Abstract
We have previously shown that the Nonomuraea flexuosa Xyn11A polypeptides devoid of the carbohydrate binding module (CBM) have better thermostability than the full-length xylanase and are effective in bleaching of pulp. To produce an enzyme preparation useful for industrial applications requiring high temperature, the region encoding the CBM was deleted from the N. flexuosa xyn11A gene and the truncated gene was expressed in Trichoderma reesei. The xylanase sequence was fused to the T. reesei mannanase I (Man5A) signal sequence or 3′ to a T. reesei carrier polypeptide, either the Man5A core/hinge or the cellulose binding domain (CBD) of cellobiohydrolase II (Cel6A, CBHII). The gene and fusion genes were expressed using the cellobiohydrolase 1 (cel7A, cbh1) promoter. Single-copy isogenic transformants in which the expression cassette replaced the cel7A gene were cultivated and analyzed. The transformants expressing the truncated N. flexuosa xyn11A produced clearly increased amounts of both the xylanase/fusion mRNA and xylanase activity compared to the corresponding strains expressing the full-length N. flexuosa xyn11A. The transformant expressing the cel6A CBD-truncated N. flexuosa xyn11A produced about 1.9 g liter−1 of the xylanase in laboratory-scale fermentations. The xylanase constituted about 25% of the secreted proteins. The production of the truncated xylanase did not induce the unfolded protein response (UPR) pathway. However, the UPR was induced when the full-length N. flexuosa xyn11A with an exact fusion to the cel7A terminator was expressed. We suggest that the T. reesei folding/secretion machinery is not able to cope properly with the bacterial CBM when the mRNA of the full-length N. flexuosa xyn11A is efficiently translated.
The filamentous fungus Trichoderma reesei is used as an industrial production organism for both homologous and heterologous proteins (19, 26). In order to produce thermostable xylanases in T. reesei, we previously isolated two xylanase genes, N. flexuosa xyn11A and N. flexuosa xyn10A, from the actinomycete Nonomuraea flexuosa DSM43186 (previously Microtetraspora flexuosa or Actinomadura flexuosa [45]) (16). The corresponding xylanases were shown to be thermostable enzymes, thus being suitable candidates for several industrial applications, e.g., enzymatic bleaching of kraft pulp (reviewed in reference 39). The N. flexuosa xyn11A gene was expressed in the filamentous fungus Trichoderma reesei by using the strong T. reesei cellobiohydrolase 1 (cbh1, cel7A) promoter and a fusion of the gene 3′ to a selection of T. reesei carrier polypeptides (24). About 0.8 g liter−1 of the recombinant N. flexuosa Xyn11A protein was produced by single-copy strains in laboratory-scale fermentations. This amount of recombinant xylanase was higher than the previously published amounts of bacterial enzymes from filamentous fungi, which do not exceed about 0.1 g liter−1 even after optimization of the codon usage for fungal expression (12, 22, 43). Even though a reasonable amount of the N. flexuosa Xyn11A protein was obtained from the T. reesei transformants, a rate-limiting factor for the production of this heterologous xylanase was observed at the transcriptional stage. The amount of the fusion mRNA detected from the mycelia of the transformants was only 10 to 12% of the amount of the cel7A mRNA in the host (24).
The N. flexuosa Xyn11A xylanase consists of a catalytic module, a GNP-rich linker (“hinge”), and a C-terminal carbohydrate binding module (CBM) (“tail”). Based on sequence homology comparisons, the CBM is a member of subfamily 2b, consisting of true xylan-binding domains (16). When the recombinant N. flexuosa Xyn11A was purified from the T. reesei culture supernatant, 23.8-kDa and 22-kDa xylanase forms devoid of the tail domain and the tail and hinge domains, respectively, were detected (16). These shorter xylanase polypeptides were purified and characterized, in addition to the full-length 33.4-kDa xylanase. All these three recombinant xylanase polypeptides were shown to be similar in terms of specific activity, pH, and temperature dependence. They were also effective in the pretreatment of kraft pulp, confirming that the CBM was not obligatory in the bleaching application. Interestingly, the thermostabilities of the recombinant 23.8-kDa and 22.0-kDa forms were both better than that of the full-length recombinant N. flexuosa Xyn11A. Thus, the truncated xylanase polypeptides were attractive candidates as enzymes for industrial applications requiring high temperatures, e.g., bleaching of kraft pulp.
In the present study, the region encoding the tail (CBM) was deleted from the N. flexuosa xyn11A gene and the truncated gene was expressed in T. reesei. It was fused to the T. reesei Man5A signal sequence and 3′ to sequences encoding carrier polypeptides, the T. reesei Man5A core/hinge, and the Cel6A cellulose binding domain (CBD). The genetic constructions were expressed in T. reesei from the strong cel7A promoter. The effects of the truncation on the expression and production of the recombinant xylanase were studied using isogenic single-copy strains in which the expression cassettes replaced the host's native cel7A gene. The results obtained were compared to those obtained with the transformants carrying the corresponding expression cassettes with the full-length N. flexuosa xyn11A. Also, the effect of the expression of the bacterial xylanase on the induction of the unfolded protein response (UPR) was studied.
MATERIALS AND METHODS
Microbial strains, plasmids, growth media, and growth conditions.
Plasmids were propagated in Escherichia coli XL1-Blue and XL10-Gold (Stratagene, La Jolla, CA). The vector backbones used in constructing the plasmids were pUC18 (EMBL database accession no. L09136), pUC19 (L09137) and pBluescript II KS+ (Stratagene). The E. coli cultivations were performed overnight at 37°C in Luria-Bertani medium (35) into which ampicillin had been added (50 to 100 μg/ml).
The T. reesei strain ALKO3620 was used as a parent in the transformations. T. reesei ALKO3620 is a low-protease mutant in which the egl2 gene (cel5A, originally named egl3 in reference 32) has been replaced with the phleomycin resistance-encoding marker gene ble from Streptoalloteichus hindustanus (8), as described in reference 24.
T. reesei strains were sporulated on Difco PD agar slants (potato dextrose broth; BD Diagnostics, Sparks, MD). The transformants were selected on Trichoderma minimal medium containing acetamide as a nitrogen source (27). The fungal mycelia for DNA isolations were obtained after the strains were grown for 2 days on Trichoderma minimal medium containing 2% proteose peptone (BD Diagnostics). Culture supernatants from 50-ml shake flask cultivations (grown for 7 days at 30°C, 250 rpm) were used in screening the transformants. A complex lactose-based cellulase-inducing medium (13) was used for enzyme production in the shake flasks. The medium contained (per liter) 40 g of whey extract, 15 g of complex nitrogen source derived from grain, 15 g KH2PO4, and 5 g (NH4)2SO4 (pH 5.5). For the fermentations, a particle-free extract was prepared from the above medium, to enable the determination of the mycelium dry mass from the cultivations. The extract was prepared by first heating the medium with the complex nitrogen source (30 g per liter) at 121°C for 15 min, letting it settle at 4°C overnight, and then decanting it. After that, lactose (30 g per liter) and the salts were added. The laboratory-scale fermentor cultivations were performed for 4 to 5 days in 2-liter Braun Biostat M fermentors (B. Braun, Melsungen, Germany). The fermentor cultivations were started by inoculating them with the fungal strains, which had been pregrown for 3 days (28°C, 230 rpm) in 200 ml of the above culture medium.
DNA techniques.
Standard DNA methods (35, 36) were used in constructing plasmids and performing Southern and Northern blotting. Each enzyme and kit was used according to the instructions from the supplier. The enzymes for DNA modifications were purchased from Fermentas GmbH (St. Leon-Rot, Germany), Finnzymes (Espoo, Finland), New England Biolabs (Beverly, MA), and Roche Diagnostics GmbH (Mannheim, Germany). QIAGEN columns (QIAGEN GmbH, Hilden, Germany) were used in the plasmid isolations. The oligonucleotides were from Sigma Genosys (Haverhill, United Kingdom). Sequencing reactions were performed and analyzed according to the instructions from Applied Biosystems and using either an ABI 373A or an ABI PRISM 310 genetic analyzer (Applied Biosystems, Foster City, CA). PCRs were performed with DyNAzyme II or DyNAzyme EXT (Finnzymes) using a PTC-100 programmable thermal controller (MJ Research Inc., Watertown, MA) or an Eppendorf Mastercycler (Eppendorf UK Ltd., Cambridge, United Kingdom). DNA fragments for subcloning and transformations were isolated by using the QIAEX II gel extraction kit (QIAGEN GmbH).
The genomic DNAs were isolated as described in reference 30. Digoxigenin-labeled (Roche Diagnostics GmbH) expression cassettes were used as probes in the Southern blot hybridizations.
Construction of the expression cassettes used in the study.
The construction of the expression cassettes pALK1118, pALK1022, and pALK1285 has been described in reference 24. These cassettes encode the full-length mature N. flexuosa Xyn11A from the N-terminal D44 to the C-terminal N344 (16) (EMBL accession no. AJ508952). The region encoding the CBM (S264 to N344) was deleted from the N. flexuosa xyn11A gene in the expression cassettes pALK1276, pALK1131, and pALK1502 by using PCR. The overall structure of all the expression cassettes was as described in reference 24. Expression of the xylanase gene or the fusion gene was from the T. reesei cel7A promoter and termination of the transcription was ensured by the cel7A terminator sequence. The above xylanase genes were fused 3′ to the Man5A signal sequence (amino acids M1 to A19; pALK1118 and pALK1276), the Man5A core/hinge carrier polypeptide (M1 to G410; pALK1022, pALK1309, pALK1692, pALK1131), or the Cel6A CBD carrier polypeptide (regions “A” and “B”; M1 to S86; pALK1285, pALK1502). For the man5A and cel6A sequences, see references 38 and 40, respectively. A synthetic linker, encoding a Kex2 protease target site (RDKR) (5), was inserted between the sequences encoding the carrier polypeptide and the xylanase. The sequence encoding the Kex2 site was not included between the signal and xylanase sequences in pALK1118 and pALK1276. The Aspergillus amdS (acetamidase) gene was used as a marker in all the genetic constructions. The amdS gene was isolated from p3SR2 (15) as a 3.1-kb SpeI-XbaI fragment that also includes the amdS promoter. The amdS sequence was ligated 3′ to the cel7A terminator in the expression cassettes. In addition, the cel7A 3′-flanking sequence (14) was inserted 3′ to the amdS. It was used, together with the cel7A promoter, to target the expression cassette to the cel7A locus.
The fusion of the xylanase or truncated xylanase genes to the cel7A terminator sequences differed in the expression cassettes as follows. The truncated N. flexuosa xyn11A gene was, in each case, exactly fused from its 3′ end to the cel7A stop codon and terminator (0.6 kb). The only full-length N. flexuosa xyn11A gene that was exactly fused to the cel7A stop codon and terminator was in the expression cassette pALK1692. In this respect, pALK1692 was identical to the genetic constructions with the truncated xylanase gene, whereas the other expression cassettes with the full-length gene contained about 250 bp of the N. flexuosa xyn11A 3′ untranslated region (UTR) sequence (to the MluI site) after the stop codon, followed by a cel7A terminator (a 0.7-kb AvaII fragment that includes 0.6 kb of the cel7A terminator preceded by a 113-bp sequence from the end of the cel7A coding region [14]). The PCR primers were designed to allow exact fusions of the xylanase sequences to the terminator sequence, to the signal sequence, or to the sequence encoding the linker between the fusion polypeptides.
The codon usage of N. flexuosa xyn11A was optimized in the expression cassettes pALK1309, pALK1131, and pALK1502, based on the most-preferred codons in the major cellulase and xylanase genes of T. reesei. In total, nine codons were changed, as follows: Gly53 GGG was changed to GGC, Ala66 GCG to GCC, Gly68 GGG to GGC, Arg85 CGG to CGC, Gly88 GGG to GGC, Gly100 GGA to GGC, Arg101 CGG to CGC, Arg102 CGG to CGC, and Val104 GTG to GTC (the numbering of the amino acids is according to reference 16).
Transformation of T. reesei and analysis of the transformants.
T. reesei protoplasts were transformed with linear expression cassettes cleaved from the vector backbones by EcoRI digestion. All the expression cassettes were transformed into T. reesei strain ALKO3620 (24). Also, the pALK1118 cassette, which had previously been transformed into ALKO4468 (24), was transformed into ALKO3620. Transformations were performed as described in reference 27 with the modifications described in reference 14. The transformants were purified on selection plates through single conidia prior to being sporulated on PD. The transformants with a Cel7A-negative phenotype were screened by performing dot blotting from the culture supernatants. The monoclonal antibody CI-258 (1) was used in the detection of Cel7A. The replacement of the cel7A gene by the expression cassette was confirmed from the Cel7A-negative strains by Southern blot assay, as described in reference 24. Transformants in which the cel7A gene was replaced with one copy of the expression cassette were chosen for fermentor cultivations.
Protein and enzyme assays and analysis of the mycelium dry mass.
Samples of the culture supernatants were run on 15% Criterion bis-Tris sodium dodecyl sulfate (SDS)-polyacrylamide slab gels (Bio-Rad, Hercules, CA). The proteins were stained with Coomassie G-250 (SimplyBlue SafeStain; Invitrogen BV, Leek, The Netherlands). A polyclonal rabbit antibody raised against the native purified Xyn11A (16) and a Protoblot AP system (Promega, Madison, WI) were used for the detection of the full-length and truncated N. flexuosa Xyn11A proteins in the Western blots. Purified recombinant N. flexuosa Xyn11A proteins of 33.4 kDa and 23.8 kDa (16) were used as controls in SDS-polyacrylamide gel electrophoresis (PAGE) gels and Western blot filters. The BenchMark and MagicMark protein standards (Invitrogen) were used as molecular mass markers for the SDS-polyacrylamide gels and Western blots, respectively. The amounts of the proteins secreted to the culture supernatants were assayed after trichloroacetic acid precipitation by using a detergent-compatible protein assay (Bio-Rad, Hercules, CA), based on the method described in reference 17. Bovine serum albumin was used as the standard protein. Xylanase activity was assayed using birch xylan (Roth no. 7500; Roth, Karlsruhe, Germany) as a substrate (2). The reactions were performed in 50 mM McIlvaine's citrate-phosphate buffer at pH 7, 70°C, for 5 min. Mannanase activity was assayed at 50°C, pH 5.3, using locust bean gum (Sigma G-0753; Oslo, Norway) as a substrate, as described in reference 38. The dry masses of mycelia were analyzed from 10-ml samples. The samples were filtered through predried and preweighed 3.0-μm and 0.45-μm filters (Millipore Oy, Espoo, Finland). The mycelia on the filters were washed with 20 ml of 0.7% NaCl, dried overnight at 80°C, and weighed.
Isolation of RNA and analysis of the transcripts.
Total RNA was isolated from samples taken 54 to 55 h (exponential phase) and 70 to 78 h (mycelium dry mass maximum) after the inoculation of the fermentor cultivations, with the exception of the transformants carrying the expression cassettes pALK1692 and pALK1285. These strains reached the stage of the mycelium dry mass maximum clearly later than the other strains, and RNA was isolated from them after 77 and 94 h of cultivation. An RNeasy plant mini kit (QIAGEN GmbH) was used in the isolations. The Northern blot gels (1× MOPS [morpholinepropanesulfonic acid], 6% formaldehyde, 1.2% agarose) were run according to standard methods (35). A total of 2.0 μg of RNA was loaded per lane. An 81-mer oligonucleotide complementary to the transcribed region of the cel7A promoter (from nucleotides +3 to −78) (10) was used to detect the mRNAs transcribed from the cel7A promoter. In addition, the N. flexuosa xyn11A transcripts were detected using a 518-bp SmaI fragment from the N. flexuosa xyn11A gene (nucleotides 504 to 1021) (16). The induction of the UPR was analyzed by using two probes, the hac1 (EcoRI-XhoI fragment from pMS119) (34) and the pdi1 (EcoRI-XhoI fragment from pMS67) (33). The 1.9-kb KpnI fragment containing the T. reesei actin gene (20, 44) was used to normalize the signals obtained. The oligonucleotide was labeled with [γ-32P]ATP by using a 5′-end-labeling kit (Amersham Biosciences, Uppsala, Sweden). The DNA fragments were labeled using [α-32P]dCTP and a Rediprime II DNA labeling system (Amersham Biosciences). The hybridizations with all the probes were performed at 42°C in 50% formamide, 6× SSPE (1× SSPE is 0.15 M NaCl, 10 mM NaH2PO4, and 1 mM EDTA [pH 7.4]), 5× Denhardt's, 0.5% SDS, 100 μg/ml salmon sperm DNA. The filters were washed using 2× SSC (1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate [pH 7.0])-0.5% SDS for 5 min at room temperature and 2× SSC-0.1% SDS for 15 min at room temperature followed by either one 15-min wash at 42°C and two 15-min washes at 52°C, for the filters probed with the oligonucleotide, or two washes at 42°C, for the filters probed with labeled DNA fragments. The filters probed with the labeled fragment were further washed twice for 15 min at 52°C with 0.1× SSC-0.1% SDS. The signals in the Northern blot filters were analyzed by using a Typhoon 8600 variable mode imager and ImageQuant 5.2 software (Molecular Dynamics, Sunnyvale, CA).
RESULTS
Production of truncated N. flexuosa Xyn11A in Trichoderma reesei.
The expression cassettes for producing the truncated N. flexuosa Xyn11A and the full-length N. flexuosa Xyn11A (Fig. 1 and Materials and Methods) were isolated from the vector backbones and transformed into T. reesei ALKO3620. Transformants in which one copy of the expression cassette replaced the cel7A gene were screened for further studies. Several parallel single-copy strains from each genetic construction were grown in shake flasks and were found to be similar in terms of the amounts of protein and xylanase activity in the culture supernatants. One single-copy representative from each genetic construction was chosen to be studied in fermentors. The growth of the strains in fermentors was followed by on-line determinations of the pH (not shown) and CO2 concentration (Fig. 2A to C). In addition, the sugars reactive to 3,5-dinitrosalisylic acid (not shown) and the mycelium dry mass (Fig. 2 D to F) were analyzed. The amounts of secreted proteins and xylanase and mannanase activities were determined from the culture supernatants (Table 1). The cleavage of the fusion proteins and the amounts of the recombinant N. flexuosa Xyn11A proteins were confirmed from SDS-polyacrylamide gels and from Western blot filters (Fig. 3).
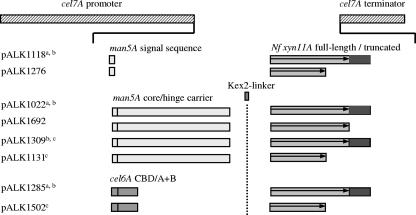
FIG. 1.
Expression cassettes. The amdS marker gene and a cel7A 3′-flanking region (not shown in Fig. 1) were ligated 3′ to the cel7A terminator, as described in reference 24. The full-length and truncated N. flexuosa xyn11A coding sequences (from D44 to N344 and from D44 to S263, respectively) are marked with arrows. a, the construction of the expression cassette has been described in reference 24. b, an approximately 250-bp N. flexuosa xyn11A 3′ UTR sequence (dark gray) precedes the fragment with the cel7A terminator sequence. c, nine codons of the N. flexuosa xyn11A sequence encoding the core protein have been changed according to the preferred T. reesei codon usage. For a more detailed description, see Materials and Methods.
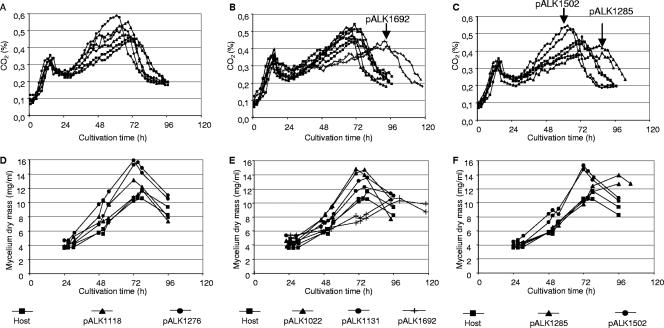
FIG. 2.
Growth of T. reesei transformants in laboratory fermentors. Two cultivations were performed for each strain. The changes in CO2 concentrations (A to C) and mycelium dry mass (D to F) during the cultivations are shown. The results from strains with fusion of xylanase to the Man5A signal sequence are shown in panels A and D, from strains with fusion to Man5A core/hinge in panels B and E, and from strains with fusion to the Cel6A CBD in panels C and F. Results from the ALKO3620 host strain are shown in all the graphs for comparison. The CO2 curves of strains with growth clearly different from that of the host are marked by arrows on graphs A to C. The growth of the strain with the expression cassette pALK1309 was similar to that of the strain with pALK1022 (not shown in panels B and E).
TABLE 1.
Protein and activity data from the cultivations of single-copy Trichoderma reesei transformants producing recombinant N. flexuosa Xyn11A (35 kDa) or the truncated N. flexuosa Xyn11A polypeptide (24 kDa)a
| Strain | Expression cassette | Amt or activity
|
|||||
|---|---|---|---|---|---|---|---|
| Protein (mg/ml ± SD) | Xylanase (nkat/ml ± SD) | Mannanase (nkat/ml ± SD) | rXyn11A (mg/ml)b | rXyn11A (nmol/ml)b | Man5A carrier (nmol/ml)c | ||
| ALKO3620 | None | 7.9 ± 0.5 | 240 ± 140 | 110 ± 20 | 0 | 0.0 | |
| RF5724 | pALK1118 | 6.5 ± 0.3 | 1,950 ± 30 | 160 ± 30 | 0.12 | 3.7 | |
| RF5024 | pALK1276 | 5.4 ± 0.3 | 7,620 ± 620 | 120 ± 30 | 0.44 | 18.4 | |
| ALKO4405 | pALK1022 | 7.1 ± 0.3 | 9,590 ± 540 | 2,890 ± 660 | 0.60 | 18.2 | 32.5 |
| RF5745 | pALK1692 | 3.3 ± 1.1 | 8,660 ± 340 | 2,850 ± 710 | 0.54 | 16.4 | 32.1 |
| RF5510 | pALK1309 | 7.2 ± 0.4 | 9,230 ± 420 | 3,020 ± 710 | 0.58 | 17.5 | 34.0 |
| RF5725 | pALK1131 | 7.2 ± 0.1 | 21,800 ± 3,460 | 5,730 ± 230 | 1.25 | 52.7d | 64.5d |
| RF5013 | pALK1285 | 3.2 ± 0.1 | 4,480 ± 530 | 80 ± 0.7 | 0.28 | 8.5 | |
| RF5139 | pALK1502 | 7.4 ± 0.5 | 32,230 ± 890 | 130 ± 3.7 | 1.85 | 77.9 | |
The results are averages from two fermentations each. The cultivations were finished 94 to 96 h after inoculation, with the exception of the RF5013 (pALK1285) and RF5745 (pALK1692) cultivations that were finished after 104 and 118 h of fermentation, respectively, due to slower growth of the strains. SD, standard deviation.
rXyn11A, recombinant 35-kDa full-length Xyn11A or the truncated 24-kDa Xyn11A polypeptide. The specific activities used in the calculations were 16,000 nkat/mg for the 35-kDa and 17,400 nkat/mg for the 24-kDa recombinant Xyn11A, according to Leskinen et al. (16). The calculated molecular masses used were 32.9 kDa and 23.8 kDa for the 35-kDa and 24-kDa xylanases, respectively. Calculated from the average activity of the two cultivations.
A specific activity of 2,120 nkat/mg and a molecular mass of 41.9 kDa were used in the calculations for the Man5A core-hinge (the specific activity has been determined for the Man5A core protein) (24). Calculated from the average activity of the two cultivations.
The fusion protein was only partially cleaved from the Kex2 linker.
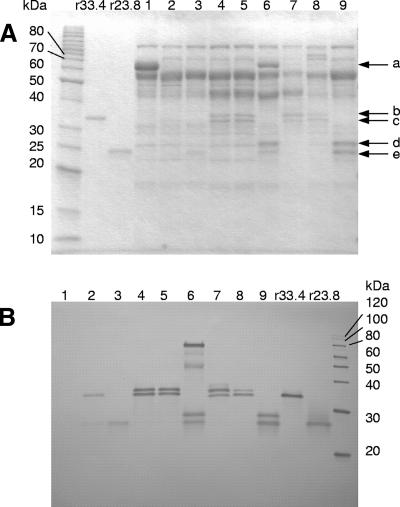
FIG. 3.
SDS-PAGE and Western blot analysis of the fermentation supernatants. (A) SDS-polyacrylamide gel and (B) Western blot analysis. Molecular mass markers are shown on the left and the right, respectively. Lanes: 1, T. reesei ALKO3620; 2 to 9, transformants containing the expression cassettes pALK1118, pALK1276, pALK1022, pALK1309, pALK1131, pALK1692, pALK1285, and pALK1502, respectively. r33.4, purified 33.4-kDa full-length recombinant N. flexuosa Xyn11A (1 μg in A and 70 ng in B). r23.8, purified 23.8-kDa recombinant truncated N. flexuosa Xyn11A (1 μg in panel A and 70 ng in panel B). Loads of 2 μl of undiluted or 6 μl of 1:100-diluted culture supernatants were applied, respectively, to SDS-polyacrylamide gels for Coomassie blue staining (A) or Western blotting (B). A rabbit polyclonal antibody synthesized against the native N. flexuosa Xyn11A was used to detect the recombinant N. flexuosa Xyn11A protein. Marked with arrows: the Man5A core/hinge-truncated N. flexuosa Xyn11A fusion protein (a), the full-length N. flexuosa Xyn11A protein forms (b, c), and the truncated N. flexuosa Xyn11A proteins (d, e). The native T. reesei Cel7A (lane 1) has a molecular mass of about 60 kDa and runs at about the same position as the Man5A core/hinge-truncated N. flexuosa Xyn11A protein (lane 6). The native T. reesei cellulases Cel7B and Cel6A run in the region of about 55 kDa.
The concentrations of CO2 in the host cultivation and in the cultivations of the transformants with pALK1118, pALK1276, pALK1022, pALK1309, and pALK1131 were at their maxima after 68 to 72 h after inoculation (Fig. 2A to C). The transformant with the expression cassette pALK1502 reached the CO2 maximum about 64 h after inoculation (Fig. 2C), suggesting that this strain grew somewhat quicker than the host. The growth of the transformants with the expression cassettes pALK1692 (man5A core/linker, full-length N. flexuosa xyn11A, exact fusion to cel7A terminator) and pALK1285 (cel6A CBD, full-length N. flexuosa xyn11A) was clearly slower than the growth of the host and the other transformants. These strains reached their CO2 maxima after 93 h (pALK1692) and 86 h (pALK1285) of growth (Fig. 2B and C, respectively). The slower growth of these two strains was also demonstrated by slower decreases of pH and concentrations of sugars reactive to 3,5-dinitrosalisylic acid in the cultivations (not shown). In addition, these strains reached their mycelium dry mass maxima about 1 day later (after 100 and 96 h of growth) than the other strains (after 70 to 78 h of growth) (Fig. 2E and F). The mycelium dry mass maxima of the transformants were higher (average, 12.5 to 15.8 mg/ml) than that of the host (average, 11.1 mg/ml), with the exception of the transformant with the pALK1692 cassette (average, 10.5 mg/ml) (Fig. 2D to F).
The amounts of the secreted proteins and the relevant enzyme activities (xylanase and mannanase) measured from the final samples of the cultivations are shown in Table 1. The host secreted, on average, 7.9 g liter−1 proteins into its culture supernatant. The transformants with expression cassettes pALK1022, pALK1309, pALK1131, and pALK1502 secreted slightly less protein (7.1 to 7.4 g liter−1). The strains with a signal sequence fusion, pALK1118 and pALK1276, produced smaller amounts of proteins, 6.5 and 5.4 g liter−1, respectively. The strains containing pALK1692 or pALK1285 produced only 3.2 to 3.3 g liter−1 of proteins in their culture supernatants.
The xylanase activity was higher in the genetic constructions using a carrier polypeptide than in the genetic constructions with a fusion of the xylanase/truncated xylanase gene to the man5A signal sequence (Table 1). The strains expressing the truncated N. flexuosa Xyn11A produced clearly higher xylanase activities than the strains with the corresponding genetic constructions containing the full-length gene. The highest xylanase activity, 32,230 nkat/ml, was produced by the strain that expressed the truncated N. flexuosa xyn11A gene fused to the sequence encoding the Cel6A CBD carrier (pALK1502). The activity from this strain corresponded to about 1.9 g liter−1 of the recombinant xylanase protein. The truncated N. flexuosa Xyn11A constituted about 25% of the proteins secreted by the pALK1502 strain. The increases in xylanase activity from the strains expressing the truncated N. flexuosa Xyn11A compared to the activities of the strains with the corresponding full-length genetic constructions were 3.9-fold for the signal sequence fusion (pALK1276 versus pALK1118), 2.4-fold for the Man5A core/linker carrier (pALK1131 versus pALK1309) and 7.2-fold for the Cel6A CBD carrier (pALK1502 versus pALK1285). The calculated molar increases of the recombinant xylanase proteins corresponding to these values are 5.0-, 3.0- and 9.2-fold for the signal sequence fusion, Man5A core/linker, and Cel6A CBD genetic constructions, respectively. Similar to the increase in xylanase activity, the mannanase activity was increased 1.9- to 2.0-fold in the pALK1131 strain compared to the activities of the pALK1022, pALK1692, and pALK1309 transformants. The increase in mannanase activity measured from the culture supernatants of the above strains resulted from the action of the Man5A core/hinge polypeptide that was used as a carrier in these strains. The nine changes made to the codons of the N. flexuosa xyn11A gene did not increase the xylanase activity in the culture supernatants (pALK1309 versus pALK1022).
Interestingly, the xylanase activity produced by the pALK1276 transformant carrying the expression cassette with the truncated N. flexuosa xyn11A fused to the man5A signal sequence (7,620 nkat/ml) was close to the activities obtained from the pALK1022, pALK1309, and pALK1692 transformants which express the full-length N. flexuosa xyn11A fused to the sequence encoding the Man5A core/hinge carrier (8,660 to 9,590 nkat/ml). The pALK1692 transformant produced xylanase activity similar to the activities of the pALK1309 and pALK1022 transformants, even though the total proteins detected from its culture supernatant were less than half of the amounts found for the pALK1309 and pALK1022 strains (Table 1).
Samples from the culture supernatants were run on an SDS-PAGE gel (Fig. 3A), and Western blotting was performed (Fig. 3B). The increase in the amount of the recombinant truncated N. flexuosa Xyn11A from T. reesei, compared to the amount of the full-length N. flexuosa Xyn11A, was clearly seen on the Coomassie blue-stained gel (Fig. 3A). The molecular masses of the recombinant N. flexuosa Xyn11A and the truncated N. flexuosa Xyn11A, when cleaved from the carrier polypeptides, were as expected and corresponded to those of the purified recombinant xylanase forms (33.4 kDa and 23.8 kDa) (16). In addition to these protein bands, protein bands of about 2- to 3-kDa-higher molecular masses were detected with the antibody. These polypeptides represented N-glycosylated forms of the recombinant xylanases, as confirmed by endo-β-N-acetylglucosaminidase Hf and peptide-N-glycosidase F treatments (not shown). The relative amounts of the nonglycosylated and the glycosylated N. flexuosa Xyn11A forms varied depending on the transformant. Culture supernatants from transformants carrying pALK1118, pALK1276, and pALK1285 contained only the nonglycosylated form or mostly this form of xylanase. About equal amounts of the two protein forms were detected in culture supernatants of the pALK1022, pALK1309, and pALK1502 transformants and more of the glycosylated form in culture supernatants of the pALK1131 and pALK1692 transformants.
The truncated recombinant xylanase was efficiently cleaved from the Cel6A CBD carrier (pALK1502). However, part of the xylanase produced by the pALK1131 transformant (Man5A core/linker as carrier) remained in an unprocessed fusion protein with a molecular mass of about 80 kDa (the theoretical mass of the fusion protein is 65.7 kDa). In addition, protein bands of about 50 to 70 kDa were detected from the pALK1131 culture supernatant. These bands most probably represent fusion proteins that have been proteolytically cleaved from N- and/or C-terminal regions.
An enzyme product containing the shortened N. flexuosa Xyn11A protein was tested in the pretreatment of oxygen-delignified birch pulp at pH 7, using a high temperature (82°C) and a reaction time of 20 min. After the enzymatic reaction, two-stage bleaching was performed using ClO2. A saving of 12% in the amount of ClO2 was obtained when the final brightness of the pulp was the same as in the control bleaching performed without the enzyme (data not shown).
Effect of truncation of N. flexuosa xyn11A on gene expression and induction of the UPR.
Total cellular RNAs were isolated from samples of mycelia taken from the fermentor cultivations in exponential growth and when the mycelium dry mass reached its maximum. The Northern blot filters were probed with an oligonucleotide that hybridizes to the untranslated region of the cel7A transcript. This made it possible to analyze both the amounts of the N. flexuosa xyn11A/fusion mRNAs from the transformants and the cel7A mRNA from the host. The results obtained from the transformants with the oligonucleotide probe were confirmed by using a fragment from the truncated N. flexuosa xyn11A gene as a probe. The pdi1 and hac1 probes were used to detect whether the UPR signal transduction pathway was induced in the transformants. The signals obtained from the Northern blot filters with the oligonucleotide, N. flexuosa xyn11A fragment, hac1, and pdi1 probes were normalized using an actin probe.
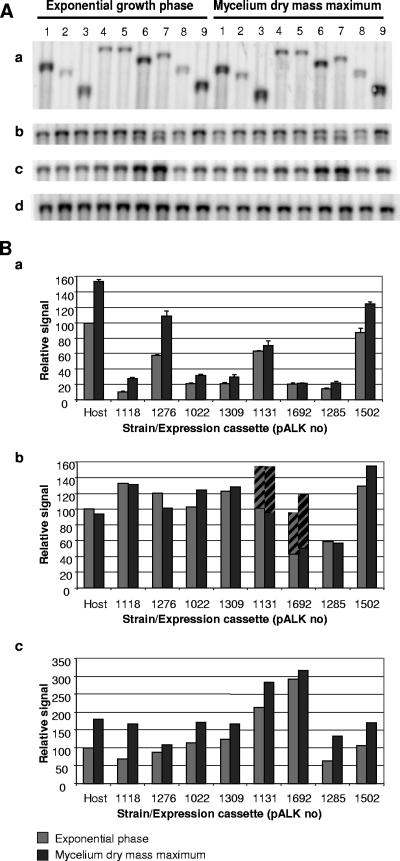
The results from the Northern blot filters and the normalized signals are shown in Fig. 4. The relative amounts of the xylanase mRNA/fusion mRNAs were clearly larger in the strains expressing the truncated N. flexuosa xyn11A gene (pALK1276, pALK1131, and pALK1502) than in the strains expressing the corresponding cassettes with the full-length N. flexuosa xyn11A. At the exponential phase, the increases were 5.7-, 3.0-, and 6.1-fold for the genetic constructions with the signal sequence fusion, Man5A core/hinge, and Cel6A CBD fusion, respectively. The amounts of the mRNA/fusion mRNA, compared to the amount of the cel7A mRNA in the host (100%), were 58% in the pALK1276, 63% in the pALK1131, and 87% in the pALK1502 transformant, whereas the strains carrying the corresponding expression cassettes with the full-length N. flexuosa xyn11A accumulated only about 10% (signal sequence fusion) and 20 to 21% (genetic constructions with a carrier) of the xylanase/fusion mRNA. The strains expressing the truncated N. flexuosa xyn11A also produced more xylanase/fusion mRNA at the stage of the mycelium dry mass maximum. The increases of the signals were 3.9-, 2.2-, and 5.6-fold for the signal sequence fusion, Man5A core/hinge, and Cel6A CBD fusion, respectively, compared to the signals for the strains expressing the full-length N. flexuosa xyn11A. The relative increase of the signal at the stage of the mycelium dry mass maximum, compared to that in the exponential phase, was the highest in the strains with xylanase fused to the signal sequence, pALK1118 (2.8-fold) and pALK1276 (1.9-fold). The corresponding relative increase was 1.4- to 1.5-fold in the host strain and in the pALK1022, pALK1309, pALK1285, and pALK1502 transformants but only 1.1-fold in the pALK1131 and pALK1692 transformants.
FIG. 4.
Northern blots. Two-microgram samples of total RNA were loaded per lane. (A) Northern blot filter hybridized with (a) oligonucleotide hybridizing to the untranslated region of the cel7A promoter, (b) hac1, (c) pdi1, and (d) actin. Lanes: 1, host; 2 to 9, transformants with the expression cassettes pALK1118, pALK1276, pALK1022, pALK1309, pALK1131, pALK1692, pALK1285, and pALK1502, respectively. (B) Relative signals, normalized with an actin probe. The gray and black bars represent the samples taken from the exponential growth phase and from the mycelium dry mass maximum, respectively. (a) Oligonucleotide probe. The averages from two parallel loadings are shown. The variations between the two loadings are shown with vertical lines. (b) hac1 probe. The filled bars represent the relative amounts of the full-length signals and the bars with diagonal lines represent the relative signals from the truncated hac1 mRNAs. (c) pdi1 probe.
The signals from the filters probed with the N. flexuosa xyn11A gene fragment (not shown) confirmed the results obtained with the oligonucleotide probe.
The transcripts from the pALK1022 and pALK1309 expression cassettes were longer than the transcript from pALK1692 (Fig. 4A), which suggests that the transcription terminated in these strains at the fungal terminator, not at the bacterial terminator. The nine changes made to codons did not have an effect on the amount of the fusion mRNA, and similar amounts of the transcripts were detected from strains carrying either expression cassette pALK1022 or pALK1309.
The UPR has been shown to be activated in various organisms, including T. reesei, when increased levels of unfolded or misfolded proteins accumulate in the endoplasmic reticulum (ER). Upon induction, transcriptional up-regulation of the ER-resident proteins, e.g., protein disulfide isomerase, is triggered (33). The hac1 gene has been shown to be the UPR pathway regulator in T. reesei (34). The hac1 mRNA is truncated at the 5′ flanking region, which removes an upstream open reading frame and results in a shorter form of hac1 mRNA. The UPR pathway was induced in the mycelia of the strains with pALK1692 (man5A core/linker, N. flexuosa Xyn11A with an exact terminator fusion) and pALK1131 (man5A core/linker, truncated xyn11A) expression cassettes, as shown by an increased amount of the pdi signal and the appearance of the truncated form of the hac1 transcript in the mycelia of these two strains. The amount of the pdi1 signal was 2.4- to 4.7-fold larger in the mycelium of the pALK1692 transformant and 1.7- to 3.3-fold larger in the mycelium of the pALK1131 transformant than in the other transformants. At the exponential phase, about half of the hac1 mRNA in the pALK1692 transformant and one-third in the pALK1131 transformant consisted of the truncated hac1 transcript. The cleaved form was also detected from the mycelia of these strains at the stage of dry mass maximum.
DISCUSSION
The N. flexuosa xyn11A gene devoid of the region encoding the CBM was expressed in T. reesei in order to produce an enzyme preparation with enhanced thermostability compared to the preparations with the full-length N. flexuosa Xyn11A (24). T. reesei successfully produced the truncated N. flexuosa Xyn11A protein and the enzyme preparations containing the truncated xylanase performed well in the bleaching of kraft pulp at a high temperature. The positive result obtained from the bleaching test was expected, because the purified recombinant N. flexuosa Xyn11A polypeptides devoid of the CBM (23.8 kDa and 22.0 kDa) have previously been shown to be efficient in this application (16). Also, several other studies have described the carbohydrate binding domains of modular endoxylanases as nonessential both in the hydrolysis of soluble xylan and in the pulp bleaching application (e.g., references 4, 11, 21, and 31).
Interestingly, the transformants expressing the truncated N. flexuosa xyn11A gene produced 2.4- to 7.2-fold more xylanase activity in their culture supernatants than the corresponding strains expressing the full-length gene (Table 1). The increases in xylanase activities correlated with increases in the mRNA signals in these strains (Fig. 4). Thus, by deleting the region encoding the CBM, we were able partly to overcome the bottleneck in transcription detected when the full-length N. flexuosa xyn11A was expressed in T. reesei (24). We are not aware of any previously published results in which deletion of the CBM/CBD from an enzyme would have increased gene expression and enzyme production in this way. Of the two T. reesei carriers tested, the Cel6A CBD was more suitable for the production of the truncated N. flexuosa Xyn11A. The xylanase was efficiently cleaved from the fusion protein, and about 1.9 g liter−1 of the recombinant xylanase was produced under the cultivation conditions used. Quite unexpectedly, the truncated N. flexuosa Xyn11A was only partially cleaved from the Man5A core/hinge, even though the full-length N. flexuosa Xyn11A was efficiently cleaved from this carrier (24) (Fig. 3). The deletion of the CBM presumably altered the fusion protein structure so that the Kex2 site in the linker region was not efficiently recognized by the Kex2 protease. As suggested in reference 7 and demonstrated in reference 37, the processing of fusion proteins is more dependent on the structural context at the processing region than on the actual amino acid sequence.
The xylanase activities produced by the strains with the expression cassettes pALK1118 (1,950 nkat/ml; full-length N. flexuosa xyn11A fused to man5A signal sequence) and pALK1022 (9,590 nkat/ml; man5A core/hinge-full-length N. flexuosa xyn11A) corresponded to the previously published results (2,460 and 9,200 nkat/ml, respectively) (24). However, the strain with the expression cassette pALK1285 (cel6A CBD-full-length N. flexuosa xyn11A) produced, unexpectedly, less xylanase activity (4,480 nkat/ml) than we previously reported (11,500 nkat/ml). This strain also grew slowly (Fig. 3) and secreted fewer proteins than the other transformants (except the pALK1692 transformant). The reason for the slow growth and low production of proteins remains unsolved but is, most probably, due to differences in the cultivation media used in the fermentations. In the previous study (24), we used a rich, complex cellulase-inducing medium. In the present study, a particle-free extract was used to enable the determination of the mycelium dry masses from the cultivations.
A carrier polypeptide has been suggested to stabilize the recombinant mRNA, to facilitate the translocation of the heterologous product in the secretory pathway, and to protect it from proteolytic degradation, as reviewed in reference 9. Our results confirm the importance of the carrier polypeptide in the posttranscriptional stages. The pALK1276 strain (truncated N. flexuosa Xyn11A, no carrier) produced about three times more mRNA at the exponential phase but less xylanase activity than the pALK1022, pALK1309, and pALK1692 strains (full-length N. flexuosa Xyn11A, Man5A core/hinge carrier) (Fig. 4 and Table 1). The pALK1502 strain (truncated N. flexuosa Xyn11A, Cel6A CBD carrier) produced only about 1.5-fold more xylanase mRNA but 4.2-fold more xylanase activity than pALK1276 (Fig. 4 and Table 1).
The nine changes in codons that were made in the N. flexuosa xyn11A sequence did not affect either the expression or the production of the recombinant xylanase. However, even without these changes, the codon usage of the N. flexuosa xyn11A resembled the codon usage of the T. reesei cel7A (3) in having a strong bias towards C at the wobble position (16). Thus, the changes did not improve the codon bias already present in the native N. flexuosa xyn11 gene. The change of codons of the Dictyoglomus thermophilum xylanase (xynB) to better resemble the codon usage of the host has been shown to result in increased expression of xynB in T. reesei (41). However, the codon usage of the native xynB differed remarkably from that of T. reesei in having a preference for A and T bases at the wobble position. Further studies, using a synthetic N. flexuosa xyn11A gene with overall optimized codon usage, would be needed to confirm whether we could further increase the expression and production of the recombinant N. flexuosa Xyn11A.
When unfolded proteins accumulate in the ER, the cell enhances its efficiency in protein folding, transport, and quality control. The transcription of the genes involved in these functions is up-regulated. This signal transduction pathway, UPR, has been characterized in detail in Saccharomyces cerevisiae and in mammalian cells (reviewed in references 18 and 25). During the last 10 years, UPR signaling has been intensively studied and has been shown to be active in the filamentous fungi T. reesei, Aspergillus nidulans, and Aspergillus niger (reviewed in reference 22). According to our results, the production of even large amounts of the truncated N. flexuosa Xyn11A did not induce the UPR pathway when the fusion protein was efficiently cleaved from the Kex2 site (pALK1502) (Fig. 4). However, the UPR was induced in two strains used in this study, the pALK1131 strain (Man5A core/hinge-truncated N. flexuosa Xyn11A) and the pALK1692 strain (Man5A core/hinge-full-length N. flexuosa Xyn11A with exact fusion to the cel7A terminator). The putative incorrect folding of the fusion protein/protein partners produced by the pALK1131 strain could be a reason for the induction of the UPR pathway. However, the growth and the amount of proteins secreted by the pALK1131 strain were not affected (Fig. 2 and Table 1), which suggests that T. reesei is able to cope with folding/secretion of the pALK1131 product by enhancing the production of foldases and proteins involved in transport/quality control.
The pALK1692 and pALK1022 transformants produced identical fusion proteins, but the UPR was induced only in the pALK1692 strain. The fusion proteins were cleaved from the Kex2 site (Fig. 3), which indicates a correct folding of the fusion partners. Also, the amounts of the fusion mRNAs detected from these two transformants were similar at the exponential growth phase (Fig. 4). The only difference between the expression cassettes pALK1692 and pALK1022 was in the 3′ UTR region that follows the full-length N. flexuosa xyn11A gene. In pALK1692, the N. flexuosa xyn11A gene was exactly fused to the T. reesei cel7A terminator, whereas in pALK1022, an approximately 250-bp 3′ UTR sequence of N. flexuosa xyn11A followed the full-length N. flexuosa xyn11A gene sequence before the cel7A terminator fragment (Fig. 1 and Materials and Methods). The 3′ UTR of the N. flexuosa xyn11A contains a very strict loop structure (40 to 50 bp) after about 40 nucleotides from the N. flexuosa xyn11A stop codon (not shown). We were not able to sequence this region even though several attempts were made, several primers were synthesized, and deletions were prepared from this region. The T. reesei transcription machinery reads through the bacterial terminator sequence, as shown by the mRNA from the pALK1022 strain being longer than that from the pALK1692 strain (Fig. 4A). However, we suggest that the bacterial 3′ UTR has an effect on the mRNA structure that decreases the efficiency of translation. The mRNA structure might not be optimal for the T. reesei translation machinery because of an improper spatial distance between the termination codon and the fungal terminator sequence and/or because of the very strong loop structure in the 3′ UTR of the N. flexuosa xyn11A. Because of the resulting slow synthesis and small amount of the full-length N. flexuosa Xyn11A in the pALK1022 mycelium, the T. reesei folding/secretion machinery copes with the heterologous full-length bacterial xylanase and there is no need to induce the UPR pathway. According to our hypothesis, the pALK1692 transformant would encounter difficulties in folding/secretion of the heterologous xylanase, because the mRNA of the full-length N. flexuosa xyn11A gene is translated more efficiently. In the yeast Saccharomyces cerevisiae, it has been shown that, when a strong hairpin-loop structure was positioned in the 3′ UTR of the pyruvate kinase gene, it had a deleterious effect upon translation, but only a small influence on the transcript stability (28). To confirm our hypothesis, both the half-lives and the translation efficiencies of the mRNAs from different N. flexuosa Xyn11A transformants should be studied.
In addition to the clear induction of the UPR in the pALK1692 transformant, this strain also grew slowly (Fig. 3) and secreted a clearly smaller amount of proteins into the culture supernatant than the amounts secreted by the host and the other transformants (Table 1). These results indicate that the strain is under secretion stress. T. reesei has been described as responding to secretion stress by decreasing the level of transcription from genes that encode the major extracellular proteins (e.g., the cellulase genes). This phenomenon has been named as a “RESS response” (repression under secretion stress) (23). We did not study the expression of the cellulase genes by Northern blotting. However, we measured the endoglucanase activity from the culture supernatants of the transformants by using hydroxyethylcellulose as a substrate (not shown). The endoglucanase activity in the culture supernatant of the pALK1692 strain was clearly lower, 190 nkat/ml, than the activities produced by the pALK1022 and pALK1309 strains, which were 820 and 800 nkat/ml, respectively. The endoglucanase activity produced by the pALK1131 strain, which shows UPR induction but not a severe secretion stress, was 510 nkat/ml.
According to our results, T. reesei is not able to properly fold/secrete large amounts of the bacterial CBM (the full-length N. flexuosa Xyn11A with the CBM). The folding of the CBM could be a challenge to the T. reesei folding/secretion machinery. Based on a sequence homology, the N. flexuosa Xyn11A CBM is a member of CBM family 2 (subfamily 2b) (16). Most CBMs in family 2 belong to bacterial hydrolases, and there are no fungal CBM members of this family (42 and http://afmb.cnrs-mrs.fr/CAZY/fam/CBM2.html). Also, there could be an interaction between the CBM and the core of the N. flexuosa Xyn11A, resulting in a structure that is recognized as an unfolded protein at the ER of T. reesei. The CBD of a bacterial protein, Erwinia chrysanthemi Cel5 cellulase, is proposed to be involved in the secretion of the Cel5 protein via the bacterial type II secretory pathway by forming a transient intramolecular interaction with the Cel5 core domain (6). The deletion of the CBD from the E. chrysanthemi Cel5 cellulase has been shown to impair the secretion of Cel5 from E. chrysanthemi (29). Similar to this result, the expression of the truncated N. flexuosa xyn11A gene in a bacterial host, Bacillus subtilis, resulted in xylanase activity several times lower than the activity from the B. subtilis transformants expressing a corresponding genetic construction with the full-length N. flexuosa xyn11A (J. Vehmaanperä, unpublished results).
In conclusion, we successfully produced a truncated N. flexuosa Xyn11A protein devoid of the CBM in T. reesei. The enzyme product was effective in the bleaching of kraft pulp. The deletion of the region encoding the CBM from the N. flexuosa xyn11A gene resulted in severalfold increases in the amounts of both the xylanase/fusion mRNA and the heterologous xylanase. The production of the truncated N. flexuosa Xyn11A did not induce the UPR pathway in T. reesei. However, the expression of the full-length N. flexuosa xyn11A, when exactly fused to the cel7A terminator, resulted in induction of the UPR, slower growth of the strain, and a decrease in the amount of the protein secreted into the culture supernatant. We suggest that the T. reesei folding/secretion machinery is not able to cope properly with large amounts of the full-length N. flexuosa Xyn11A protein due to the bacterial CBM.
Acknowledgments
We thank Varpu Backman, Anne Halonen, Merja Helanterä, Outi Könönen, Kirsti Leskinen, Outi Nikkilä, and Jaana Oksanen for skillful technical assistance. Sanna Hiljanen-Berg, Sirpa Okko, and Teemu Halonen are acknowledged for performing the laboratory-scale fermentations. Tarja Lahtinen, Fei Liu, and Pasi Taipalus are thanked for testing the truncated N. flexuosa Xyn11A in pulp bleaching. Tiina Pakula is thanked for her valuable help in analyzing the signals from the Northern blot filters. Christian P. Kubicek is acknowledged for providing us with the actin gene probe, and Markku Saloheimo is acknowledged both for providing us with the hac1 and pdi1 probes and for useful discussions. John Londesborough is acknowledged for critically reading the manuscript and for correcting the language.
Footnotes
Published ahead of print on 23 March 2007.
REFERENCES
- 1.Aho, S., V. Olkkonen, T. Jalava, M. Paloheimo, R. Bühler, M.-L. Niku-Paavola, E. H. Bamford, and M. Korhola. 1991. Monoclonal antibodies against core and cellulose-binding domains of Trichoderma reesei cellobiohydrolases I and II and endoglucanase I. Eur. J. Biochem. 200:643-649. [DOI] [PubMed] [Google Scholar]
- 2.Bailey, M. J., P. Biely, and K. Poutanen. 1992. Interlaboratory testing of methods for assay of xylanase activity. J. Biotechnol. 23:257-270. [Google Scholar]
- 3.Bergquist, P., V. Te′o, M. Gibbs, A. Czifersky, F. P. de Faria, M. Azevedo, and H. Nevalainen. 2002. Expression of xylanase enzymes from thermophilic microorganisms in fungal hosts. Extremophiles 6:177-184. [DOI] [PubMed] [Google Scholar]
- 4.Black, G. W., G. P. Hazlewood, S. J. Millward-Sadler, J. I. Laurie, and H. J. Gilbert. 1995. A modular xylanase containing a novel non-catalytic xylan-specific binding domain. Biochem. J. 307:191-195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Calmels, T. P. G., F. Martin, H. Durand, and G. Tiraby. 1991. Proteolytic events in the processing of secreted proteins in fungi. J. Biotechnol. 17:51-66. [DOI] [PubMed] [Google Scholar]
- 6.Chapon, V., H. D. Simpson, X. Morelli, E. Brun, and F. Barras. 2000. Alteration of a single tryptophan residue on the cellulose-binding domain blocks secretion of the Erwinia chrysanthemi Cel5 cellulase (ex-EGZ) via the type II system. J. Mol. Biol. 303:117-123. [DOI] [PubMed] [Google Scholar]
- 7.Contreras, R., D. Carrez, J. R. Kinghorn, C. A. M. J. J. van den Hondel, and W. Fiers. 1991. Efficient Kex2-like processing of a glucoamylase-interleukin-6 fusion protein by Aspergillus nidulans and secretion of mature interleukin-6. Bio/Technology 9:378-381. [DOI] [PubMed] [Google Scholar]
- 8.Drocourt, D., T. Calmels, J.-P. Reynes, M. Baron, and G. Tiraby. 1990. Cassettes of the Streptoalloteichus hindustanus ble gene for transformation of lower and higher eukaryotes to phleomycin resistance. Nucleic Acids Res. 18:4009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gouka, R. J., P. J. Punt, and C. A. M. J. J. van den Hondel. 1997. Efficient production of secreted proteins by Aspergillus: progress, limitations and prospects. Appl. Microbiol. Biotechnol. 47:1-11. [DOI] [PubMed] [Google Scholar]
- 10.Ilmén, M. 1997. Molecular mechanisms of glucose repression in the filamentous fungus Trichoderma reesei. VTT Publications 315, VTT Offsetpaino, Espoo, Finland.
- 11.Irwin, D., E. D. Jung, and D. B. Wilson. 1994. Characterization and sequence of a Thermomonospora fusca xylanase. Appl. Environ. Microbiol. 60:763-770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Jeenes, D. J., D. A. MacKenzie, I. N. Roberts, and D. B. Archer. 1991. Heterologous protein production by filamentous fungi. Biotechnol. Genet. Eng. Rev. 9:327-367. [PubMed] [Google Scholar]
- 13.Joutsjoki, V. V., T. K. Torkkeli, and K. M. H. Nevalainen. 1993. Transformation of Trichoderma reesei with the Hormoconis resinae glucoamylase P (gamP) gene: production of a heterologous glucoamylase by Trichoderma reesei. Curr. Genet. 24:223-228. [DOI] [PubMed] [Google Scholar]
- 14.Karhunen, T., A. Mäntylä, K. M. H. Nevalainen, and P. L. Suominen. 1993. High frequency one-step gene replacement in Trichoderma reesei. I. Endoglucanase I overproduction. Mol. Gen. Genet. 241:515-522. [DOI] [PubMed] [Google Scholar]
- 15.Kelly, J. M., and M. J. Hynes. 1985. Transformation of Aspergillus niger by the amdS gene of Aspergillus nidulans. EMBO J. 4:475-479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Leskinen, S., A. Mäntylä, R. Fagerström, J. Vehmaanperä, R. Lantto, M. Paloheimo, and P. Suominen. 2005. Thermostable xylanases, Xyn10A and Xyn11A, from the actinomycete Nonomuraea flexuosa: isolation of the genes and characterization of recombinant Xyn11A polypeptides produced in Trichoderma reesei. Appl. Microbiol. Biotechnol. 67:495-505. [DOI] [PubMed] [Google Scholar]
- 17.Lowry, O. H., N. J. Roseborough, A. L. Farr, and R. J. Randall. 1951. Protein measurement with the folin phenol reagent. J. Biol. Chem. 193:265-275. [PubMed] [Google Scholar]
- 18.Ma, Y., and L. M. Hendershot. 2001. The unfolding tale of the unfolded protein response. Cell 107:827-830. [DOI] [PubMed] [Google Scholar]
- 19.Mäntylä, A., M. Paloheimo, and P. Suominen. 1998. Industrial mutants and recombinant strains of Trichoderma reesei, p. 291-309. In C. P. Kubicek and G. E. Harman (ed.), Trichoderma and Gliocladium, vol. 2. Taylor and Francis Ltd., London, United Kingdom. [Google Scholar]
- 20.Matheucci, E., F. Henrique-Silva, S. el-Gogary, C. H. Rossini, A. Leite, J. E. Vera, J. C. Urioste, O. Crivellaro, and H. el-Dorry. 1995. Structure, organization and promoter expression of the actin-encoding gene in Trichoderma reesei. Gene 8:103-106. [DOI] [PubMed] [Google Scholar]
- 21.Morris, D. D., M. D. Gibbs, C. W. J. Chin, M.-H. Koh, K. K. Y. Wong, R. W. Allison, P. J. Nelson, and P. L. Bergquist. 1998. Cloning of the xynB gene from Dictyoglomus thermophilum Rt46B.1 and action of the gene product on kraft pulp. Appl. Environ. Microbiol. 64:1759-1765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Nevalainen, H., V. Te′o, M. Penttilä, and T. Pakula. 2005. Heterologous gene expression in filamentous fungi: a holistic view, p. 211-237. In D. K. Adora and R. M. Berka (ed.), Applied mycology and biotechnology, vol. 5. Genes and genomics. Elsevier B. V., Amsterdam, The Netherlands. [Google Scholar]
- 23.Pakula, T. M., M. Laxell, A. Huuskonen, J. Uusitalo, M. Saloheimo, and M. Penttilä. 2003. The effect of drugs inhibiting protein secretion in the filamentous fungus Trichoderma reesei. J. Biol. Chem. 278:45011-45020. [DOI] [PubMed] [Google Scholar]
- 24.Paloheimo, M., A. Mäntylä, J. Kallio, and P. Suominen. 2003. High-yield production of a bacterial xylanase in the filamentous fungus Trichoderma reesei requires a carrier polypeptide with an intact domain structure. Appl. Environ. Microbiol. 69:7073-7082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Patil, C., and P. Walter. 2001. Intracellular signaling from the endoplasmic reticulum to the nucleus: the unfolded protein response in yeast and mammals. Curr. Opin. Cell Biol. 13:349-356. [DOI] [PubMed] [Google Scholar]
- 26.Penttilä, M. 1998. Heterologous protein production in Trichoderma, p. 365-382. In C. P. Kubicek and G. E. Harman (ed.), Trichoderma and Gliocladium, vol. 2. Taylor and Francis Ltd., London, United Kingdom. [Google Scholar]
- 27.Penttilä, M., H. Nevalainen, M. Rättö, E. Salminen, and J. Knowles. 1987. A versatile transformation system for the cellulolytic filamentous fungus Trichoderma reesei. Gene 61:155-164. [DOI] [PubMed] [Google Scholar]
- 28.Purvis, I. J., A. J. E. Bettany, L. Loughlin, and A. J. P. Brown. 1987. The effects of alterations within the 3′ untranslated region of the pyruvate kinase messenger RNA upon its stability and translation in Saccharomyces cerevisiae. Nucleic Acids Res. 15:7951-7962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Py, B., M. Chippaux, and F. Barras. 1993. Mutagenesis of cellulase EGZ for studying the general protein secretory pathway in Erwinia chrysanthemi. Mol. Microbiol. 7:785-793. [DOI] [PubMed] [Google Scholar]
- 30.Raeder, U., and P. Broda. 1985. Rapid preparation of DNA from filamentous fungi. Lett. Appl. Microbiol. 1:17-20. [Google Scholar]
- 31.Rixon, J. E., J. H. Clarke, G. P. Hazlewood, R. W. Hoyland, A. J. McCarthym, and H. J. Gilbert. 1996. Do the non-catalytic polysaccharide-binding domains and linker regions enhance the biobleaching properties of modular xylanases? Appl. Microbiol. Biotechnol. 46:514-520. [DOI] [PubMed] [Google Scholar]
- 32.Saloheimo, M., P. Lehtovaara, M. Penttilä, T. T. Teeri, J. Stålberg, G. Johansson, G. Pettersson, M. Clayessens, P. Tomme, and J. K. C. Knowles. 1988. EGIII, a new endoglucanase from Trichoderma reesei: characterization of both the gene and enzyme. Gene 63:11-21. [DOI] [PubMed] [Google Scholar]
- 33.Saloheimo, M., M. Lund, and M. E. Penttilä. 1999. The protein disulphide isomerase gene of the fungus Trichoderma reesei is induced by endoplasmic reticulum stress and regulated by the carbon source. Mol. Gen. Genet. 22:35-45. [DOI] [PubMed] [Google Scholar]
- 34.Saloheimo, M., M. Valkonen, and M. Penttilä. 2003. Activation mechanisms of the HACI-mediated unfolded protein response in filamentous fungi. Mol. Microbiol. 47:1149-1161. [DOI] [PubMed] [Google Scholar]
- 35.Sambrook, J., and D. W. Russell. 2001. Molecular cloning, a laboratory manual. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- 36.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning, a laboratory manual. Cold Spring Harbor Laboratory, Cold Spring Harbor, NY.
- 37.Spencer, J. A., D. J. Jeenes, D. A. MacKenzie, D. T. Haynie, and D. B. Archer. 1998. Determinants of the fidelity of processing glucoamylase-lysozyme fusions by Aspergillus niger. Eur. J. Biochem. 258:107-112. [DOI] [PubMed] [Google Scholar]
- 38.Stålbrand, H., A. Saloheimo, J. Vehmaanperä, B. Henrissat, and M. Penttilä. 1995. Cloning and expression in Saccharomyces cerevisiae of a Trichoderma reesei β-mannanase gene containing a cellulose binding domain. Appl. Environ. Microbiol. 61:1090-1097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Suurnäkki, A. M. Tenkanen, J. Buchert, and L. Viikari. 1997. Hemicellulases in the bleaching of chemical pulps. Adv. Biochem. Eng. Biotechnol. 57:261-287. [DOI] [PubMed] [Google Scholar]
- 40.Teeri, T., P. Lehtovaara, S. Kauppinen, I. Salovuori, and J. Knowles. 1987. Homologous domains in Trichoderma reesei cellulolytic enzymes: gene sequence and expression of cellobiohydrolase II. Gene 51:43-52. [DOI] [PubMed] [Google Scholar]
- 41.Te′o, V. S. J., A. E. Cziferszky, P. L. Bergquist, and K. M. H. Nevalainen. 2000. Codon optimization of xylanase gene xynB from the thermophilic bacterium Dictyoglomus thermophilum for expression in the filamentous fungus Trichoderma reesei. FEMS Microbiol. Lett. 190:13-19. [DOI] [PubMed] [Google Scholar]
- 42.Tomme, P., R. A. J. Warren, R. C. Miller, Jr., D. G. Kilburn, and N. R. Gilkes. 1995. Cellulose-binding domains: classification and properties, p. 142-163. In J. N. Saddler and M. Penner (ed.), Enzymatic degradation of insoluble polysaccharides, ACS Symp. Ser., vol. 618. American Chemical Society, Washington, DC. [Google Scholar]
- 43.van den Hondel, C. A. M. J. J., P. Punt, and R. F. M. van Gorcom. 1991. Heterologous gene expression in filamentous fungi, p. 241-250. In J. W. Bennet and L. L. Lasure (ed.), More gene manipulations in fungi. Academic Press, San Diego, CA.
- 44.Zeilinger, S., R. L. Mach, and C. P. Kubicek. 1998. Two adjacent protein binding motifs in the cbh2 (cellobiohydrolase II-encoding) promoter of the fungus Hypocrea jecorina (Trichoderma reesei) cooperate in the induction by cellulose. J. Biol. Chem. 273:34463-34471. [DOI] [PubMed] [Google Scholar]
- 45.Zhang, Z., Y. Wang, and J. Ruan. 1998. Reclassification of Thermomonospora and Microtetraspora. Int. J. Syst. Bacteriol. 48:411-422. [DOI] [PubMed] [Google Scholar]