The cells were then fixed in formalin and stained with crystal violet, and the optical density at 570 nm was measured with a Titertek Multiskan MC microplate reader (Flow/ICN Biomedicals, Costa Mesa, CA) to assess cell death.
Fluorescent-actin staining (FAS) assay.
The adherence pattern of clinical isolates was analyzed by a modification of the method described by Vial et al. (46). Briefly, 70% confluent HeLa cells were infected with a bacterial suspension that had been preincubated for 2 h at 37°C in 10% CO2 (1:20 dilution of an overnight culture in Dulbecco modified Eagle medium), followed by 5 min of centrifugation (250 × g), and afterwards incubated for 3 h at 37°C in a 10% CO2 atmosphere. The cells were washed three times with prewarmed D-PBS (10 mM phosphate, 140 mM NaCl, 0.8 mM MgCl2, 0.9 mM CaCl2, pH 7.4) to remove nonadherent bacteria and subsequently fixed for 15 min in 4% (wt/vol) paraformaldehyde. The fixed cells were washed with D-PBS, quenched in 0.2% (wt/vol) glycine for 10 min, and permeabilized with 0.1% (wt/vol) Triton X-100 in D-PBS plus 4% (wt/vol) paraformaldehyde for 3 min. The cells were blocked with 3% (wt/vol) bovine serum albumin in D-PBS for 30 min. For FAS, phalloidin-Texas Red was used at a 1:100 dilution, and for DNA staining, 4′,6′-diamidino-2-phenylindole (DAPI) in dimethyl sulfoxide was used at a 1:1,000 dilution. After three washing steps, cells were mounted with 1,4-diazabicyclo[2.2.2]octane (DABCO)-Mowiol (DAKO, Hamburg, Germany).
Analysis of hemolytic activities.
The hemolytic phenotype was determined with Columbia blood agar and enterohemolysin agar (Sifin, Berlin, Germany) containing 5% defibrinated and washed human erythrocytes and 10 mM CaCl2. After 5 h of incubation, a wide, clear lysis zone becomes visible around α-hemolysin (α-hly)-producing colonies, whereas EHEC hemolysin (EHEC-hly) only produces a narrow, turbid zone after 16 h.
Analysis by electron microscopy.
To visualize BFP on the bacterial surface, single colonies were grown in 2 ml V-SIF medium according to Lockwood and Randall (26) at 37°C overnight without shaking. A 15-μl bacterial suspension was applied to a Formvar-coated copper grid and flattened by osmotic shock by incubation in distilled water for 20 s. Bacteria were contrasted with 1% phosphotungstic acid (pH 7.4) for 50 s. Excess fluid was removed with filter paper, and the grid was air dried. Analysis and evaluation of the samples were performed with a Phillips 410 electron microscope at a magnification of ×21,000.
RESULTS
Development of a single MPCR for simultaneous detection of major E. coli virulence factors.
For the simultaneous identification and differentiation of the seven currently established E. coli pathotypes causing gastrointestinal disease, we developed a one-step MPCR incorporating 12 primer pairs that have been designed and optimized to have compatible temperature-related properties and to give rise to DNA fragments of sufficiently different sizes to be unequivocally resolved by standard agarose gel electrophoresis (Table 1). Sequences of primers were chosen according to gene-specific consensus sequences derived from several accessible sequences for each gene. Thereby, the specificity of the primers and the spectrum of detectable variations of each gene were improved. In accordance with a recently developed MPCR for the detection of LEE-harboring E. coli strains (29), we used the highly conserved LEE gene escV, which exhibits a minimum of 96% identity on the DNA level, as a marker for the specific identification of typical EPEC, ATEC, and LEE-positive STEC. LEE-harboring pathotypes were differentiated by the presence or absence of the bfpB gene that is located on the EAF plasmid of typical EPEC and the bacteriophage-encoded Stx-encoding genes stx1 and/or stx2 of STEC, respectively. The specificity of these genes for each pathotype has been established previously (4, 12). For the detection of ETEC, we designed primer pairs that specifically recognize LT toxin and heat-stable toxin variants STIa and STIb. EIEC can be identified by the presence of the intermediary regulator InvE of the Ipa proteins that are located on the 140-MDa plasmid harbored by all EIEC strains. Since EAEC strains have been found to be quite heterogeneous, we selected three EAEC marker genes, namely, astA, aggR, and pic, that are predominantly present in diarrhea-causing EAEC (6, 33, 45). The design of the pic primer pair was optimized for higher binding to the pic gene of EAEC rather than to the pic gene of uropathogenic E. coli, based on sequence alignments of the respective genes of uropathogenic E. coli CFT073 (accession no. AE014075) and EAEC strain O42 (accession no. AF097644). To unambiguously identify a particular isolate as a bona fide E. coli strain, we included a primer pair for the detection of the E. coli-specific uidA gene in the MPCR.
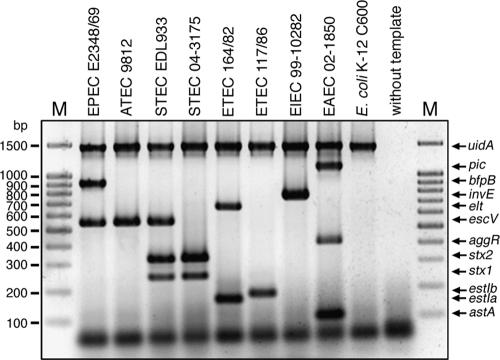
In a first approach, we validated the MPCR by using as reference strains EPEC strain E2348/69, ATEC strain 9812, STEC strain EDL933, STEC strain 04-3175, ETEC strain 164/82, ETEC strain 117/86, EIEC strain 99-10282, EAEC strain 02-1850, and, as an example of an apathogenic E. coli strain, E. coli C600. As demonstrated in Fig. 1, the specific DNA fragments corresponding to genes defining the appropriate pathotypes, EPEC (escV positive, bfp positive, stx negative), ATEC (escV positive, bfp negative, stx negative), STEC (escV positive/negative, bfp negative, stx positive), ETEC (elt positive and/or estIa positive, estIb positive), EIEC (invE positive), and EAEC (astA positive and/or aggR positive and/or pic positive), were easily detected by MPCR in a single reaction mixture. No cross-priming was observed for any other pathotype. The identities of the PCR products obtained with the reference strains were verified by nucleotide sequence analysis.
FIG. 1.
MPCR analysis of reference strains. Twelve primer pairs were designed for the specific detection of the seven major categories of intestinal pathogenic E. coli. All of the primer pairs yield specific gene products indicating the appropriate pathotypes and generate no unspecific products. The amplicons can be differentiated by agarose gel electrophoresis (2%). Lanes M contained molecular size markers. The other lanes contained EPEC strain E2348/69 (uidA positive, LEE positive, bfp positive), ATEC strain 9812 (uidA positive, LEE positive), STEC strain EDL933 (uidA positive, LEE positive, stx1 positive, stx2 positive), STEC strain 04-3175 (uidA positive, stx1 positive, stx2 positive), ETEC strain 164/82 (uidA positive, elt positive, estIa positive), ETEC strain 117/86 (uidA positive, estIb positive), EIEC strain 99-10282 (uidA positive, invE positive), EAEC strain 02-1850 (uidA positive, astA positive, aggR positive, pic positive), E. coli C600 (uidA positive), and a negative control lacking template DNA.
Distribution of Afa/Dr or AIDA adhesins in ATEC.
The Afa/Dr adhesins represent a major family of adhesins in diffusely adhering E. coli strains (39). Therefore, we investigated whether AfaE might be involved in the diffuse-adherence phenotype of ATEC and thus might be used as a potential marker for the differentiation of ATEC in the MPCR. For this, we analyzed 246 E. coli isolates from patients with diarrhea with specific primers for afaE (data not shown). However, only one ATEC strain (3431-4/86), control uropathogenic E. coli strain KS52, and seven LEE-negative E. coli strains possess the afaE gene. Therefore, we conclude that afaE is rarely associated with the presence of the LEE PAI and, consequently, is not involved in the commonly observed diffuse-adherence pattern of ATEC strains. In addition, we examined ATEC strains for the presence of the aidA gene encoding AIDA (adhesin involved in diffuse adherence), the adhesin of the clinical isolate E. coli 2787 (3). This analysis revealed that only control strain 2787 was aidA positive. As a consequence, aidA does not qualify as a specific marker for ATEC. Therefore, we used the escV-positive, bfpB- and stx-negative genotypic pattern for the identification of ATEC strains.
Validation of the novel MPCR with clinical isolates exhibiting different pathotypes.
To further evaluate the newly developed MPCR and demonstrate its diagnostic utility, we analyzed 246 isolates of our strain collection derived from various geographic regions. The analysis by MPCR revealed 20 typical EPEC, 24 ATEC, 19 LEE-positive STEC, 20 LEE-negative STEC, 21 ETEC, 6 EIEC, and 57 EAEC strains (Tables 2 to 6). The remaining 74 strains were either commensals (51 strains) or extraintestinal pathogenic (ExPEC) strains (23 strains) (Table 7). In addition, with the novel MPCR, we detected five strains that could not be associated with one of the currently accepted pathogroups of intestinal pathogenic E. coli. Hence, these strains were designated intermediate strains.
TABLE 2.
Detection of virulence genes in E. coli by MPCR
| Pathogroup and serotype (no. of strains) | No. of strains detected by:
|
||||||
|---|---|---|---|---|---|---|---|
| MPCR targeting
|
Single-gene-specific PCR targeting
|
||||||
| uidA | escV | bfpB | astA | lifA/efa1 | ent | hly | |
| Classical EPEC | |||||||
| O55:H6 (3) | 3 | 3 | 3 | 3 | 3 | ||
| O86:H− (2) | 2 | 2 | 2 | ||||
| O86:H(rough) (1) | 1 | 1 | 1 | ||||
| O111:H− (3) | 3 | 3 | 3 | 2 | 3 | ||
| O111:H2 (2) | 2 | 2 | 2 | 2 | 2 | ||
| O111:H(NT)a (1) | 1 | 1 | 1 | 1 | 1 | ||
| O114:H2 (2) | 2 | 2 | 2 | 2 | 2 | 2 | |
| O127:H6 (1) | 1 | 1 | 1 | 1 | 1 | ||
| O128:H2 (3) | 3 | 3 | 3 | 3 | 3 | ||
| O142:H6 (2) | 2 | 2 | 2 | 2 | 2 | ||
| ATEC | |||||||
| O8:H− (1) | 1 | 1 | 1 | 1 | 1 (α-hly) | ||
| O26:H− (1) | 1 | 1 | 1 | 1 | 1 (α-hly) | ||
| O26:H11 (2) | 2 | 2 | 2 | 2 | 1 (EHEC-hly) | ||
| O26:K60 (1) | 1 | 1 | 1 | 1 | 1 (α-hly) | ||
| O55:H− (1) | 1 | 1 | 1 | 1 | |||
| O55:H6 (1) | 1 | 1 | 1 | ||||
| O55:H7 (8) | 8 | 8 | 8 | 6 | 1 (α-hly) | ||
| O86:H8:K61 (1) | 1 | 1 | |||||
| O119:H9:K61 (1) | 1 | 1 | 1 | 1 (α-hly) | |||
| O125:H− (1) | 1 | 1 | |||||
| O127:H40 (2) | 2 | 2 | 2 | ||||
| O128:H2 (3) | 3 | 3 | |||||
| O128:H8 (1) | 1 | 1 | 1 | ||||
NT, nontypeable.
TABLE 6.
Detection of virulence genes in E. coli by MPCR
| Serotype (no. of strains) of pathogroup EAEC | No. of strains detected by:
|
||||||
|---|---|---|---|---|---|---|---|
| MPCR targeting
|
Single-gene-specific PCR targeting
|
||||||
| uidA | aggR | pic | astA | lifA/efa1 | ent | hly | |
| O2:H6 (2) | 2 | 1 | 1 | 1 (α-hly) | |||
| O3:H2 (1) | 1 | 1 | 1 | 1 | 1 (α-hly) | ||
| O5:H12 (1) | 1 | 1 | 1 | ||||
| O6:H1 (1) | 1 | 1 | 1 (α-hly) | ||||
| O6:H(NT)a (1) | 1 | 1 | 1 (α-hly) | ||||
| O8:H− (1) | 1 | 1 | |||||
| O8:H25 (1) | 1 | 1 | |||||
| O17:H− (1) | 1 | 1 | 1 | ||||
| O17:H37(1) | 1 | 1 | |||||
| O18:H− (1) | 1 | 1 | 1 (α-hly) | ||||
| O21:H− (1) | 1 | 1 | |||||
| O21:H2 (1) | 1 | 1 | 1 | 1 (α-hly) | |||
| O23:H− (1) | 1 | 1 | |||||
| O25:H4 (2) | 2 | 2 | 1 (α-hly) | ||||
| O40:H18 (1) | 1 | 1 | |||||
| O44:H12:K74 (1) | 1 | 1 | 1 | ||||
| O44:H18 (1) | 1 | 1 | 1 | ||||
| O44:H(NT) (4) | 4 | 3 | 3 | 3 | 1 (α-hly) | ||
| O52:H45 (1) | 1 | 1 | |||||
| O77:H− (1) | 1 | 1 | 1 | 1 (α-hly) | |||
| O77:H11 (1) | 1 | 1 | 1 | ||||
| O78:H10 (1) | 1 | 1 | 1 | ||||
| O78:H(NT) (2) | 2 | 2 | |||||
| O86:H(NT) (3) | 3 | 3 | 1 | ||||
| O88:H4 (1) | 1 | 1 | 1 (α-hly) | ||||
| O88:H6 (1) | 1 | 1 | |||||
| O89:H− (2) | 2 | 2 | 2 | ||||
| O89:H(NT) (1) | 1 | 1 | 1 | ||||
| O92:H(NT) (1) | 1 | 1 | 1 | ||||
| O106:H− (1) | 1 | 1 | 1 | 1 | |||
| O111:H(NT) (1) | 1 | 1 | 1 | 1 | |||
| O126:H27 (1) | 1 | 1 | 1 | ||||
| O126:K71 (1) | 1 | 1 | |||||
| O126:H(NT) (3) | 3 | 1 | 1 | 3 | |||
| O127a:H4 (1) | 1 | 1 | 1 | ||||
| O127:H21 (1) | 1 | 1 | 1 | ||||
| O128:H(NT) (1) | 1 | 1 | |||||
| O130:H26 (1) | 1 | 1 | |||||
| O133:H29 (2) | 2 | 2 | |||||
| O142:K86 (1) | 1 | 1 | |||||
| O(rough):H− (1) | 1 | 1 | 1 | ||||
| O(rough):H(NT) (1) | 1 | 1 | |||||
| O(NT):H4 (1) | 1 | 1 | |||||
| O(NT):H6 (2) | 2 | 2 | |||||
NT, nontypeable.
TABLE 7.
Detection of virulence genes in E. coli by MPCR
| Pathogroup and serotype (no. of strains) | No. of strains detected by:
|
|||||||
|---|---|---|---|---|---|---|---|---|
| MPCR targeting
|
Single-gene-specific PCR targeting
|
|||||||
| uidA | bfpB | stx2 | estla | astA | lifA/efa1 | ent | hly | |
| Intermediate | ||||||||
| O127:H− (1) | 1 | 1 | 1 | 1 | ||||
| O(rough):H− (1) | 1 | 1 | 1 | 1 | ||||
| O100:H− (1) | 1 | 1 | 1 | |||||
| O175:H28 (1) | 1 | 1 | 1 | 1 | ||||
| O169:H41 (1) | 1 | 1 | 1 | |||||
| ExPEC | ||||||||
| NDa (23) | 23 | 2 | 10 (α-hly) | |||||
| Commensal | ||||||||
| ND (51) | 50 | 2 | ||||||
ND, not determined.
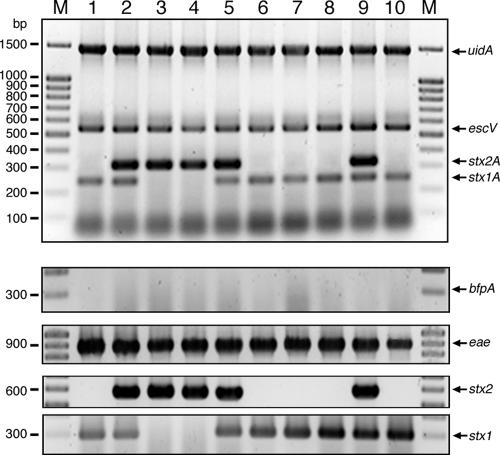
By different approaches, including comparative PCR analysis, adherence assays, cytotoxicity assays, invasion assays, and detection of specific metabolic pathways, the pathotype of each strain was validated independently. The comparative PCR analysis with established single primer pairs specific for each gene [SK1/SK2, eae; CesT(+)/CesT(−), cesT; CesD+new/CesD−new, cesD; EP1/EP2, bfpA; PerF/PerR, per; KS7/KS8, stx1; LP43/LP44, stx2; STIaprimer1/STIaprimer2, estIa; STIbprimer1/STIbprimer2, estIb] (37, 38, 43) yielded the same virulence gene patterns as the one-step MPCR analysis for all of the clinical isolates analyzed. Figure 2 shows a representative example of the comparative PCR analysis of LEE-positive STEC strains. As expected, STEC strain Hi8 [O(rough):H−] harboring Stx2 subtype f, which, with rare exceptions (41), is frequently present in pigeons (27), tested negative with primers MP3-stx2A-F and -R. This is probably due to the relatively low sequence identity of stx2f to other stx2 subtypes (63.4 and 57.4% identity to the A and B subunits of Stx2 of E. coli O157:H7 strain EDL933) (36).
FIG. 2.
Comparative PCR analysis of STEC strains. Strains were analyzed by MPCR (top) and single-PCR approaches (bottom; EP1/EP2, bfp; SK1/SK2, eae; LP43/LP44, stx2; KS7/KS8, stx1). All strains exhibited the same gene pattern in the MPCR and single PCR assays and were positive for uidA; LEE (escV, eae); and stx1, stx2, or both. Lanes: M, marker; 1, 11062; 2, Y113; 3, 493/88; 4, 5720/96; 5, EDL933; 6, E. coli 04-2936; 7, E. coli 04-2938; 8, E. coli 04-3313; 9, E. coli 04-3453; 10, E. coli 04-4080.
Phenotypic assays, including adherence pattern analysis of LEE-harboring and EAEC strains, induction of actin polymerization, cytotoxicity assays of STEC, gentamicin protection assays, and actin comet staining of EIEC, as well as the analysis of metabolic pathways with indicator media for the verification of species identity as E. coli, were conducted and confirmed, in effect, the pathotypes determined for all of the strains by the novel MPCR, again demonstrating its specificity.
Sole exceptions were attributed to the heterogeneous EAEC pathogroup. The EAEC pathogroup comprises strains that all adhere to host cells with the aggregative phenotype, but as they exhibit highly diverse virulence factor profiles, their unequivocal identification and classification represent a particular challenge. Therefore, we performed adherence assays to screen for the characteristic stacked-brick aggregative-adherence pattern in tissue culture that is regarded the “gold standard” for the classification of E. coli isolates as EAEC strains. These experiments demonstrated that 85% of the MPCR-designated EAEC strains did possess the EAEC characteristic stacked-brick adherence pattern. The remaining 15% false positives obtained by MPCR harbored the marker genes astA and/or pic that had been previously often used for EAEC identification (6, 33, 45). However, the detection of aggR or a combination of two of the three marker genes (astA, aggR, pic) used yielded 100% agreement of the MPCR classification and the observed adherence pattern. These findings demonstrate that the novel MPCR is able to detect all intestinal pathotypes of E. coli.
As described previously, we could confirm the presence of the EAEC marker genes astA and pic in other E. coli pathotypes (21). Since these strains harbor at least one other major virulence factor indicative of a particular pathotype, they have been grouped into the associated pathogroup. In contrast to astA, which was detected in members of all other E. coli pathotypes, the Pic serine protease-encoding gene was only present in one EIEC strain (E. coli 02-10479 [O:124:H−]); however, as it also harbors the pInv plasmid and invaded target cells, it was easily designated an EIEC strain.
Detection of novel intermediate pathotypes.
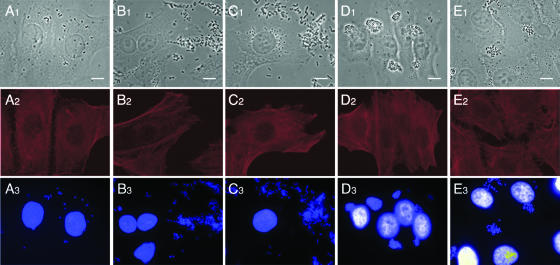
Besides the 167 well-defined intestinal pathogenic E. coli strains belonging to one of the seven major pathotypes, we identified five strains that—in addition to sharing the E. coli-specific uidA gene—exhibited mixed virulence patterns indicative of two or three distinctly defined pathovars, namely, strains 2771/97 [O100:H−, uidA positive, stx2 positive, estIa positive], 04-3908 (O:175:H28, uidA positive, stx2 positive, estIa positive, astA positive), 03-7355 (O169:H41, uidA positive, estIa positive, astA positive), 4932-53 [O(rough):H−, uidA positive, bfp positive], and 265-1 (O127:H−, uidA positive, bfp positive) (Fig. 3). The identities of the specific amplicons obtained by the MPCR were confirmed by DNA sequencing. Genotypes were additionally confirmed by single PCRs with established primer pairs as described earlier in this report. FAS assays with HeLa cells revealed that strain 2771/97 exhibits a diffuse adherence pattern, whereas strains 04-3908 and 03-7355 adhered in an aggregative fashion and strains 4932-53 and 265-1 formed microcolonies and exhibited localized adherence (Fig. 4). None of these strains, including both EAF-positive but LEE-negative strains, was able to induce actin polymerization or to invade target cells.
FIG. 3.
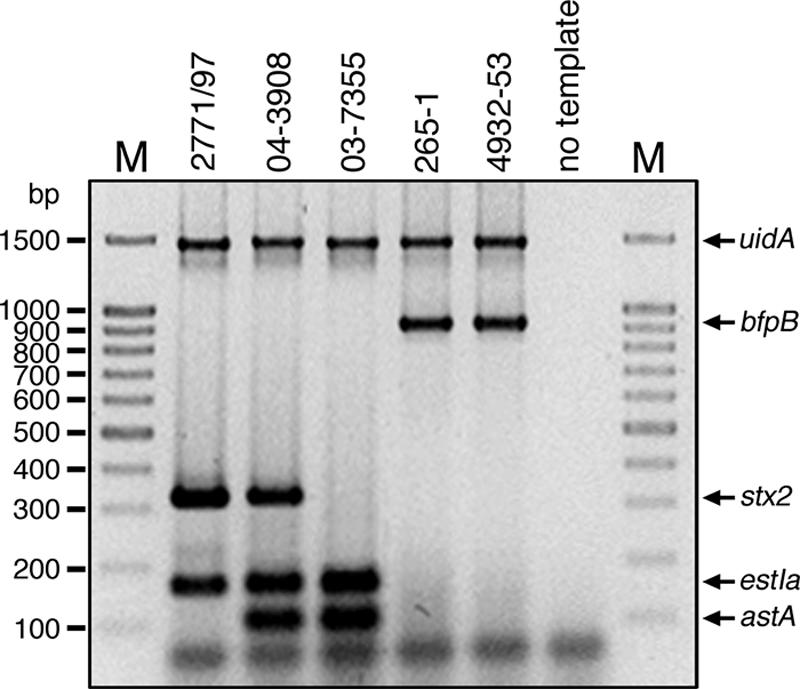
MPCR analysis of intermediate strains. Single colonies of intermediate strains were directly incubated in the PCR mixture, lysed by the initial denaturation step, and analyzed by MPCR as described in Materials and Methods. All intermediate strains, i.e., 2771/97 (uidA positive, stx2 positive, estIa positive), 04-3908 (uidA positive, stx2 positive, estIa positive, astA positive), 265-1 (uidA positive, bfpB positive), and 4932-53 (uidA positive, bfpB positive), exhibit unusual genotypes. The negative control was tested with the MPCR set of primer pairs but lacked template DNA. Lanes M contained molecular size markers.
FIG. 4.
Adherence patterns of intermediate intestinal pathogenic E. coli strains. Adherence behavior of unconventional E. coli strains to HeLa cells and induction of actin polymerization were monitored by FAS assay. Strain 2771/97 (A; stx2 positive, estIa positive) adheres in a diffuse pattern to target cells, whereas strains 04-3908 (B; stx2 positive, estIa positive, astA positive) and 03-7355 (C; estIa positive, astA positive) exhibit aggregative adherence. Strains 4932-53 (D; LEE negative, bfp positive) and 265-1 (E; LEE negative, bfp positive) show localized adherence patterns and develop microcolonies. All strains are not able to induce actin polymerization in HeLa cells (A2, B2, C2, D2, E2). Actin filaments were labeled with phalloidin-Texas Red (red), and DNA was visualized with DAPI (blue). Bars = 10 μm.
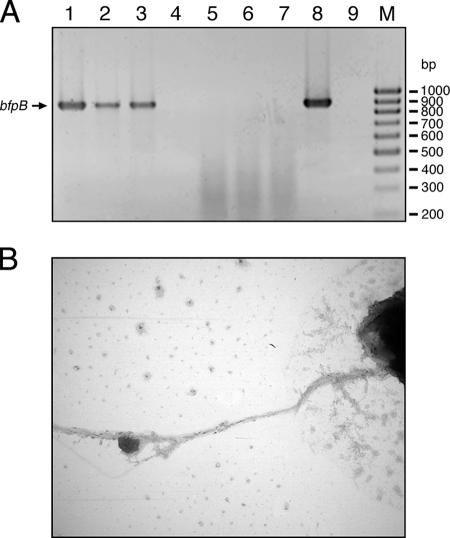
To further confirm the expression of BFP of the LEE-negative, EAF-positive, diarrhea-associated isolates, we performed reverse transcription (RT)-PCR and electron microscopic analysis. For that purpose, we isolated the total RNAs of EAF plasmid-harboring strains and demonstrated bfpA and bfpB mRNA transcription by detection of the appropriate cDNAs by PCR analysis. Furthermore, both strains 4932-53 and 265-1 form pili that attach to each other by forming bundles, as visualized by electron microscopy (Fig. 5). This clearly indicates that strains 4932-53 and 265-1 contain the EAF plasmid and also synthesize BFP, although they apparently do not harbor the LEE PAI. To our knowledge, this is the first time strains with this genetic makeup have been detected and described.
FIG. 5.
Verification of BFP pheno- and genotypes of intermediate pathogens. BFP expression was confirmed by detection of bfpB cDNA by RT-PCR (A) and by electron microscopy analysis (B). Whole RNAs of strains 265-1 (LEE negative, EAF positive), 4932-53 (LEE negative, EAF positive), E2348/69 (LEE positive, EAF positive), and 3431-4/86 (LEE positive, EAF negative) were isolated and transcribed to cDNAs. Transcription of bfpB could be detected for strain 265-1 (A, lane 1), 4932-53 (A, lane 2), and E2348/69 (A, lane 3). Strain 3431-4/86 cDNA (A, lane 4) did not yield a PCR product, like the negative controls with mRNAs of strains 265-1 (A, lane 5), 4932-53 (A, lane 6), and E2348/69 (A, lane 7) as the template and the sample without a template (A, lane 9). The positive control with genomic DNA of strain E2348/69 (A, lane 8) as the template produced the expected 910-bp PCR product. Lane M contained molecular size markers. (B) Production of BFP was visualized by electron microscopy analysis of strain 265-1 (magnification, ×21,000).
Detection of major virulence factors.
To further evaluate the distribution of virulence factors in intestinal pathogenic E. coli, we performed PCR analysis of the 246 clinical isolates of our strain collection with specific primer pairs for the detection of α-hly, EHEC-hly, lymphostatin (LifA/Efa1), and the ShET2 (Shigella enterotoxin 2) homologue enterotoxin encoded by ent (30) (Tables 2 to 7). These factors are all associated with an enhanced virulence of different pathogenic E. coli strains. α-hly, first described in uropathogenic E. coli, was detected in typical EPEC (5%), ATEC (29%), ETEC (14%), EAEC (17%), and ExPEC (43%) (Tables 2 to 7). α-hly was absent in all STEC (n = 52) and EIEC (n = 6) strains and the five newly identified intermediate strains. In contrast, EHEC-hly was solely present in STEC strains. Interestingly, EHEC-hly was identified with a higher frequency in LEE-positive (89%) than in LEE-negative (35%) STEC strains. PCR results were verified by measuring the hemolytic activities of representative strains on Columbia blood agar and enterohemolysin agar. In addition to the above-listed STEC strains, we analyzed 11 stx2f-positive STEC isolates from pigeons that were all positive for astA but negative for all other virulence factors.
LifA/Efa1 was described in EPEC E2348/69 as lymphocyte inhibitory factor A (LifA), which inhibits mitogen-activated proliferation of peripheral blood lymphocytes and lamina propria mononuclear cells and the synthesis of proinflammatory cytokines (1). On the other hand, the protein has also been described as EHEC factor for adherence 1 (Efa1), which is present on the cell surface and directly mediates cell-cell contact with target cells (28). The lifA/efa1 gene is part of a 22-kb PAI designated O122 in STEC strain EDL933 and SpLE3 in STEC strain Sakai. The lifA/efa1 gene has also been found in mosaic LEE PAIs of bovine STEC strains 413/89-1 and RW1374 and rabbit EPEC strains RDEC-1 and 83/39. It is mostly flanked by the enterohemolysin encoded by ent, which has similarities to Shigella enterohemolysin ShET2. ShET2 exhibits enterotoxic activity, manifested by a significant increase in the transepithelial electrical potential difference in short-circuit current without changes in tissue conductance and is not associated with tissue damage (22). We found that these virulence factors are linked to LEE-associated pathogens and were not found in LEE-negative STEC, ETEC, EIEC, EAEC, and ExPEC strains or apathogenic E. coli (Tables 2 to 7). Eighty percent of 20 typical EPEC strains were positive for the lifA/efa1 and ent genes. Interestingly, one strain was found to be ent positive but lifA/efa1 negative. A lower frequency of lifA/efa1-positive strains (50%, n = 24) was found among ATEC strains. However, also in this group 8% of the strains were lifA/efa1 positive but ent negative and a further 12.5% were lifA/efa1 negative but ent positive. Excluding the stx2f-positive STEC strains, 67% of the 18 LEE-positive STEC strains contained the full-length lifA/efa1 and ent genes, whereas 33% were positive for the ent gene combined with the ent-adjacent part of the lifA/efa1 gene. The truncated version of lifA/efa1 was present in three STEC O157:H7 strains, one STEC O157:H− strain, one STEC O145:H−strain, and one STEC O5:H− strain. Regarding the serotype of LEE-positive pathogens, lifA/efa1 and ent were predominantly found in O26, O55, O103, and O111 strains. The intermediate strains were negative for lifA/efa1 and ent, except those bfp-positive, LEE-negative strains that contained both full-length lifA/efa1 and ent, indicating that they are related to LEE-harboring intestinal pathogens.
DISCUSSION
For the simultaneous and rapid identification and differentiation of diarrheagenic E. coli strains belonging to the seven major pathotypes (EPEC, ATEC, [LEE positive and LEE negative] STEC, ETEC, EIEC, and EAEC), we set up a single-step MPCR. The design and development of the MPCR were monitored with nine reference strains. All of the reference strains exhibited the expected gene pattern, as confirmed by DNA sequencing, and no cross-priming by the MPCR primer pairs was observed. Furthermore, all PCR amplicons showed comparable band intensities and are of sufficiently different sizes to be unequivocally resolved by standard agarose gel electrophoresis.
The specificity of the MPCR was validated with a subset of reference strains and further evaluated with 246 clinical E. coli isolates derived from patients from different geographic regions. Classification of all detected pathogens by the MPCR was confirmed by comparative PCR analysis performed independently and by a phenotypic analysis that included adherence assays, FAS tests, cytotoxicity assays, gentamicin protection assays, screening for actin comets, and analysis of growth on metabolic media to verify the identity of a specific strain as E. coli. With very few exceptions, all of the strains tested showed the expected reactions characteristic of their particular pathotypes. Only in the EAEC group were minor variations due to the remarkable heterogeneity of this pathogroup observed. Previous studies had shown that the astA, aggR, and pic genes appear to represent common genes in diarrhea-associated EAEC (6, 33, 45). Therefore, specific primers for these EAEC genes were included in the MPCR. Aggregative-adherence assays revealed a high reliability (total 85%) of the novel MPCR, with 100% agreement for the detection of aggR or a combination of at least two of the three EAEC marker genes astA, aggR, and pic.
Five noncategorizable intestinal pathogenic E. coli strains were identified during this investigation. These strains, 2771/97 [O100:H−, stx2 positive, estIa positive], 04-3908 (O175:H28, stx2 positive, estIa positive, astA positive), 03-7355 (O169:H41, estIa positive, astA positive), 4932-53 [O(rough):H−, EAF positive, LEE negative], and 265-1 (O127:H−, EAF positive, LEE negative), exhibited rather unconventional patterns of virulence genes. To our knowledge, this is the first time these particular virulence gene profiles have been identified in intestinal pathogenic E. coli. For the purpose of this study, we designated these strains intermediate strains as they contain two or three characteristic major virulence factors that have been established as molecular markers for distinct pathovars. Consequently, these strains cannot be assigned unambiguously to an established pathogroup. Nevertheless, we categorized strains 04-3908 and 03-7355 as EAEC, as both are astA positive and—more importantly—adhere in an aggregative pattern to epithelial cells or inert surfaces such as glass coverslips. Interestingly, their virulence factor repertoire is complemented by additional virulence factors, such as the ETEC-defining heat-stable toxin and the STEC-characteristic Stxs. Strain 2771/97 can be categorized as a STEC strain in combination with ETEC heat-stable toxin as an additional virulence factor or, alternatively, as an ETEC strain plus Stx2. Otherwise, 2771/97 might also represent a member of a new pathogroup.
Moreover, these results also indicate that the presence of different virulence genes is apparently associated with a specific genetic environment of their host strains. Some virulence genes are widely distributed among the seven major intestinal pathogenic E. coli groups, like astA or α-hly; others are restricted to a certain genetic background, like EHEC-hly to STEC or lifA/efa1 and ent to LEE-associated strains. Interestingly, some major virulence factors appear not to be restricted to one pathovar, as Stx2 and STIa were found in one and the same strain.
These findings provide additional evidence and emphasize that—as exemplified in intestinal pathogenic E. coli—a particular profile of virulence genes is not static and restricted to a particular index pathogroup but instead represents a dynamic matrix of genes whose constituents can also be identified in other host strains, thereby complementing their virulence profiles to potentially enhance fitness and pathogenicity. Moreover, harboring more than their established set of virulence factors might represent an additional advantage for pathogens against host defenses (16). Emergence of new pathogenic strains through the successive acquisition of virulence factors was previously shown by phylogenetic analyses, suggesting that gain and loss of mobile virulence elements have frequently occurred in separate lineages of pathogenic E. coli and might enable the combination of different major virulence patterns (11).
Newly identified EAF-positive and LEE-negative strains 4932-53 and 265-1 may play an important role in the transmission of virulence factors like BFP, LifA/Efa1, or the ShET2 homologue Ent. To further characterize these intermediate strains, we used different approaches such as PCR analysis, FAS assays, and electron microscopy. By PCR with established primers directed against the LEE genes eae, cesT, and cesD and against the EAF plasmid genes bfpA and per, the designated genotype was confirmed. Subsequently, we verified the transcription of bfpA and bfpB by RT-PCR and additionally visualized BFP by electron microscopy. Furthermore, the formation of microcolonies on HeLa cells without the induction of actin polymerization or pedestal formation clearly demonstrated the EAF-positive but LEE-negative nature of strains 4932-53 and 265-1. These strains might represent originally typical EPEC strains that upon subculture lost the LEE PAI. Although some PAIs, like PAI I and PAI II of uropathogenic E. coli strains, are frequently lost (35), the LEE PAI of EPEC is known to be quite stable when integrated into the chromosome. Moreover, loss of the EAF plasmid would have been a more likely event, as it has been shown to be relatively unstable. Therefore, we conclude that these newly identified strains did not (yet) carry the LEE PAI but rather had acquired the EAF plasmid. Therefore, these strains might represent an evolutionary link for the evolvement of EPEC and might even be regarded as emerging pathogens en route to EPEC or ATEC. This implies that EPEC strains might be derived from coexisting ATEC- and LEE-negative, EAF-positive strains with the result of EAF plasmid transfer to ATEC strains or vice versa. The relationship of these strains to typical EPEC and ATEC is underlined by the presence of the lifA/efa1 and ent genes, which are linked to LEE-harboring intestinal pathogens, as shown previously (1, 11) and confirmed in this study. ATEC bacteria might play a central role in the evolution of intestinal pathogenic E. coli, as they provide an excellent scaffold for basic infection involving the LEE PAI-encoded type III secretion system. Pathogenicity might be enhanced by complementation with other virulence factors such as LifA/Efa1, Ent, α-hly, EAST1, or Stxs, grouping them as STEC strains, or the EAF plasmid, designating them typical EPEC strains. Previous studies of STEC support this hypothesis in that STEC bacteria have probably arisen from a simple genetic event, where Stx moved from Shigella via a bacteriophage into EPEC (24).
Further examination revealed that some genes, like astA or α-hly, were observed in a broad variety of pathotypes, in contrast to the above-mentioned LEE-associated lifA/efa1 and ent genes, and EHEC-hly, which was exclusively found in STEC strains. This reflects the fact that different virulence genes require different genetic environments of their host strains (10, 20). Some pathogenic lineages, such as E. coli O157:H7, may be more likely than others to acquire foreign DNA because of an enhanced recombination ability, a secondary effect of defective mismatch repair (24). Nevertheless, some virulence factors are strictly associated with other virulence factors. In Vibrio cholerae, cholera toxin is acquired from a phage that requires pili for successful adherence, which are also critical for host colonization and are encoded by a second phage (19). The parallel pattern of evolution suggests that there is a selective advantage favoring the buildup of specific combinations of virulence factors enabling the establishment and transmission of new virulent clones. These combinations might, to some degree, also occur in between the seven major categories of intestinal pathogenic E. coli that were previously clearly separated. This is shown by the detection of strains with mixed gene patterns of stx2, estIa, and astA.
In summary, this study shows that index virulence factors are not, in general, restricted to one pathogroup but instead can also occur in other pathotypes, where they might contribute to their virulence potential. However, there are hints that not all virulence factor combinations are realized and that certain factors appear to require a specific genetic background. This also implies that classification of a particular isolate in a specific pathogroup should be based on as large a genetic profile as possible and—in a practical setting—also feasible. In this respect, we demonstrated that the newly developed MPCR represents a highly specific, robust, and cost-efficient diagnostic tool for the differentiation of diarrheagenic E. coli and therefore might have the potential to be introduced into routine diagnostic testing in clinical microbiology laboratories.
TABLE 3.
Detection of virulence genes in E. coli by MPCR
| Pathogroup and serotype (no. of strains) | No. of strains detected by:
|
|||||||
|---|---|---|---|---|---|---|---|---|
| MPCR targeting
|
Single-gene-specific PCR targeting
|
|||||||
| uidA | escV | stx1 | stx2 | astA | lifA/efa1 | ent | hly | |
| LEE-positive STEC | ||||||||
| O5:H− (1) | 1 | 1 | 1 | 1 (5′) | 1 | 1 (EHEC-hly) | ||
| O26:H− (1) | 1 | 1 | 1 | 1 | 1 | 1 (EHEC-hly) | ||
| O26:H11 (4) | 4 | 4 | 2 | 3 | 1 | 4 | 4 | 4 (EHEC-hly) |
| O103:H2 (2) | 2 | 2 | 2 | 2 | 2 | 2 (EHEC-hly) | ||
| O111:H− (3) | 3 | 3 | 3 | 2 | 3 | 3 | 2 (EHEC-hly) | |
| O118:H16 (1) | 1 | 1 | 1 | 1 | 1 | |||
| O145:H− (1) | 1 | 1 | 1 | 1 (5′) | 1 | 1 (EHEC-hly) | ||
| O157:H− (2) | 2 | 2 | 2 | 2 (1 [5′]) | 2 | 2 (EHEC-hly) | ||
| O157:H7 (3) | 3 | 3 | 2 | 3 | 3 (5′) | 3 | 3 (EHEC-hly) | |
| O(rough):H− (1) | 1 | 1 | stx2f | 1 | ||||
| LEE-negative STEC | ||||||||
| O8:H21 (1) | 1 | 1 | ||||||
| O23:H15 (1) | 1 | 1 | 1 | 1 | 1 (EHEC-hly) | |||
| O40:H6 (1) | 1 | 1 | 1 | |||||
| O60:H− (1) | 1 | 1 | 1 | |||||
| O76:H19 (2) | 2 | 2 | 2 | 2 (EHEC-hly) | ||||
| O77:H− (1) | 1 | 1 | 1 | |||||
| O91:H− (3) | 3 | 2 | 3 | 1 | ||||
| O91:H14 (2) | 2 | 2 | ||||||
| O101:H9 (1) | 1 | 1 | ||||||
| O113:H4 (1) | 1 | 1 | 1 | 1 | 1 (EHEC-hly) | |||
| O126:H29 (1) | 1 | 1 | ||||||
| O128:H2 (1) | 1 | 1 | 1 | 1 (EHEC-hly) | ||||
| O146:H− (1) | 1 | 1 | 1 | |||||
| O146:H21 (1) | 1 | 1 | 1 (EHEC-hly) | |||||
| O178:H19 (1) | 1 | 1 | ||||||
| O(rough):H25(1) | 1 | 1 | 1 | 1 (EHEC-hly) | ||||
TABLE 4.
Detection of virulence genes in E. coli by MPCR
| Serotype (no. of strains) of pathogroup ETEC | No. of strains detected by:
|
|||||||
|---|---|---|---|---|---|---|---|---|
| MPCR targeting
|
Single-gene-specific PCR targeting
|
|||||||
| uidA | estla | estlb | elt | astA | lifA/efa1 | ent | hly | |
| O6:H− (1) | 1 | 1 | ||||||
| O25:H42 (1) | 1 | 1 | ||||||
| O114:H− (1) | 1 | 1 | ||||||
| O128:H− (2) | 2 | 2 | ||||||
| O128:H21 (2) | 2 | 2 | ||||||
| O147:H19:K88(1) | 1 | 1 | 1 | |||||
| O148:H28 (1) | 1 | 1 | 1 | |||||
| O149:H− (1) | 1 | 1 | 1 | |||||
| O149:K88 (1) | 1 | 1 | 1 | 1 (α-hly) | ||||
| O159:H21 (1) | 1 | 1 | ||||||
| O167:H(rough) (1) | 1 | 1 | ||||||
| O(rough):H− (1) | 1 | 1 | 1 | 1 (α-hly) | ||||
| O(rough):H21 (4) | 4 | 4 | ||||||
| O(rough):H(NT)a (1) | 1 | 1 | 1 | |||||
| O(NT):H− (1) | 1 | 1 | ||||||
| O(NT):H18 (1) | 1 | |||||||
NT, nontypeable.
TABLE 5.
Detection of virulence genes in E. coli by MPCR
| Serotype (no. of strains) of pathogroup EIEC | No. of strains detected by:
|
||||||
|---|---|---|---|---|---|---|---|
| MPCR targeting
|
Single-gene-specific PCR targeting
|
||||||
| uidA | invE | pic | astA | lifA/efa1 | ent | hly | |
| O124:H− (1) | 1 | 1 | 1 | ||||
| O143:H− (1) | 1 | 1 | |||||
| O144:H− (1) | 1 | 1 | 1 | ||||
| O152:H− (1) | 1 | 1 | |||||
| O164:H− (2) | 2 | 2 | |||||
Acknowledgments
We are indebted to our colleagues A. Cravioto, H.-G. Sonntag, C. Jallat, K. Jann, S. Knutton, C. Le Bouguénec, L. W. Riley, G. Peters, S. L. Moseley, F. Ørskov and I. Ørskov, G. Schmidt, L. R. Trabulsi, and K. Wachsmuth for generous donations of clinical E. coli isolates. Furthermore, we thank Jennifer Schilling and Pia Schedewig for technical help.
We acknowledge partial support of this study by grants from the Deutsche Forschungsgemeinschaft (DFG SFB293 TP B5) and the Bundesministerium für Bildung and Forschung [BMBF Project Network of Competence Pathogenomics Alliance functional genomics research on enterohemorrhagic, enteropathogenic and enteroaggregative Escherichia coli (EHEC, EPEC, EAEC) PTJ-BIO/03U213B VBIIIPG3 and ERA Net PathoGenoMics PTJ-BIO/0313937C].
Footnotes
Published ahead of print on 30 March 2007.
REFERENCES
- 1.Badea, L., S. Doughty, L. Nicholls, J. Sloan, R. M. Robins-Browne, and E. L. Hartland. 2003. Contribution of Efa1/LifA to the adherence of enteropathogenic Escherichia coli to epithelial cells. Microb. Pathog. 34:205-215. [DOI] [PubMed] [Google Scholar]
- 2.Beinke, C., S. Laarmann, C. Wachter, H. Karch, L. Greune, and M. A. Schmidt. 1998. Diffusely adhering Escherichia coli strains induce attaching and effacing phenotypes and secrete homologs of Esp proteins. Infect. Immun. 66:528-539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Benz, I., and M. A. Schmidt. 1989. Cloning and expression of an adhesin (AIDA-I) involved in the diffuse adherence of enteropathogenic Escherichia coli. Infect. Immun. 57:1506-1511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bielaszewska, M., A. W. Friedrich, T. Aldick, R. Schurk-Bulgrin, and H. Karch. 2006. Shiga toxin activatable by intestinal mucus in Escherichia coli isolated from humans: predictor for a severe clinical outcome. Clin. Infect. Dis. 43:1160-1167. [DOI] [PubMed] [Google Scholar]
- 5.Bielaszewska, M., W. Zhang, P. I. Tarr, A. K. Sonntag, and H. Karch. 2005. Molecular profiling and phenotypic analysis of Escherichia coli O26:H11 and O26:NM: secular and geographic consistency of enterohemorrhagic and enteropathogenic isolates. J. Clin. Microbiol. 43:4225-4228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cerna, J. F., J. P. Nataro, and T. Estrada-Garcia. 2003. Multiplex PCR for detection of three plasmid-borne genes of enteroaggregative Escherichia coli strains. J. Clin. Microbiol. 41:2138-2140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chen, H. D., and G. Frankel. 2005. Enteropathogenic Escherichia coli: unravelling pathogenesis. FEMS Microbiol. Rev. 29:83-98. [DOI] [PubMed] [Google Scholar]
- 8.Dean, P., M. Maresca, and B. Kenny. 2005. EPEC's weapons of mass subversion. Curr. Opin. Microbiol. 8:28-34. [DOI] [PubMed] [Google Scholar]
- 9.Donnenberg, M. S., and J. B. Kaper. 1992. Enteropathogenic Escherichia coli. Infect. Immun. 60:3953-3961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Escobar-Páramo, P., O. Clermont, A. B. Blanc-Potard, H. Bui, C. Le Bouguénec, and E. Denamur. 2004. A specific genetic background is required for acquisition and expression of virulence factors in Escherichia coli. Mol. Biol. Evol. 21:1085-1094. [DOI] [PubMed] [Google Scholar]
- 11.Finlay, B. B., and S. Falkow. 1997. Common themes in microbial pathogenicity revisited. Microbiol. Mol. Biol. Rev. 61:136-169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Friedrich, A. W., J. Borell, M. Bielaszewska, A. Fruth, H. Tschäpe, and H. Karch. 2003. Shiga toxin 1c-producing Escherichia coli strains: phenotypic and genetic characterization and association with human disease. J. Clin. Microbiol. 41:2448-2453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Garmendia, J., G. Frankel, and V. F. Crepin. 2005. Enteropathogenic and enterohemorrhagic Escherichia coli infections: translocation, translocation, translocation. Infect. Immun. 73:2573-2585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gentry, M. K., and J. M. Dalrymple. 1980. Quantitative microtiter cytotoxicity assay for Shigella toxin. J. Clin. Microbiol. 12:361-366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gomes, T. A. T., K. Irino, D. M. Girão, V. B. C. Girão, B. E. C. Guth, T. M. I. Vaz, F. C. Moreira, S. H. Chinarelli, and M. A. M. Vieira. 2004. Emerging enteropathogenic Escherichia coli strains? Emerg. Infect. Dis. 10:1851-1855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hacker, J., G. Blum-Oehler, I. Muhldorfer, and H. Tschäpe. 1997. Pathogenicity islands of virulent bacteria: structure, function and impact on microbial evolution. Mol. Microbiol. 23:1089-1097. [DOI] [PubMed] [Google Scholar]
- 17.Harrington, S. M., E. G. Dudley, and J. P. Nataro. 2006. Pathogenesis of enteroaggregative Escherichia coli infection. FEMS Microbiol. Lett. 254:12-18. [DOI] [PubMed] [Google Scholar]
- 18.Kaper, J. B., J. P. Nataro, and H. L. T. Mobley. 2004. Pathogenic Escherichia coli. Nat. Rev. Microbiol. 2:123-140. [DOI] [PubMed] [Google Scholar]
- 19.Karaolis, D. K., S. Somara, D. R. Maneval, Jr., J. A. Johnson, and J. B. Kaper. 1999. A bacteriophage encoding a pathogenicity island, a type-IV pilus and a phage receptor in cholera bacteria. Nature 399:375-379. [DOI] [PubMed] [Google Scholar]
- 20.Karmali, M. A., M. Mascarenhas, S. Shen, K. Ziebell, S. Johnson, R. Reid-Smith, J. Isaac-Renton, C. Clark, K. Rahn, and J. B. Kaper. 2003. Association of genomic O island 122 of Escherichia coli EDL 933 with verocytotoxin-producing Escherichia coli seropathotypes that are linked to epidemic and/or serious disease. J. Clin. Microbiol. 41:4930-4940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kerényi, M., H. E. Allison, I. Batai, A. Sonnevend, L. Emody, N. Plaveczky, and T. Pal. 2005. Occurrence of hlyA and sheA genes in extraintestinal Escherichia coli strains. J. Clin. Microbiol. 43:2965-2968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Klapproth, J. M., I. C. Scaletsky, B. P. McNamara, L. C. Lai, C. Malstrom, S. P. James, and M. S. Donnenberg. 2000. A large toxin from pathogenic Escherichia coli strains that inhibits lymphocyte activation. Infect. Immun. 68:2148-2155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lan, R., M. C. Alles, K. Donohoe, M. B. Martinez, and P. R. Reeves. 2004. Molecular evolutionary relationships of enteroinvasive Escherichia coli and Shigella spp. Infect. Immun. 72:5080-5088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.LeClerc, J. E., B. Li, W. L. Payne, and T. A. Cebula. 1996. High mutation frequencies among Escherichia coli and Salmonella pathogens. Science 274:1208-1211. [DOI] [PubMed] [Google Scholar]
- 25.Levine, M. M. 1987. Escherichia coli that cause diarrhea: enterotoxigenic, enteropathogenic, enteroinvasive, enterohemorrhagic, and enteroadherent. J. Infect. Dis. 15:276-280. [DOI] [PubMed] [Google Scholar]
- 26.Lockwood, J. S., and H. T. Randall. 1949. The place of electrolyte studies in surgical patients. Bull. N. Y. Acad. Med. 25:228-239. [PMC free article] [PubMed] [Google Scholar]
- 27.Morabito, S., G. Dell'Omo, U. Agrimi, H. Schmidt, H. Karch, T. Cheasty, and A. Caprioli. 2001. Detection and characterization of Shiga toxin-producing Escherichia coli in feral pigeons. Vet. Microbiol. 82:275-283. [DOI] [PubMed] [Google Scholar]
- 28.Morabito, S., R. Tozzoli, E. Oswald, and A. Caprioli. 2003. A mosaic pathogenicity island made up of the locus of enterocyte effacement and a pathogenicity island of Escherichia coli O157:H7 is frequently present in attaching and effacing E. coli. Infect. Immun. 71:3343-3348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Müller, D., P. Hagedorn, S. Brast, G. Heusipp, M. Bielaszewska, A. W. Friedrich, H. Karch, and M. A. Schmidt. 2006. Rapid identification and differentiation of clinical isolates of enteropathogenic Escherichia coli (EPEC), atypical EPEC, and Shiga toxin-producing Escherichia coli by a one-step multiplex PCR method. J. Clin. Microbiol. 44:2626-2629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Nataro, J. P., J. Seriwatana, A. Fasano, D. R. Maneval, L. D. Guers, F. Noriega, F. Dubovsky, M. M. Levine, and J. G. Morris, Jr. 1995. Identification and cloning of a novel plasmid-encoded enterotoxin of enteroinvasive Escherichia coli and Shigella strains. Infect. Immun. 63:4721-4728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Nataro, J. P., and J. B. Kaper. 1998. Diarrheagenic Escherichia coli. Clin. Microbiol. Rev. 11:142-201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ørskov, F., and I. Ørskov. 1992. Escherichia coli serotyping and disease in man and animals. Can. J. Microbiol. 38:699-704. [PubMed] [Google Scholar]
- 33.Piva, I. C., A. L. Pereira, L. R. Ferraz, R. S. Silva, A. C. Vieira, J. E. Blanco, M. Blanco, J. Blanco, and L. G. Giugliano. 2003. Virulence markers of enteroaggregative Escherichia coli isolated from children and adults with diarrhea in Brasilia, Brazil. J. Clin. Microbiol. 41:1827-1832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Prager, R., A. Fruth, and H. Tschäpe. 2003. Subtyping of pathogenic E. coli strains using flagellar (H) antigens: serotyping vs. fliC-polymorphism. Int. J. Med. Microbiol. 292:477-486. [DOI] [PubMed] [Google Scholar]
- 35.Reid, S. D., C. J. Herbelin, A. C. Bumbaugh, R. K. Selander, and T. S. Whittam. 2000. Parallel evolution of virulence in pathogenic Escherichia coli. Nature 406:64-67. [DOI] [PubMed] [Google Scholar]
- 36.Savarino, S. J., A. McVeigh, J. Watson, A. Cravioto, J. Molina, P. Echeverria, M. K. Bhan, M. M. Levine, and A. Fasano. 1996. Enteroaggregative Escherichia coli heat-stable enterotoxin is not restricted to enteroaggregative E. coli. J. Infect. Dis. 173:1019-1022. [DOI] [PubMed] [Google Scholar]
- 37.Schmidt, H., J. Scheef, S. Morabito, A. Caprioli, L. H. Wieler, and H. Karch. 2000. A new Shiga toxin 2 variant (Stx2f) from Escherichia coli isolated from pigeons. Appl. Environ. Microbiol. 66:1205-1208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Schultsz, C., G. J. Pool, R. van Ketel, B. de Wever, P. Speelman, and J. Dankert. 1994. Detection of enterotoxigenic Escherichia coli in stool samples by using nonradioactively labeled oligonucleotide DNA probes and PCR. J. Clin. Microbiol. 32:2393-2397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Servin, A. 2005. Pathogenesis of Afa/Dr diffusely adhering Escherichia coli. Clin. Microbiol. Rev. 18:264-292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Sonntag, A. K., M. Bielaszewska, A. Mellmann, N. Dierksen, P. Schierack, L. H. Wieler, M. A. Schmidt, and H. Karch. 2005. Shiga toxin 2e-producing Escherichia coli isolates from humans and pigs differ in their virulence profile and interactions with intestinal epithelial cells. Appl. Environ. Microbiol. 71:8855-8863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Sonntag, A. K., E. Zenner, H. Karch, and M. Bielaszewska. 2005. Pigeons as a possible reservoir of Shiga toxin 2f-producing Escherichia coli pathogenic to humans. Berl. Münch. Tierärztl. Wochenschr. 118:464-470. [PubMed] [Google Scholar]
- 42.Tarr, P. I., C. A. Gordon, and W. I. Chandler. 2005. Shiga-toxin-producing Escherichia coli and haemolytic uraemic syndrome. Lancet 365:1073-1086. [DOI] [PubMed] [Google Scholar]
- 43.Tornieporth, N. G., J. John, K. Salgado, P. de Jesus, E. Latham, M. C. Melo, S. T. Gunzburg, and L. W. Riley. 1995. Differentiation of pathogenic Escherichia coli strains in Brazilian children by PCR. J. Clin. Microbiol. 33:1371-1374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Trabulsi, L. R., R. Keller, and T. A. T. Gomes. 2002. Typical and atypical enteropathogenic Escherichia coli. Emerg. Infect. Dis. 8:508-513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Tsai, C. C., S. Y. Chen, and H. Y. Tsen. 2003. Screening the enteroaggregative Escherichia coli activity and detection of the aggA, aafA, and astA genes with novel PCR primers for the Escherichia coli isolates from diarrhea cases in Taiwan. Diagn. Microbiol. Infect. Dis. 46:159-165. [DOI] [PubMed] [Google Scholar]
- 46.Vial, P. A., J. J. Mathewson, H. L. DuPont, L. Guers, and M. M. Levine. 1990. Comparison of two assay methods for patterns of adherence to HEp-2 cells of Escherichia coli from patients with diarrhea. J. Clin. Microbiol. 28:882-885. [DOI] [PMC free article] [PubMed] [Google Scholar]