Abstract
The immunoglobulin G (IgG)-binding streptococcal protein G is often used for immunoprecipitation or immunoadsorption-based assays, as it exhibits binding to a broader spectrum of host species IgG and IgG subclasses than the alternative, Staphylococcus aureus protein A. Caulobacter crescentus produces a hexagonally arranged paracrystalline protein surface layer (S-layer) composed of a single secreted protein, RsaA, that is notably tolerant of heterologous peptide insertions while maintaining the surface-attached crystalline character. Here, a protein G IgG-binding domain, GB1, was expressed as an insertion into full-length RsaA on the cell surface to produce densely packed immunoreactive particles. GB1 insertions at five separate sites were expressed, and all bound rabbit and goat IgG, but expression levels were reduced compared to those of wild-type RsaA and poor binding to mouse IgG was noted. To remedy this, we used the 20-amino-acid Muc1 peptide derived from human mucins as a spacer, since insertions of multiple tandem repeats were well tolerated for RsaA secretion and assembly. This strategy worked remarkably well, and recombinant RsaA proteins, containing up to three GB1 domains, surrounded by Muc1 peptides, not only were secreted and assembled but did so at wild-type levels. The ability to bind IgG (including mouse IgG) increased as GB1 units were added, and those with three GB1 domains bound twice as much rabbit IgG per cell as S. aureus cells (Pansorbin). The ability of recombinant protein G-Caulobacter cells to function as immunoactive reagents was assessed in an immunoprecipitation assay using a FLAG-tagged protein and anti-FLAG mouse monoclonal antibody; their performance was comparable to that of protein G-Sepharose beads. This work demonstrates the potential for using cells expressing recombinant RsaA/GB1 in immunoassays, especially considering that protein G-Caulobacter cells are more cost-effective than protein G beads and exhibit a broader species and IgG isotype binding range than protein A.
The immunoglobulin G (IgG)-binding protein G, found on the surface of streptococcal cells, acts as a bacterial Fc receptor (9). It contains three highly similar regions of 55 amino acids (aa), each separated by a short 15-aa spacer sequence, that enable binding of IgG from many species, including humans, mice, rabbits, and goats (1, 9). As such, protein G is frequently used for immunoprecipitation or immunoadsorption-based assays, including the purification of antibodies from protein mixtures, the formation of antibody-adsorbent particles for specific antigen immunoprecipitation (e.g., protein G-agarose), and the analysis of protein fractions (10). It is particularly useful for applications in which mouse monoclonal antibodies are used since it exhibits superior binding affinity for all isotypes of these antibodies compared to the other commonly used IgG-binding proteins, such as Staphylococcus aureus protein A (15). In fact, antibodies from species or subclasses that do not bind well to protein A generally bind to protein G, making it the immunobinding protein of choice for many commonly used research applications. It is, however, considerably more expensive, partly because it is marketed as conjugated to agarose beads rather than simply as killed whole cells, as is the case for protein A.
RsaA is the sole component of the paracrystalline protein surface layer (S-layer) of Caulobacter crescentus (30, 31), a nonpathogenic, dimorphic, gram-negative, aquatic bacterium, with related species commonly inhabiting the freshwater biosphere in North America (25, 27). RsaA monomers are secreted from the bacterium via a type I ABC transporter system (2), and approximately 40,000 subunits are assembled into a lattice array with hexagonal symmetry on the cell surface (30). RsaA is the predominant protein of C. crescentus, accounting for 10 to 12% of the total protein synthesis (35). The 1,026-aa RsaA protein has an N-terminal cellular adhesion signal and a C-terminal secretion signal (5-7). The C. crescentus genome has been sequenced, a significant asset for genetic manipulation. Further, Caulobacter can be easily and inexpensively cultured to high density in simple defined media. All of these considerations have implications for the development of biotechnological applications based on the expression of foreign proteins using the Caulobacter S-layer protein. Indeed, we have exploited the S-layer secretion apparatus in order to express heterologous proteins as large as 600 aa in size (23) by fusing them to portions of RsaA containing the C-terminal secretion signal (5-7). The resulting protein products are secreted from C. crescentus at yields ranging up to 250 mg/liter (7).
We have also explored the possibility of using the S-layer to display foreign peptides on the Caulobacter cell surface in the dense, highly ordered S-layer structure (5, 35). This presentation system has many potential applications, such as the development of whole-cell vaccines, tumor suppressors, cellular adsorbents, and peptide display libraries and the screening of antibody libraries (3, 13, 23). We have identified sites where insertions of nucleotides that maintain reading frames into the full-length rsaA gene can be made without impeding the synthesis, folding, secretion, and self-assembly of the corresponding protein on the bacterial cell surface (5, 6, 35). Peptide sequence insertions of more than 650 aa have been made while maintaining secretion, attachment, and crystallization (23). In general, the display of such sizes has not been accomplished with other outer membrane proteins or appendage structures in gram-negative bacteria and rivals the lower-density display mechanisms of some gram-positive bacteria, such as the sortase-mediated pathway (18).
The SbpA S-layer protein of Bacillus sphaericus has also been used to make heterologous proteins that can be recrystallized on peptidoglycan-containing sacculi and secondary cell wall polymer-coated planar surfaces and microbeads. A recent study describes the development of affinity microparticles using recombinant S-layer-expressing tandem copies of the IgG-binding domain from Staphylococcus aureus protein A for use in extracorporeal blood purification (37). These studies carried out with another model system serve to demonstrate, along with the present study, that regularly structured protein lattices can exhibit the functionality of the foreign protein.
In this report, we investigated the possibility of constructing functional immunoreactive reagents using the Caulobacter S-layer. A single recombinant RsaA/GB1 protein was detectable on the cell surface, and Caulobacter cells expressing a chimeric RsaA/GB1 S-layer were capable of binding IgG. However, the expression levels of RsaA/GB1 protein were low and IgG binding was deficient in several respects. To resolve these issues, a second generation of constructs was engineered in which multiple copies of the GB1 segment were inserted in conjunction with a spacer peptide. This facilitated secretion and allowed for the display of multiple GB1 domains. The constructs performed well in every respect, including the use of these bacteria in an immunoprecipitation assay with mouse monoclonal antibodies.
MATERIALS AND METHODS
Bacterial strains, growth conditions, plasmids, and reagents.
The C. crescentus strains and plasmids used in this study are listed in Table 1. The oligonucleotides used in this study are listed in Table 2. Escherichia coli strain DH5α (Invitrogen, Carlsbad, CA) was grown at 37°C in LB medium (26). C. crescentus strains were propagated in liquid peptone-yeast extract at 30°C as previously described (4). For growth on solid medium, agar was added at 1.2% (wt/vol). Where necessary, media were supplemented with ampicillin (AP) at 50 μg/ml and chloramphenicol (CM) at 20 μg/ml (E. coli) or 2 μg/ml (C. crescentus). Electroporation of C. crescentus was performed as previously described (14).
TABLE 1.
C. crescentus strains and intermediate plasmids used in this study
| Strain or plasmid | Relevant characteristicsa | Reference |
|---|---|---|
| Strains | ||
| JS4019 | recA repBAC+ derivative of JS4011 | 36 |
| JS4015 | Sap− (point mutation) recA variant of JS4000 | 34 |
| JS4022 | Sap− (point mutation) recA repBAC+ derivative of JS4015, constructed as described for JS4019 (36) | This study |
| Plasmids | ||
| pBSKII ESH | Modified pBluescript SKII; Ap | 33 |
| PUC8 | Small high-copy plasmid; Ap | 20 |
| pUC9CXS | Epitope carrier vector; Ap, Cm | 6 |
| pUC9CXSGB1 | Epitope carrier vector containing the GB1 segment; Ap, Cm | This study |
| pTZ18U:rsaAΔP(AciI574BamHI) | rsaA gene without its native promoter (ΔP) carrying a BamHI linker insertion at a site corresponding to aa 574 of RsaA; Ap | 6 |
| pTZ18U:rsaAΔP(HinPI622BamHI) | rsaAΔP carrying a BamHI linker insertion at a site corresponding to aa 622 of RsaA; Ap | 6 |
| pTZ18U:rsaAΔP(HinPI690BamHI) | rsaAΔP carrying a BamHI linker insertion at a site corresponding to aa 690 of RsaA; Ap | 6 |
| pTZ18U:rsaAΔP(HinPI723BamHI) | rsaAΔP carrying a BamHI linker insertion at a site corresponding to aa 723 of RsaA; Ap | 6 |
| pTZ18U:rsaAΔP(HinPI944BamHI) | rsaAΔP carrying a BamHI linker insertion at a site corresponding to aa 944 of RsaA; Ap | 6 |
| p4A | pUC8-type vector containing oriV with a modified rsaA promoter region; Cm | This study |
| p4ArsaA(723Δ)GSCCΔ | p4A containing rsaAΔP with a segment containing several unique restriction sites inserted at the BamHI linker site corresponding to aa 723 of RsaA; Cm | This study |
| p4ArsaA(723Δ)GB1SN | p4A containing rsaAΔP with modified GB1 inserted at the BamHI linker site corresponding to aa 723 of RsaA; Cm | This study |
| p4ArsaA(723Δ)Muc1B | p4A containing rsaAΔP with Muc1B segment inserted at the BamHI linker site corresponding to aa 723 of RsaA; Cm | This study |
Ap, ampicillin; Cm, chloramphenicol.
TABLE 2.
Oligonucleotides used in this study
| Oligonucleotide | Sequence |
|---|---|
| IDGB1F | GCTCGAGGGAAGACCGACACGTATAAGCTGATCCTCAACGGCAGACGCTGAAGGGCGAGACCACGACCGAAGCGGTGGACGCCGCGACGGCGGAAAAGGTGTTCAAGCAGTAT |
| IDGB1R | AGGCCTCGGGCTTCTCCGTGACCGTGAACGTCTTCGTGGCGTCATCGTACGTCCACTCGCCGTCGACGCCATTGTCGTTCGCATACTGCTTGAACACCTTTTCCGCCGTCGC |
| 1060 | GCCTACTCTTCCTTTTTCAATATTATTGAA |
| 1920 | GCCTAGTACTCTGTCAGACCAAGTTTACTCATA |
| JNCHE-1 | GGAAGATCTGTTAACTTTTCAGGAGCTAAGGAAGCT |
| JNCHE-2 | GGAAGATCTGTTAACACAATAACTGCCTTAAAAAAATTA |
| GSCC-1 | AATTCGGATCCAGATCTGTTAACAATGCATCAGAGCAGAAGCTGATCTCGGAAGAGGACCTCAGGCCTTCTGCAGATGGATCCA |
| GSCC-2 | AGCTTGGATCCATCTGCAGAAGGCCTGAGGTCCTCTTCCGAGATCAGCTTCTGCTCTGATGCATTGTTAACAGATCTGGATCCG |
| KBPGBS-1 | GGAAGATCTACTAGTTATAAGCTGATCCTCAACGGC |
| KBPGNP-2 | GGCTGCAGCGCTAGCCTCCGTGACCGTGAACGTCTT |
| MUC1B-3 | GATCTACTAGTCCGCCCGCCCACGGCGTGACCTCGGCGCCGGACACGCGCCCCGCCCCGGGTAGCACCGCTAGCGCTGCA |
| MUC1B-4 | GCGCTAGCGGTGCTACCCGGGGCGGGGCGCGTGTCCGGCGCCGAGGTCACGCCGTGGGCGGGCGGACTAGTA |
The plasmids used to construct the protein G-RsaA display constructs are described in Table 1. Plasmid DNA was isolated using a QIAprep miniprep plasmid isolation kit (QIAGEN), and DNA fragments were recovered from agarose gels using a QIAEX II gel extraction kit (QIAGEN). The Nucleic Acid Protein Service Unit of the University of British Columbia performed oligonucleotide synthesis and DNA sequencing. PCR was done under standard conditions using either Taq or Pfx polymerase (Invitrogen). Annealing of complementary oligonucleotides was done by mixing 100 ng of each oligonucleotide, heating the oligonucleotides in a boiling water bath for 2 min, and allowing them to cool slowly to room temperature.
Cloning the GB1 domain.
A portion of the streptococcal protein G coding sequence (aa 299 to 360), including the IgG-binding domain GB1 (aa 303 to 357, underlined) KTDTYKLILNGKTLKGETTTEAVDAATAEKVFKQYANDNGVDGEWTYDDATKTFTVTEKPE, was selected for insertion into RsaA. The oligonucleotides IDGB1F and IDGB1R were designed with a 30-bp overlap for annealing, taking into account C. crescentus codon usage and incorporating 5′ XhoI and 3′ StuI restriction sites. They were annealed and extended by Taq polymerase. Vent polymerase was used to polish the ends prior to blunt ligation into the StuI site of the intermediate plasmid pBSKIIESH (33).
After sequence confirmation, the 189-bp GB1 DNA fragment was directionally cloned into pUC9CXS (6) by using XhoI/StuI, resulting in pUC9CXSGB1. The XhoI/StuI insert (termed GB1XS) was the source of the GB1 coding sequence used to make the RsaA/GB1XS constructs.
Construction of the RsaA/GB1 display constructs.
To insert the GB1XS coding sequence into the full-length rsaA gene, an approximately 1-kb BamHI fragment, including the 189-bp XhoI/StuI GB1 DNA fragment as well as a promoterless CM resistance gene, was excised from pUC9CXSGB1 and ligated into each of the five pTZ18U:rsaAΔP vectors (Table 1), which contain a BamHI linker site at aa positions 574, 622, 690, 723, and 944. The expression of CM resistance as well as diagnostic restriction enzyme digests was used to identify clones with the correct insert orientation. After excision of the CM resistance gene by BglII digestion and religation, the EcoRI/HindIII RsaA/GB1XS-encoding regions were transferred to the peptide display vector p4A.
p4A is a pUC8-based vector that was used to express full-length variants of the rsaA gene under the control of a modified rsaA promoter region. In addition to a colicin E1 replicon, it contains a second origin of replication, oriV, derived from RSF1010, which allows for replication in both E. coli strains and in those C. crescentus strains that contain the repBAC genes of RSF1010 (36). p4A was constructed as follows. The oligonucleotides 1060 and 1920 were used for an inverse PCR on pUC8 (20). This PCR product was digested with StuI and ScaI and ligated to the HpaI-digested modified CM resistance gene called CHE (see below), resulting in pUC8CX, essentially replacing the AP resistance gene with a CM resistance gene. The CHE gene (modified to remove an EcoRI site) was obtained by PCR amplification, using oligonucleotides JNCHE-1 and JNCHE-2 and pMMB206 as the target (21). oriV was inserted by digesting pUC8CX with EcoO109, blunting with Klenow polymerase, and then digesting it with HindIII. The fragment was replaced with the 521-bp HindIII-XmnI oriV fragment from pCR2.1oriV (36), resulting in pUC8CVX. The lac promoter was then replaced with a modified rsaA promoter region. The EcoRI-HindIII fragment from plasmid pSSa49ΔSD (8) containing the modified promoter region was ligated into pUC8, resulting in pUC8ΔSD. Then, pUC8ΔSD and pUC8CVX were digested with EcoRI and ligated together, and E. coli transformants were selected using AP and CM. Correct orientation was determined with an NdeI digest, and the plasmid fusion was digested with SapI and PstI, blunted with Klenow polymerase, and religated, selecting for CM resistance. This last manipulation removed nearly all of pUC8ΔSD except the modified rsaA promoter region, which was now upstream of the EcoRI-HindIII multiple cloning site; this plasmid was termed p4A.
As previously described (35), peptide insert constructs will be referred to herein as RsaA(insert location in terms of amino acid position)/segment, for example, RsaA(690)/GB1XS indicates that the GB1XS domain has been inserted at aa position 690 of RsaA. All peptide display constructs were expressed in the S-layer-associated protease (Sap)-deficient C. crescentus strain JS4022.
To aid some clone constructions, a fragment was engineered with several unique restriction sites to enable directional cloning of DNA segments into rsaA. This segment was constructed by annealing the oligonucleotides GSCC-1 and GSCC-2 and ligating the overhanging EcoRI/HindIII ends into digested pUC8, followed by installing the BglII-framed promoterless CM resistance gene from pUC9CXS into the segment's BglII site. This GSCC/CM segment was isolated by digesting the flanking BamHI sites and then directionally cloned into the linker site in pTZ18U:rsaAΔP(HinP1723 BamHI) (Table 1) by selecting for CM resistance. The CM resistance gene was removed by BglII digestion and religation. Prior to this last cloning step, the DNA region downstream of rsaA was altered by digestion with FseI and SphI, blunting with Klenow polymerase, and religation. This removed several restriction sites, including PstI. The EcoRI/HindIII RsaA region was transferred to p4A, now called p4ARsaA(723Δ)GSCCΔ; this plasmid has unique BglII and PstI sites that can be used for directional cloning.
The GB1SN and Muc1B segments were cloned into these sites. For GB1SN, oligonucleotides KBPGBS-1 and KBPGNP were used for PCR on GB1XS DNA to engineer GB1 with BglII and SpeI sites on the 5′ end and NheI and PstI sites on the 3′ end. This segment, termed GB1SN, was digested with BglII/PstI and directionally cloned into p4ARsaA(723Δ)/GSCCΔ, replacing GSCCΔ and resulting in p4ARsaA(723Δ)/GB1SN. The Muc1B segment was constructed in a similar fashion, except with annealing of oligonucleotides Muc1B-3 and Muc1B-4 and ligating, resulting in p4ARsaA(723Δ)/Muc1B.
Tandem repeats of GB1SN and Muc1B were made in the following manner: to construct an Muc1B segment upstream of a GB1SN segment, the larger fragment from an NheI/HindIII digest of p4ARsaA(723Δ)/Muc1B was isolated and ligated to the smaller fragment of an SpeI/HindIII digest of p4ARsaA(723Δ)/PGSN. The resulting plasmid, p4ARsaA(723Δ)/Muc1B-PGSN, was renamed p4ARsaA(723Δ)/ MG for simplicity. Using this general cloning method, any arrangement of repeats could be constructed and multimerized, exponentially if desired.
Protein isolation, separation, and Western immunoblotting.
RsaA and RsaA/GB1 recombinant proteins were recovered from the cell surface by low-pH extraction, as described previously (38). Sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) using 7.5% gels separated soluble proteins, which were visualized by staining with Coomassie brilliant blue R. For immunoblotting, proteins were transferred to BioTrace NT nitrocellulose membranes (Pall Life Sciences). Except as described for the pull-down assay (below), membranes were blocked with Tris-buffered saline (TBS)-milk, (20 mM Tris base, 154 mM NaCl, pH 7.4, containing 3% [wt/vol] nonfat milk) and incubated with 1:1,000 to 1:5,000 horseradish peroxidase (HRP)-conjugated goat anti-mouse or goat anti-rabbit antiserum (Antibodies Inc.) in TBS-milk. Blots were washed with TBS and visualized with 4-chloro-1-naphthol as previously described (29).
Evaluating the cell surface expression and IgG-binding functionality of RsaA/GB1 constructs.
To directly assess the functional surface expression of the GB1 domain for RsaA/GB1XS display constructs, colloidal gold labeling was performed in conjunction with electron microscopy. Briefly, cells were first incubated with normal rabbit immune serum followed by 5 nm protein A-colloidal gold labeling, as described previously (28). Both the S-layer-deficient JS4019 strain and JS4022 cells expressing RsaA(723) were used as negative controls. Cells were visualized using a Siemens 101A transmission electron microscope operated at 60 kV.
To directly assess the IgG-binding capability of the GB1 domain when expressed as part of a recombinant protein on the surface of Caulobacter cells, cells expressing RsaA(723Δ)/Muc1B/GB1SN constructs were labeled with a randomly selected Cy3-coupled monoclonal antibody (directed to the c-myc peptide) by the addition of antibody to cells in peptone-yeast extract medium, followed by processing as previously described (19), and then examined using epifluorescence and a rhodamine filter set on a Zeiss photomicroscope III. In addition, a spectrofluorometric assay was developed for a semiquantitative evaluation of Cy3-coupled anti-c-myc monoclonal antibody binding. Briefly, 4 × 108 cells were labeled with the Cy3-coupled mouse monoclonal IgG for 30 min at 4°C. The cells were centrifuged (14,000 × g for 4 min), the supernatant completely removed, and the cells suspended in 1 ml of 10 mM HEPES, pH 7.5, and 10 mM EGTA to effectively remove the S-layer from the cell surface (22, 38). After 5 min of incubation at room temperature, the preparation was centrifuged to eliminate the light-scattering effects of the bacteria and the supernatant fraction was examined in a spectrofluorometer (550-nm excitation and 570-nm emission wavelengths).
Measuring the experimental IgG-binding capacities of protein G-expressing C. crescentus and Pansorbin cells.
Overnight cultures of C. crescentus strain JS4022 expressing RsaA(723Δ)/Muc1B/GB1SN constructs were pelleted for 5 min at 14,000 × g at 4°C, washed twice with phosphate buffer-Tween 20 (PB-T) (2 mM KH2PO4, 10 mM Na2HPO4, 0.05% Tween 20), and resuspended in PB-T in 1/10 of the original culture volume. Optical density readings at 600 nm were obtained, and the approximate cell culture concentration was calculated according to the following empirically determined conversion factor: optical density at 600 nm = 3.1 × 109 cells/ml. Pansorbin cells (Calbiochem) are heat-killed, formalin-fixed Staphylococcus aureus cells (Cowan I strain) that bear a high cell surface density of protein A which, according to the manufacturer's package insert, allows them to bind 19 μg human IgG/mg of cells.
To determine binding capacity, 1 × 108 JS4022 cells expressing RsaA(723Δ)/Muc1B/GB1SN constructs or 5 μl of a 1:10 dilution (5.95 μg) of Pansorbin cells (supplied at 11.9 mg/ml) were incubated for 30 min at room temperature on a rotator in 500 μl PB-T with 1 μg of HRP-conjugated rabbit anti-guinea pig IgG (supplied at 7.8 μg/μl; Sigma-Aldrich). Cells were then pelleted by centrifugation for 5 min at 14,000 × g, washed with 1 ml PB-T, and resuspended in 100 μl of PB-T. To determine the amount of IgG bound to the cells, an enzyme-linked immunosorbent assay (ELISA)-type assay was performed using the HRP substrate o-phenylenediamine (Sigma) as previously described (32). A standard curve was generated using twofold serial dilutions (1:1,250 to 1:20,000) of the HRP-conjugated antibody. Briefly, 100-μl volumes of diluted IgG (1:25) and cell pellets [1:10 for RsaA(723Δ)/Muc1B/GB1SN constructs and 1:25 for Pansorbin cells] were mixed with 100 μl of the colorimetric substrate solution (3 mg o-phenylenediamine dissolved in 5 ml 0.1 M citric acid and 5 ml 0.1 M Na2HPO4, supplemented with 6 μl 30% H2O2 [immediately before use]) in a 96-well plate and the reaction was allowed to progress for approximately 2 to 5 min before it was stopped by the addition of 100 μl 3 M HCl. For each sample, A495 minus A405 was determined using a SpectraMAX 190 plate reader (Molecular Devices) and SoftMax Pro 4.3.1 software. The amount of IgG bound to the cells was calculated relative to the HRP IgG standard curve. For the Caulobacter cells, the corresponding dry weight of the cells was calculated by weighing lyophilized cell pellets containing a known number of cells. All reactions were performed in triplicate. An average was determined for IgG-binding efficiency, and the standard deviation was calculated using Microsoft Excel 2001 software.
Immunoprecipitation (pull-down) assay.
Whole-cell lysates from murine B-lymphocyte WEHI-231 cell lines stably transfected with a gene specifying RapGapII-FLAG (17), a protein of about 100 kDa, or mock transfected with empty vector were generously provided by Michael Gold (Department of Microbiology and Immunology, University of British Columbia). Protein concentration was determined using a Micro bicinchoninic acid protein assay reagent kit (Pierce). Lysate with 300 μg total protein was mixed with 1.5 μl of anti-FLAG M2 mouse monoclonal antibody (Sigma) in a total volume of 300 μl PB-T. This sample was incubated overnight at 4°C on a rotator and then divided in thirds. To each 100 μl was added either 1 × 109 JS4022 cells expressing p4ARsaA(723Δ) or p4ARsaA(723Δ)/MGMGMGM (prepared as described above) or 20 μl protein G-Sepharose beads (50:50 slurry; Calbiochem). Samples were incubated for 1 h at 4°C on a rotator. Cell-bead-immune complexes were pelleted by centrifugation at 14,000 × g for 5 min, suspended in 1 ml PB-T, and pelleted again. Recombinant S-layer protein was extracted by suspending cells in 15 μl 10 mM EGTA, 10 mM HEPES, pH 7.5, for 10 min at room temperature and pelleting the cells. The recovered supernatants or protein G beads were mixed with 10 μl of SDS-PAGE sample buffer (26), boiled for 5 min, separated by 7.5% SDS-PAGE, and transferred to a nitrocellulose membrane. The membrane was blocked for 1 h with TBS-5% milk and then for an additional hour with TBS-3% milk-3% goat serum (Invitrogen) and treated with anti-FLAG antibody (Sigma) at 10 μg/ml in TBS-3% milk-0.5% goat serum for 2 h at 4°C. The blot was washed four times with TBS-T (TBS and 0.1% Tween 20), incubated with 1:2,000 peroxidase-labeled anti-mouse antibody (Amersham Pharmacia Biotech) in TBS-3% milk-0.5% goat serum, and washed three times with TBS-T. Labeling was visualized using the enhanced chemiluminescence Western blotting detection system and Kodak X-OMAT LS film.
RESULTS
Expression and S-layer display of GB1XS as a single copy.
Protein G contains three nearly identical domains, GB1, GB2, and GB3, separated from each other by two identical 15-aa spacers. For this study, we chose to clone GB1 by inserting the corresponding DNA into the rsaA gene at sites known to be capable of foreign peptide display (6), as described in Materials and Methods.
GB1 prepared in this manner (termed GB1XS) was first inserted into RsaA at the site corresponding to aa 723. The secretion levels of the modified RsaA, as monitored by a low-pH extraction method, showed a reduced expression level, estimated to be ca. 10 to 20% of normal RsaA secretion. This was lower than expected, since numerous other segments of ca. 50 aa, as well as much larger ones, have been displayed at this site with little impact on secretion levels (23). Nevertheless, when S-layer proteins were recovered from the cell surface by low-pH treatment and analyzed by Western immunoblotting using only an HRP-coupled goat-derived secondary antiserum, a positive response was obtained (Fig. 1), indicating a degree of GB1 functionality.
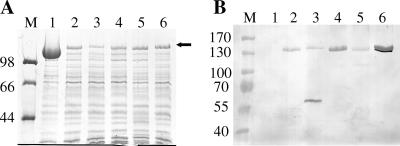
FIG. 1.
SDS-PAGE (A) and Western immunoblot (B) analyses of normalized low-pH-extracted RsaA-protein G peptide (GB1) display constructs. Equal volumes of protein preparations were loaded. Immunoblot were treated with HRP-conjugated goat anti-rabbit antibody (1:1,000) and developed with 4-chloro-1-naphthol. M, molecular mass standard (in kDa). Lanes: 1, p4ARsaA(723Δ); 2, p4ARsa(574)/GB1XS; 3, p4ARsa(622)/GB1XS; 4, p4ARsa(690)/GB1XS; 5, p4ARsa(723)/GB1XS; 6, p4ARsa(944)/GB1XS.
In an effort to improve the secretion levels or antibody binding of RsaA/GB1XS chimers, GB1XS DNA was also inserted into sites in rsaA corresponding to aa 574, 622, 690, and 944. As qualitatively estimated by SDS-PAGE and Western immunoblotting with goat antiserum (Fig. 1), the secretion levels of the 574- and 622-modified sites were comparable to that of the 723 site and resulted in low-level antibody binding; the 690 site demonstrated somewhat better antibody binding and the 944 site allowed somewhat higher expression levels and significantly improved antibody binding. This improvement was surprising in that the 944 site is very close to the minimum C-terminal secretion signal (7) and inference with secretion was anticipated.
Electron microscopy analysis of GB1XS display on intact cells by using normal rabbit serum followed by colloidal gold labeling generally mirrored the relative differences in recombinant RsaA/GB1XS expression seen by Western immunoblot analysis with goat antiserum (data not shown). Examples of GB1XS display at aa 690 and 944 (the best sites for binding as judged by Western analysis) are shown in Fig. 2, where the sample cells exemplify that the highest level of labeling occurred at the 944 position in this method of display. Note that microscopy indicated that RsaA(944)/GB1XS was distributed in patches rather than homogenously over the cell surface; this is likely a consequence of S-layer crystallization occurring even though a full complement of protein is not secreted.
FIG. 2.
Colloidal gold labeling of chimeric RsaA-protein G (GB1XS) S-layer on the surface of C. crescentus cells. Whole cells expressing chimeric constructs RsaA(690)/GB1XS (A) and RsaA(944)/GB1XS (B) were incubated with normal rabbit antiserum followed by 5 nM protein A-colloidal gold labeling.
An ELISA method was developed to evaluate the amount of rabbit antibody binding by using commercially available HRP-coupled IgG. The use of rabbit antiserum allowed a comparison to commercially available protein A-displaying staphylococcal cells (Pansorbin). In our assay, the levels of IgG binding per mg (dry weight) of cells to Pansorbin were less than the values published by the company selling Pansorbin. The Caulobacter cells displaying RsaA(944)/GB1XS bound at levels that were greater than, but comparable to, those for Pansorbin (0.57 μg/mg [dry weight] of cells for Caulobacter cells versus 0.48 μg for Pansorbin) (Table 3). As expected for RsaA(723)/GB1XS, IgG binding was somewhat less than (0.4 μg/mg [dry weight] of cells), but nevertheless comparable to, the Pansorbin standard.
TABLE 3.
Assessment of rabbit and mouse antibody binding
| Protein G display construct | Quantitative ELISA assessment of rabbit IgG-binding capacity (μg IgG/mg [dry wt] of cells)a | Qualitative assessment of mouse IgG binding
|
|
|---|---|---|---|
| Labeling result by fluorescence microscopyb | Absorbance value for spectro-fluorometric assay (550-nm excitation/ 570-nm emission)c | ||
| RsaA723 (no protein G) | 0.09 (0.03) | − | 0.29 |
| RsaA723-GXS | 0.40 (0.09) | − | 0.41 (0.12) |
| RsaA944-GXS | 0.57 (0.34) | NDe | ND |
| RsaA723-GSN | 0.64 (0.19) | ++ | 0.41 (0.12) |
| RsaA723-G-M | 0.20 (0.05) | − | 0.44 (0.15) |
| RsaA723-G-M-G | 0.64 (0.32) | + | 0.39 (0.10) |
| RsaA723-G-M-G-M-G | 0.79 (0.39) | +++ | 0.65 (0.36) |
| RsaA723-M-G-M-G-M-G-M | 0.98 (0.42) | +++++ | 0.69 (0.40) |
| Pansorbind | 0.48 (0.12) | ND | ND |
Standard deviations are indicated in parentheses. Data represents averages of five runs.
−, no detectable label; +++++, maximum possible fluorescence label.
Values in parentheses are the result of subtraction of the RsaA723 absorbance value.
Pansorbin cells display protein A.
ND, not determined.
We then tested the ability of the Caulobacter cells to perform in a typical pull-down assay by using WEHI-231 cells expressing a FLAG-tagged protein and an anti-FLAG tag mouse monoclonal antibody, comparing the Caulobacter cells displaying RsaA(944)/GB1XS with commercially available protein G coupled to Sepharose beads. The assay was performed according to a standard immunoprecipitation protocol with the exception that no EDTA was used, and instead, after retrieval of the Caulobacter cells by centrifugation, the S-layer and attached antibody were removed from the cell surface by EGTA treatment to ensure that no significant amounts of cell components would be applied to the SDS-PAGE gel. Despite numerous attempts and assay modifications, little or no activity was noted for the Caulobacter cells (data not shown). This was unexpected, as it is suggested throughout the literature that protein G is capable of comparable binding efficiencies to rabbit, goat, and mouse IgGs. We hypothesized that this discrepancy might be the result of GB1 folding incorrectly within the S-layer monomer or could be due to the fact that our constructs bear only one IgG-binding domain, GB1, as opposed to the three domains, GB1 to GB3, present in native protein G and thus were unable to bind murine IgG with any degree of efficiency. Immunofluorescence labeling of RsaA(944)/ GB1XS with a Cy3-coupled mouse monoclonal antibody supported this hypothesis, showing no detectable binding of murine IgG (data not shown).
We concluded then that the best GB1XS display construct performed in a manner that was comparable to that of Pansorbin cells but was not improved in any material way.
Development and assessment of constructs displaying multiple copies of GB1 by using Muc1 as a spacer sequence.
Though it seemed a poor prospect to expect that secretion levels of RsaA bearing multiple GB1 units would be practical, given the difficulties in the secretion of a single GB1 unit, we pursued this possibility by selecting Muc1, a 20-aa repeated sequence derived from human mucins. For this phase of the study, a slightly smaller GB1 DNA segment was flanked by alternate restriction sites (SpeI and NheI) to produce GB1SN. We chose the insertion site at aa 723 in RsaA for these studies because of numerous other successful insertions at this site (not shown) and a concern that larger insertions at aa 944 must eventually impact the nearby secretion signal.
The two GB1 units and their flanking amino acids can be compared in the context of insertion at aa 723 of RsaA: GB1XS, L721G722DGSARGKTDT(YKL- - -VTE)KPEASRSGSAA723T724; and GB1SN, L721G722DGSRSTS(YKL- - -VTE)ASADGSAA723T724, where boldface letters are RsaA amino acids and their locations, italic letters are amino acids derived from restriction sites, regular letters are the extra authentic flanking protein G amino acids in GB1XS, and the IgG-binding domain GB1 is underlined and in parentheses (see Materials and Methods for full GB1 sequence). The Muc1 spacer sequence was TSPPAHGVTSAPDTRPAPGSTAS.
The change from GB1XS to GB1SN, resulting in relatively modest changes in the amino acid sequence adjacent to the GB1 sequence, led to a notable improvement of secretion levels (Fig. 3). Interestingly, the level of secretion/display was restored to nearly wild-type levels as we assembled the GB1SN-Muc1 tandem pair (abbreviated G-M), G-M-G, and G-M-G-M-G. Only when constructs proceeded to M-G-M-G-M-G-M (a display of approximately 260 aa of heterologous sequence) was there a modest reduction in recombinant RsaA expression levels (Fig. 3).
FIG. 3.
SDS-PAGE (A) and Western immunoblot (B) analyses of normalized low-pH-extracted RsaA and various recombinant RsaA proteins containing the original GB1XS and sequential additions of GB1SN and Muc1B sequences. Immunoblots were treated with HRP-conjugated goat anti-mouse antibody (1:5,000) and developed with 4-chloro-1-naphthol. M, molecular mass standard (in kDa). Lanes: 1, p4ARsaA(723Δ); 2, p4ARsaA(723Δ)/GB1XS; 3, p4ARsaA(723Δ)/GB1SN; 4, p4ARsaA(723Δ)/GM; 5, p4ARsaA(723Δ)/GMG; 6, p4ARsaA(723Δ)/GMGMG; 7, p4ARsaA(723Δ)/MGMGMGM.
Western immunoblot analysis using goat antibody confirmed that these constructs readily bound IgG. Qualitative examination of the blots indicated that an increase in the number of GB1SN repeats correlated with a more intense labeling of the chimeric RsaA bands.
Quantitative analysis of bound rabbit IgG confirmed that increased numbers of GB1 repeats correlated with increased binding capacity (Table 3). Three GB1SN repeats resulted in 0.98 μg/mg (dry weight) of cells, fully twofold greater than that for the Pansorbin comparator. Unexpectedly, the G-M construct showed poor IgG binding, even less than that of any other single GB1 display construct.
We were interested in whether mouse IgG could also now bind to GB1SN-displaying Caulobacter cells. We were unable to identify a commercial source of HRP-coupled mouse IgG for the ELISA and so relied upon two semiquantitative assays: the evaluation of fluorescent mouse monoclonal antibody labeling by microscopy and a spectrofluorometric method we developed. The results of both approaches are detailed in Table 3. The spectrofluorometric assay confirmed minimal mouse IgG binding with a single copy of GB1 (GB1XS or GB1SN). RsaA/GB1SN expression did result in detectable labeling by fluorescence microscopy, possibly a consequence of the high level of expression compared to that of the GB1XS construct. Equally poor binding was observed for the construct bearing two copies of GB1SN separated by the Muc1 spacer (G-M-G), in direct contrast to what was observed using rabbit IgG. Further, in the case of the G-M construct, both assays parallel the anomalously poor IgG binding that we had observed previously using rabbit IgG. By contrast, we noted significant binding with three copies of GB1SN (G-M-G-M-G), especially when they were surrounded with additional copies of the Muc1 spacer peptide (M-G-M-G-M-G-M). In the latter case, we judged the labeling to be maximal, based on our previous experience with the Cy3-coupled anti-c-myc antibody when it was used to detect the c-myc epitope displayed within the S-layer (J. Smit and J. Nomellini, unpublished data).
Recovery of immune complexes from eukaryotic whole-cell lysates.
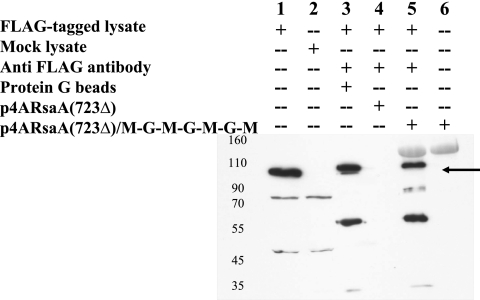
With the demonstration that mouse IgG did bind well to cells displaying the M-G-M-G-M-G-M construct, we repeated the pull-down assay, comparing this optimal construct to protein G-conjugated agarose beads. The Caulobacter cells were now readily able to precipitate anti-FLAG monoclonal/FLAG-tagged protein complexes in a manner essentially comparable to that of the protein G-Sepharose beads (Fig. 4). To control for the specificity of the RsaA/GB1-IgG-antigen interaction, negative control reactions were run using mock-transfected lysates instead of FLAG-tagged protein-expressing lysates, with C. crescentus cells expressing full-length RsaA lacking the M-G-M-G-M-G-M insert. Note that despite a blocking step with goat serum, in this Western blot some reactivity to the RsaA/M-G-M-G-M-G-M protein (Fig. 4, lanes 5 and 6) as well as the IgG heavy chains (lanes 3 and 5) persists. Together, these results demonstrate the potential of using whole recombinant Caulobacter cells as immunoreactive particles.
FIG. 4.
Immunoprecipitation of FLAG-tagged protein from eukaryotic cell lysates by using anti-FLAG antibody and protein G beads or Caulobacter cells expressing RsaA(723Δ) or recombinant RsaA(723Δ)/MGMGMGM. Molecular mass standards are indicated in kDa. The arrow indicates the position of the FLAG-tagged RapGapII protein. The reactive proteins of smaller size in lanes 3 and 5 are Ig heavy chains, and the bands above the arrow reflect antibody binding to RsaA(723Δ)/MGMGMGM.
DISCUSSION
The Caulobacter S-layer secretion system is established as a viable alternative to existing protein production systems (5, 7, 35), but it remains to be determined whether composite RsaA proteins can retain the functionality of the foreign protein segment, either when produced as secreted fusion proteins or when displayed within the intact assembled S-layer. We demonstrated success when assessing the ability of RsaA fusion proteins to act as particulate enzymes (12), but the potential for foreign peptide display within the S-layer protein to produce whole Caulobacter cells with nonnative biological activities on their surface also needs to be explored. We previously demonstrated that full-length RsaA can be modified by the insertion of in-frame foreign peptides without disrupting the normal formation of the S-layer, resulting in the stable expression of the foreign peptide on the surface of whole cells (5). In this report, we examined the display of a foreign peptide with a distinct biological function, streptococcal protein G. As such, we have been able to assess the ability of Caulobacter cells to be used directly as functional immunoreactive particles.
Muc1 is a 20-aa sequence derived from human mucins, where it is tandemly repeated many times. Because of aberrant overexpression of mucins in many types of solid tumors, Muc1 is being evaluated by several research groups (including our own) as a potential anticancer vaccine component. We have had considerable success in displaying tandem repeats of Muc1 within the Caulobacter S-layer protein; we have been able to secrete chimeric RsaA/Muc1 variants with up to 32 tandem copies of Muc1 (with some spacer amino acids; a total of 679 aa of heterologous sequence) (Bhatnagar, Das, Nomellini, Smit, and Suresh, unpublished). This unexpected tolerance of the secretion apparatus for this sequence may be due to the fact that there is little predicted secondary structure and that the generally hydrophilic amino acid composition is similar to that of RsaA (e.g., 25% of the residues are serine or threonine). Thus, Muc1 seemed to be a reasonable candidate to test whether the use of a spacer could improve GB1 secretion. Even so, we were surprised at the large improvement in secretion levels, permitting not one but several tandem copies of GB1 to be efficiently secreted in the context of RsaA.
Moreover, it should be noted that the size limit of GB1 and Muc1 combinations that can be secreted is still greater than what is reported here. We subsequently produced M-M-G-M-G-M-G-M-M and G-M-G-M-G-M-M-M-M (>300 aa of heterologous sequence) with no significant drop in secretion efficiency (data not shown). No further improvement of mouse IgG binding was observed using the immunofluorescence assay (data not shown). Although it may be that the assay detection is at saturation, it seems equally plausible that available space on the Caulobacter cell surface for binding IgG has been saturated and increased exposure or flexibility of the GB1 units cannot overcome that limitation (also see calculations below).
The improvement in terms of GB1 display was more than just high levels of recombinant RsaA secretion. There was also the indication that there was a major improvement in IgG binding by the recombinant protein, especially with regard to mouse IgG. This might be due simply to the display of multiple GB1 copies and/or to the improved ability of GB1 to fold properly to engage the IgG Fc region. We are presently devising methods to quantitatively measure the strength of binding. The role of Muc1 as a spacer may be more complex than simply increasing folding flexibility; we noted that the GB1-Muc1 combination consistently bound less IgG than all others, including the single GB1 constructs and even the original underexpressed RsaA/GB1XS constructs.
We do note that in comparing a single copy of GB1 with three tandemly displayed copies (per RsaA monomer), while a significant increase in the amount of IgG bound was observed, it is not three times the level of a single copy. This might be explained by simple steric considerations, i.e., limiting the access of more IgG molecules to the surface of Caulobacter cells. It is difficult to calculate a precise surface density of the bound IgG, but even as an approximation, it is considerably less than the density of RsaA monomers, estimated to be 20,000 copies/μm2 of surface area. It is likely that this high surface density accounts for the fact that the GB1-displaying Caulobacter cells were calculated to bind more than twice the amount of IgG than is possible for Pansorbin cells.
Exploring this area further, a fully expressed S-layer accounts for approximately 40,000 monomers per Caulobacter cell (30). We empirically determined that there are 1.3 × 109 Caulobacter cells (dry weight) per mg. Typical IgG molecules have a molecular mass of 150 to 160 kDa (15). If we assume that without regard to the number or configuration of displayed GB1 units a maximum binding capacity will not exceed one IgG molecule per RsaA/GB1 monomer, then 1 mg of cells, ignoring all steric hindrance considerations, can bind a theoretical maximum of about 14 μg of IgG (if S-layer coverage is at wild-type levels). Our experiments indicate that, with the best constructs, it was possible to bind a maximum of about 2 μg rabbit IgG/mg Caulobacter cells (or about 5,700 IgG molecules/cell). Though we have found it difficult to determine an accurate effective dimension for an IgG molecule coupled to a horseradish peroxidase label, given the likelihood of steric hindrance on the Caulobacter surface, we believe that this maximum value is quite reasonable.
Commercially available fixed protein A-expressing S. aureus cells have a reported binding capacity of 19 μg human IgG/mg cells (Pansorbin product insert). However, in our assay, we found that Pansorbin cells bind 1.24 μg of rabbit IgG/mg (dry weight) of cells. This discrepancy may be accounted for partly by the fact that we are using rabbit IgG rather than human IgG or that we are using an HRP-conjugated antibody, as the HRP moiety could cause steric hindrance and reduced binding. There may yet be some other proportionate error in our assay conditions or calculations, but it does seem clear that the best Caulobacter GB1/RsaA constructs can markedly exceed the per-cell capacity of Pansorbin for IgG.
A major application of intact Caulobacter cells displaying IgG-binding capacity is as a substitute for protein G-Sepharose beads in standard pull-down assays, and our initial assessment here suggests that they can indeed be considered as a lower-cost alternative. The method reported here is modified from typical procedures in that EDTA is not used, and the related reagent EGTA is used only at later stages, since both chemicals disrupt the integrity of the S-layer, which requires calcium to crystallize (22). The method we adopted eliminated the need to solubilize whole bacteria in SDS-PAGE sample buffer and so minimized the potential difficulties of the comigration of numerous irrelevant bacterial proteins and increased sample viscosity due to released bacterial DNA and RNA.
The RsaA/GB1-displaying Caulobacter cells have the potential for substituting for protein G-Sepharose beads in other types of immunochemistry experiments as well, especially applications in which Pansorbin cells are now used or could be used if there was a need to use mouse antiserum or monoclonal antibodies.
As a substitute for protein G-Sepharose beads, there is the expectation of a much lower cost of production. Though we have not yet done a full-scale cost evaluation of fermentation, quality assessment, and packaging, a low production cost seems reasonable. For example, the cost of the defined culture medium for Caulobacter (using glucose, glutamate, and ammonium chloride as carbon and nitrogen sources) is about five cents per liter using research-grade chemicals. Assuming a typical package size is 1 gram (wet weight) of cells (as is typical for Pansorbin), this translates to two to three cents per package. The only significant downstream processing steps are cell concentration and formalin stabilization. On this latter aspect, we are presently evaluating the long-term stability of formalin-stabilized Caulobacter cells. Preliminary results suggest that, unexpectedly, they bind still greater levels of IgG than live, unfixed cells.
Two additional advances in terms of Caulobacter peptide display technology are reported in this study. First, we previously observed significant proteolytic degradation of the recombinant S-layer proteins (5, 34, 35). Here, we used a new C. crescentus strain for recombinant protein expression, JS4022, which is enabled for shuttle vector expression (see below) and in which the protease responsible for S-layer-specific degradation, Sap (34) has been deleted. As expected, no evidence of proteolytic cleavage was observed for any of the RsaA/GB1 constructs. Second, we developed a new generation of Caulobacter expression vectors based on the pUC8 backbone that can be shuttled back and forth between E. coli and C. crescentus strains expressing the repBAC genes of RSF1010 (36). The vector has advantages over the previous generation of shuttle vector, including a smaller size (approximately 4 kb) and fewer extraneous restriction sites. In addition, we have modified the spacing between the rsaA promoter region and the predicted Shine-Dalgarno site. Without this modification, we were unable to stably introduce the vector into Caulobacter (data not shown), and following the change, the vector has proven to be quite stable; indeed, we have found it difficult to screen for Caulobacter cells “cured” of this vector. In a previous low-copy-number vector, this promoter-to-Shine-Dalgarno spacing change had the effect of reducing the levels of RsaA produced (8) and we reason that this change is somehow important for the stable maintenance of the present high-copy-number vector.
In terms of developing an IgG-binding reagent, the C. crescentus construct offers the distinct advantage of displaying protein G, thus offering a much broader spectrum of both host species and IgG subclasses (e.g., goat, sheep, and all murine isotypes) than S. aureus protein A. However, it is currently available in an expensive format, i.e., conjugated to agarose beads. Protein G-expressing Caulobacter cells could therefore represent an equally useful but cost-effective alternative, similar to commercially available S. aureus cells. Furthermore, Caulobacter cells produce minimal amounts of lipopolysaccharide endotoxin activity (23), which otherwise could interfere in some cell and immune-based applications.
This is the first published report demonstrating the use of whole Caulobacter cells displaying a fully functional foreign peptide in a research application. In another S-layer model system, B. sphaericus S-layer protein SbpA was first fused to a specific antigen and used as the basis of a dipstick-type immunoassay for the rapid determination of specific IgE in serum (11), and it was used to display tandem IgG-binding sequences from S. aureus protein A to purify autoantibodies from blood (37). These applications indicate that S-layer proteins offer a novel and effective way to immobilize foreign proteins in the context of a particular biological assay. Although the Bacillus system is complicated by the fact that, unlike with C. crescentus, recombinant proteins have to be expressed in E. coli, the distinct advantage is that recombinant SbpA can be recrystallized in an oriented fashion on a variety of supports precoated with a secondary cell wall polymer (16, 24, 37). Taken together, the successes obtained with both of these model systems suggest that S-layer fusion proteins can be used in many applications where it is necessary or advantageous to immobilize and/or isolate IgG.
Acknowledgments
We thank Ying Zhao, Andrea Pusic, Krisztina Balázs, and Corin Forrester for technical assistance during this study and Cameron Forde for assistance with the spectrofluorometric assay. We thank Michael Gold (University of British Columbia) for providing cell lysates for the pull-down assay.
This research was supported by grants from the Natural Sciences and Engineering Research Council of Canada and the Canadian Bacterial Diseases Network to J.S.
Footnotes
Published ahead of print on 23 March 2007.
REFERENCES
- 1.Akerström, B., E. Nielsen, and L. Bjorck. 1987. Definition of IgG- and albumin-binding regions of streptococcal protein G. J. Biol. Chem. 262:13388-13391. [PubMed] [Google Scholar]
- 2.Awram, P., and J. Smit. 1998. The Caulobacter crescentus paracrystalline S-layer protein is secreted by an ABC transporter (type I) secretion apparatus. J. Bacteriol. 180:3062-3069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bhatnagar, P. K., A. Awasthi, J. F. Nomellini, J. Smit, and M. R. Suresh. 2006. Anti-tumor effects of the bacterium Caulobacter crescentus in murine tumor models. Cancer Biol. Ther. 5:485-491. [DOI] [PubMed] [Google Scholar]
- 4.Bingle, W. H., K. D. Le, and J. Smit. 1996. The extreme N-terminus of the Caulobacter crescentus surface-layer protein directs export of passenger proteins from the cytoplasm but is not required for secretion of the native protein. Can. J. Microbiol. 42:672-684. [DOI] [PubMed] [Google Scholar]
- 5.Bingle, W. H., J. F. Nomellini, and J. Smit. 1997. Cell-surface display of a Pseudomonas aeruginosa strain K pilin peptide within the paracrystalline S-layer of Caulobacter crescentus. Mol. Microbiol. 26:277-288. [DOI] [PubMed] [Google Scholar]
- 6.Bingle, W. H., J. F. Nomellini, and J. Smit. 1997. Linker mutagenesis of the Caulobacter crescentus S-layer protein: toward a definition of an N-terminal anchoring region and a C-terminal secretion signal and the potential for heterologous protein secretion. J. Bacteriol. 179:601-611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bingle, W. H., J. F. Nomellini, and J. Smit. 2000. Secretion of the Caulobacter crescentus S-layer protein: further localization of the C-terminal secretion signal and its use for secretion of recombinant proteins. J. Bacteriol. 182:3298-3301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bingle, W. H., and J. Smit. 1990. High-level expression vectors for Caulobacter crescentus incorporating the transcription/translation initiation regions of the paracrystalline surface-layer-protein gene. Plasmid 24:143-148. [DOI] [PubMed] [Google Scholar]
- 9.Björck, L., and G. Kronvall. 1984. Purification and some properties of streptococcal protein G, a novel IgG-binding reagent. J. Immunol. 133:969-974. [PubMed] [Google Scholar]
- 10.Bonifacino, J. S., E. C. Dell'Angelica, and T. A. Springer. 2001. Immunoprecipitation, p. 8.3.1-8.3.28. In J. E. Coligan, A. M. Kruisbeek, D. H. Margulies, E. M. Stevach, and W. Strober (ed.), Current protocols in immunology. John Wiley and Sons, Inc., New York, NY. [DOI] [PubMed]
- 11.Breitwieser, A., C. Mader, I. Schocher, K. Hoffmann-Sommergruber, W. Aberer, O. Scheiner, U. B. Sleytr, and M. Sara. 1998. A novel dipstick developed for rapid Bet v 1-specific IgE detection: recombinant allergen immobilized via a monoclonal antibody to crystalline bacterial cell-surface layers. Allergy 53:786-793. [DOI] [PubMed] [Google Scholar]
- 12.Duncan, G., C. A. Tarling, W. H. Bingle, J. F. Nomellini, M. Yamage, I. R. Dorocicz, S. G. Withers, and J. Smit. 2005. Evaluation of a new system for developing particulate enzymes based on the surface (S)-layer protein (RsaA) of Caulobacter crescentus: fusion with the beta-1,4-glycanase (Cex) from the cellulolytic bacterium Cellulomonas fimi yields a robust, catalytically active product. Appl. Biochem. Biotechnol. 127:95-110. [DOI] [PubMed] [Google Scholar]
- 13.Georgiou, G., C. Stathopoulos, P. S. Daugherty, A. R. Nayak, B. L. Iverson, and R. Curtiss III. 1997. Display of heterologous proteins on the surface of microorganisms: from the screening of combinatorial libraries to live recombinant vaccines. Nat. Biotechnol. 15:29-34. [DOI] [PubMed] [Google Scholar]
- 14.Gilchrist, A., and J. Smit. 1991. Transformation of freshwater and marine caulobacters by electroporation. J. Bacteriol. 173:921-925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Harlow, E., and D. Lane. 1999. Antibodies: a laboratory manual. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- 16.Ilk, N., C. Völlenkle, E. M. Egelseer, A. Breitwieser, U. B. Sleytr, and M. Sára. 2002. Molecular characterization of the S-layer gene, sbpA, of Bacillus sphaericus CCM 2177 and production of a functional S-layer fusion protein with the ability to recrystallize in a defined orientation while presenting the fused allergen. Appl. Environ. Microbiol. 68:3251-3260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Krebs, D. L., Y. Yang, M. Dang, J. Haussmann, and M. R. Gold. 1999. Rapid and efficient retrovirus-mediated gene transfer into B cell lines. Methods Cell Sci. 21:57-68. [DOI] [PubMed] [Google Scholar]
- 18.Marraffini, L. A., A. C. Dedent, and O. Schneewind. 2006. Sortases and the art of anchoring proteins to the envelopes of gram-positive bacteria. Microbiol. Mol. Biol. Rev. 70:192-221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Merker, R. I., and J. Smit. 1988. Characterization of the adhesive holdfast of marine and freshwater caulobacters. Appl. Environ. Microbiol. 54:2078-2085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Messing, J., and J. Vieira. 1982. A new pair of M13 vectors for selecting either DNA strand of double-digest restriction fragments. Gene 19:269-276. [DOI] [PubMed] [Google Scholar]
- 21.Morales, V. M., A. Backman, and M. Bagdasarian. 1991. A series of wide-host-range low-copy-number vectors that allow direct screening for recombinants. Gene 97:39-47. [DOI] [PubMed] [Google Scholar]
- 22.Nomellini, J. F., S. Kupcu, U. B. Sleytr, and J. Smit. 1997. Factors controlling in vitro recrystallization of the Caulobacter crescentus paracrystalline S-layer. J. Bacteriol. 179:6349-6354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Nomellini, J. F., M. C. Toporowski, and J. Smit. 2004. Secretion or presentation of recombinant proteins and peptides mediated by the S-layer of Caulobacter crescentus, p. 477-524. In F. Baneyx (ed.), Expression technologies: current status and future trends. Horizon Scientific Press, Norfolk, United Kingdom.
- 24.Pleschberger, M., A. Neubauer, E. M. Egelseer, S. Weigert, B. Lindner, U. B. Sleytr, S. Muyldermans, and M. Sara. 2003. Generation of a functional monomolecular protein lattice consisting of an S-layer fusion protein comprising the variable domain of a camel heavy chain antibody. Bioconjug. Chem. 14:440-448. [DOI] [PubMed] [Google Scholar]
- 25.Poindexter, J. S. 1981. The caulobacters: ubiquitous unusual bacteria. Microbiol. Rev. 45:123-179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- 27.Shapiro, L. 1976. Differentiation in the Caulobacter cell cycle. Annu. Rev. Microbiol. 30:377-407. [DOI] [PubMed] [Google Scholar]
- 28.Smit, J., and N. Agabian. 1982. Cell surface patterning and morphogenesis: biogenesis of a periodic surface array during Caulobacter development. J. Cell Biol. 95:41-49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Smit, J., and N. Agabian. 1984. Cloning of the major protein of the Caulobacter crescentus periodic surface layer: detection and characterization of the cloned peptide by protein expression assays. J. Bacteriol. 160:1137-1145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Smit, J., H. Engelhardt, S. Volker, S. H. Smith, and W. Baumeister. 1992. The S-layer of Caulobacter crescentus: three-dimensional image reconstruction and structure analysis by electron microscopy. J. Bacteriol. 174:6527-6538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Smit, J., D. A. Grano, R. M. Glaeser, and N. Agabian. 1981. Periodic surface array in Caulobacter crescentus: fine structure and chemical analysis. J. Bacteriol. 146:1135-1150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Taylor, J. A., J. A. O'Brien, V. J. Lord, J. C. Meyer, and A. R. Bellamy. 1993. The RER-localized rotavirus intracellular receptor: a truncated purified soluble form is multivalent and binds virus particles. Virology 194:807-814. [DOI] [PubMed] [Google Scholar]
- 33.Toporowski, M. C., J. F. Nomellini, P. Awram, and J. Smit. 2004. Two outer membrane proteins are required for maximal type I secretion of the Caulobacter crescentus S-layer protein. J. Bacteriol. 186:8000-8009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Umelo-Njaka, E., W. H. Bingle, F. Borchani, K. D. Le, P. Awram, T. Blake, J. F. Nomellini, and J. Smit. 2002. Caulobacter crescentus synthesizes an S-layer-editing metalloprotease possessing a domain sharing sequence similarity with its paracrystalline S-layer protein. J. Bacteriol. 184:2709-2718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Umelo-Njaka, E., J. F. Nomellini, W. H. Bingle, L. G. Glasier, R. T. Irvin, and J. Smit. 2001. Expression and testing of Pseudomonas aeruginosa vaccine candidate proteins prepared with the Caulobacter crescentus S-layer protein expression system. Vaccine 19:1406-1415. [DOI] [PubMed] [Google Scholar]
- 36.Umelo-Njaka, E., J. F. Nomellini, H. Yim, and J. Smit. 2001. Development of small high-copy-number plasmid vectors for gene expression in Caulobacter crescentus. Plasmid 46:37-46. [DOI] [PubMed] [Google Scholar]
- 37.Völlenkle, C., S. Weigert, N. Ilk, E. Egelseer, V. Weber, F. Loth, D. Falkenhagen, U. B. Sleytr, and M. Sara. 2004. Construction of a functional S-layer fusion protein comprising an immunoglobulin G-binding domain for development of specific adsorbents for extracorporeal blood purification. Appl. Environ. Microbiol. 70:1514-1521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Walker, S. G., S. H. Smith, and J. Smit. 1992. Isolation and comparison of the paracrystalline surface layer proteins of freshwater caulobacters. J. Bacteriol. 174:1783-1792. [DOI] [PMC free article] [PubMed] [Google Scholar]