Abstract
Current metagenomic approaches to the study of complex microbial consortia provide a glimpse into the community metabolism and occasionally allow genomic assemblies for the most abundant organisms. However, little information is gained for the members of the community present at low frequencies, especially those representing yet-uncultured taxa, which include the bulk of the diversity present in most environments. Here we used phylogenetically directed cell separation by fluorescence in situ hybridization and flow cytometry, followed by amplification and sequencing of a fraction of the genomic DNA of several bacterial cells that belong to the TM7 phylum. Partial genomic assembly allowed, for the first time, a look into the evolution and potential metabolism of a soil representative from this group of organisms for which there are no species in stable laboratory cultures. Genomic reconstruction from targeted cells of uncultured organisms isolated directly from the environment represents a powerful approach to access any specific members of a community and an alternative way to assess the community's metabolic potential.
Over the last several years, there has been an unprecedented surge in the number and diversity of genomic approaches used to study microbial communities (24, 45). While sequence-based methods and functional screening have been used successfully over the past decade to discover specific genes and gene products from the environment (42), most of the research was focused on a few metabolic markers or was aimed primarily at biotechnological applications (29). A number of approaches have been developed to better understand the structure of microbial communities and to establish links between specific organisms and the metabolic potential encoded in their genes. Among these, fluorescence in situ hybridization (FISH) allows microscopic characterization and separation of microorganisms from environmental samples (2, 15, 29), while taxon-specific probes enable identification of cloned genomic fragments which could be sequenced and analyzed for encoded metabolic features (4). High-throughput cultivation methods have also been developed for less specific, but rapid, access to viable organisms representing a much larger fraction of the microbial community than was accessible using traditional microbiological techniques (12, 50).
Shotgun sequencing of genomic DNA mixtures representing entire microbial communities brought a new dimension to environmental microbiology. Such sequencing efforts have led to near-complete genomic and metabolic reconstruction of relatively simple consortia and have addressed important aspects of microbial biogeochemistry, bioremediation, and symbiosis (23, 31, 46, 47, 49). While the approach has allowed “gene-centric” comparative studies of complex microbial communities, generating and deconvoluting the genomic information specific to some of the less abundant taxa are still not feasible. Considering that most communities have a large number of species that are present at low abundance but may play important ecological roles, approaches that tap into their genomic information, in the absence of cultivation or gigabase-scale shotgun sequencing, would enable more-comprehensive studies of such consortia.
Whole-genome amplification has been applied in microbial studies to characterize the structure of communities from highly contaminated sites, where the amount of biomass was below standard detection levels (1), and to characterize populations of methanotrophs enriched by FISH/fluorescence-activated cell sorting (28). It has also been used for sequencing genomes from single cells of cultured bacteria to near completion and for preliminary characterization of relatives of cultured species (39, 51). Here we combined the use of taxon-specific separation of microbial cells by flow cytometry with whole-genome amplification to gain access to a low-abundance soil bacterium from the candidate TM7 division. This is the first targeted isolation and partial genomic sequencing of cells representing an uncultured group of organisms.
MATERIALS AND METHODS
Purification of bacteria and DNA from soil samples.
Soil from a rich, moist site in Ramona, CA, was used for bacterial purification. One hundred fifty grams of soil was homogenized in 250 ml ice-cold phosphate-buffered saline (PBS) in a Waring blender by three 1-min pulses (5). The soil debris was removed by centrifugation (15 min, 800 × g, 4°C) and the supernatant transferred to a fresh tube and centrifuged for 15 min at 14,000 × g (4°C). The resulting bacterial pellet was resuspended in PBS and purified by isopycnic density gradient centrifugation in Nycodenz (Sigma-Aldrich, St. Louis, MO) (5). Total microbial-community DNA was isolated from purified cells as previously described (1).
FISH.
An aliquot of the purified bacterial pellet was washed and fixed by resuspension in 100% ethanol, followed by centrifugation. Hybridization with the TM7-specific oligonucleotide probe TM7905 (labeled with AlexaFluor 546; Molecular Probes, Carlsbad, CA) was performed as originally described for environmental TM7 bacteria (27). Control hybridizations of Escherichia coli cells used the Gam42a oligonucleotide (30) labeled with AlexaFluor 488.
Flow cytometry analysis and sorting were performed with a Dako MoFlo flow cytometer (Fort Collins, CO) equipped with a Coherent Enterprise II (Santa Clara, CA) argon ion laser. The 488-nm line was used as the excitation source for forward scatter and side scatter properties. The fluorophore excitation source was a Coherent Innova 70C (Santa Clara, CA) water-cooled, mixed-gas laser tuned to 530 nm. Forward scatter, side scatter, and fluorescent properties were detected by R928 photomultiplier tubes (Hammamatsu, Shizuoka-ken, Japan). Fluorescence was detected between 550 and 590 nm. Data were collected and analyzed using DakoCytomation Summit v3.1 software. Bacterial cells displaying the fluorescent signal were sorted into 0.2-μl PCR tubes at 100, 50, 10, 5, and 1 cell per tube.
MDA.
Cells sorted in 1.2-μl-PBS droplets were lysed using a KOH lysis buffer and amplified by multiple displacement amplification (MDA) as described previously (25), with some modifications. Smaller reaction volumes were used: 1.2 μl of lysis buffer, 1.2 μl of neutralization buffer, and a 20-μl final volume. The initial amplification using phi 29 polymerase (Epicenter, Madison, WI) was done at 30°C for 4 h, followed by heat inactivation (65°C for 10 min). Following small-subunit (SSU) rRNA sequence verification, the initial product was reamplified in four separate MDA reactions and combined for library construction.
SSU rRNA gene sequencing.
Bacterial SSU rRNA genes were amplified by PCR (HotStart PCR mix; QIAGEN, Valencia, CA) from the MDA-generated DNA products using the universal primers 27F (5′-TAGAGTTTGATCCTGGCTCAG-3′) and 1492R (5′-TACGGYTACCTTGTTACGACTT-3′). Clone libraries were generated using a TOPO TA cloning kit (Invitrogen, Carlsbad, CA), and plasmid insert sequencing was performed using a 3730xl DNA analyzer (Applied Biosystems, Forster City, CA). High-quality individual clone reads were assembled using Sequencher (Gene Codes Corporation, Ann Arbor, MI), verified for potential chimeric artifacts, and classified taxonomically using the online tools at the RDP-II and Greengenes databases (10, 17). This resulted in 91 sequences for the soil environmental DNA library and 69 sequences for the MDA-amplified DNA library. A secondary-structure model of the TM7 SSU rRNA was generated using RnaViz 2.0 (16).
Genomic library construction, sequencing, and primary assembly.
MDA-amplified DNA from five sorted cells was mechanically sheared and used to generate libraries in a lambda ZAP Express cloning vector (Stratagene, La Jolla, CA) according to the manufacturer's protocol. Phagemid libraries were produced from the parental lambda clones by in vivo excision in E. coli host cells. Average insert sizes were 2 to 4 kb. Inserts from randomly picked colonies were end sequenced using a 3730xl DNA analyzer with T3 and T7 primers. FASTA-formatted sequences and corresponding Phred quality files were created from 21,497 chromatograms. Subsequently, 1,446 reads (6.7%) were removed for having low-quality scores (less than 200 bases with a Phred score of <20) and/or representing chimeras, leaving 20,051 sequences as input for an initial Phrap assembly that resulted in 714 contigs and 734 singletons, with a combined length of 1,839,704 bp.
Secondary assembly, genome annotation, and analysis.
Visualization of the assembly in Consed (21) revealed a relatively high occurrence of abnormal end pair relationships (distance and orientation violations) between forward and reverse reads from the same template clone. Also, many contigs contained regions with high similarity to other regions on the same contig or other contigs. Those discrepancies could be a result of amplification, cloning, or assembly artifacts or, alternatively, due to the nonclonal nature of the bacterial population. To improve the quality of the assembly, we first ran the sequences through the standard gene prediction and annotation pipeline at Oak Ridge National Laboratory (ORNL). The annotation process also resulted in a binning of the contigs based on GC content, which separated the TM7 genomic data from that of a Pseudomonas sp. coisolate. The gene map information obtained for every contig (including apparent fragmented or full-length genes) was used to aid a secondary-assembly process in Sequencher. During the secondary assembly, we eliminated the reads and contigs that had clear signs of chimeric artifacts (multiple truncated genes, sometimes with inverted regions) and also trimmed low-coverage contig ends containing polymorphisms which were preventing assemblies. A final gene prediction and an annotation were generated using the ORNL pipeline. Automated gene prediction was performed by using the output of Critica complemented with the output of GlimmerBlast analysis and was used to evaluate overlaps and alternative start sites. The resulting list of predicted coding sequences were translated, and these amino acid sequences were used to query and derive product descriptions by using the NCBI NR, UniProt, TIGRFam, Pfam, PRIAM, KEGG, COG, and InterPro databases. The tRNAScanSE tool was used to find tRNA genes, whereas ribosomal RNAs were found by using BLASTN versus the 16S and 23S rRNA databases.
Statistical analysis of the genome coverage.
Coverage depth for consensus positions on the contigs was determined using a Perl script (available upon request). Read coordinates were extracted from the Phrap assembly file. For each nucleotide position on a contig, the total number of reads contributing to that consensus position was determined. To estimate the progression in genome coverage, we used the accumulation of novel functional gene categories (clusters of orthologous groups [COGs]) as a function of sequence reads being generated. We compared the TM7 sequencing progress to that observed for several other completed bacterial genomes of different sizes for which we were able to obtain the sequence reads deposited in GenBank. To accomplish this, we identified all of the genes in the genomes that belong to a COG category (10−6 threshold) and used the top COG hit for every gene. We then created a BLAST database that contained those genes as nucleotide sequences and used as queries the project sequence reads in the order they were generated. Since every sequence read can potentially hit two or even more genes, we allowed that and retrieved hits that had bit scores over 50 (approximately a 50-bp overlap) and 95% minimal identity values (values determined empirically). We assigned to each pair of sequence read-COG hits a unique identifier and used the list as input into the program EstimateS (11) to generate COG accumulation curves (Mao Tau expected-richness function). To plot these curves, the number of reads was normalized to the total reads for the sequencing project, taking into account reads that hit multiple COGs and reads that did not hit any COG.
Phylogenetic analyses.
SSU rRNA sequences amplified from the MDA library were aligned with rRNA genes from representative bacterial genomes covering all major taxa and several related environmental sequences by use of the online NAST tool at Greengenes (17). The alignment was manually inspected and corrected, and regions of high variability that were not confidently aligned were masked out. The final alignment contained 56 sequences and 1,080 positions. A maximum-likelihood tree was calculated with Phyml (22) by use of empirically determined nucleotide frequencies, a generalized time-reversible substitution model, with estimated fractions of invariable sites and six substitution rates following a gamma distribution model with optimized shape parameter. Branch support was calculated using a boot-strapped data set (100 replicates) generated by Seqboot from the PHYLIP package (18) and the same parameters in Phyml.
For protein phylogenetic analysis, the amino acid sequences of 13 ribosomal protein genes identified in the TM7 genomic data (L1, L2, L3, L4, L5, L14, L22, L23, L24, S3, S8, S14, and S19) were loaded into the RibAlign database (43), aligned with the corresponding genes from organisms represented in the rRNA tree by use of MAFFT, and concatenated in a single file. Regions that were not confidently aligned or that contained large gaps/insertions were masked out. The final alignment contained 38 sequences and 1,134 positions. A maximum-likelihood tree was calculated in Phyml by use of a Whelan and Goldman amino acid substitution model, with estimated fractions of invariable sites and six substitution rates following a gamma distribution model with optimized shape parameter. Branch support was calculated using a boot-strapped data set (100 replicates generated by Seqboot) and the same parameters in Phyml.
Nucleotide sequence and project accession numbers.
Unique rRNA sequences from the MDA-amplified library were deposited in GenBank under accession numbers EF451973 to EF451974. The Whole Genome Shotgun project was deposited in DDBJ/EMBL/GenBank under the project accession number AAXS00000000. The version described in this paper is the first version, accession number AAXS01000000.
RESULTS
Separation of TM7 bacterial cells from soil and genomic DNA amplification.
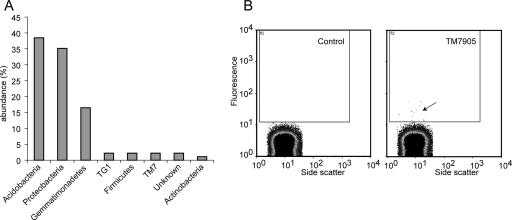
Based on SSU rRNA sequences, the soil sample under study contained mostly members of the Proteobacteria (35%), Acidobacteria (38%), and Gemmatimonadetes (16%) and, at much lower abundance levels (2% or less), representatives of several other phyla, including TM7 (Fig. 1). A cellular fraction of the soil sample was hybridized with a fluorescently labeled TM7-specific probe. Using flow cytometry, we detected a small fraction of cells (0.02%) with a fluorescence level approximately 10 times higher than that of the background, based on the unstained control populations (Fig. 1). Those cells were sorted into sterile, DNA-free tubes and used for genomic amplification.
FIG. 1.
(A) Taxonomic distribution (domain/phylum level) of SSU rRNA sequences in the PCR clone library prepared from the soil sample. (B) Flow cytometric analysis of soil bacteria hybridized with a fluorescent TM7-specific oligonucleotide probe (TM7905) or in the absence of hybridization (Control). The inner rectangle indicates the gating used for separation, and the arrow points to the low-frequency, highly fluorescent cells in the hybridized sample that were separated.
Because the oligonucleotide probes were designed to target most lineages within the TM7 division and not a specific species, the hybridization stringency conditions allowed for some nontarget cells to be sorted as well. It was determined by SSU rRNA gene amplification that the pools of larger numbers of cells (∼100) contained multiple TM7 strains and also various non-TM7 bacteria (data not shown). Based on test experiments, we determined that five was the minimal number of cells that balanced efficient genomic amplification with low levels of other bacterial coisolates. Others have also reported the difficulty of avoiding nontarget cells when separating by flow cytometry (28, 51).
Characterization of the amplified TM7 genomic DNA.
The MDA-amplified genomic DNA of five separated cells served as a template for PCR of SSU rRNA genes. Based on similarity to known environmental sequences, 61 of the 69 sequenced clones (89%) belonged to the TM7 phylum. There appeared to be no significant differences at the level of the TM7 SSU rRNA genes among the several cells sampled, as the sequences were >99.5% identical between any two clones. The rare polymorphisms did not cluster in groups of clones, suggesting they were due to PCR errors and did not represent sequence variation within the population. Therefore, while the five sorted cells were not clonal and genomic heterogeneity often occurred at the population level even though rRNA genes were identical, operationally we considered them to represent one “species,” which we refer to as TM7_GTL1. The remaining eight clone sequences were found to be nearly identical (>99.5%) to those of SSU ribosomal genes from several environmental Pseudomonas sp. isolates, including Pseudomonas rhodesiae, an organism isolated from natural mineral waters (13). These clones may therefore represent an actual Pseudomonas cell that was separated from the soil sample rather than laboratory contamination.
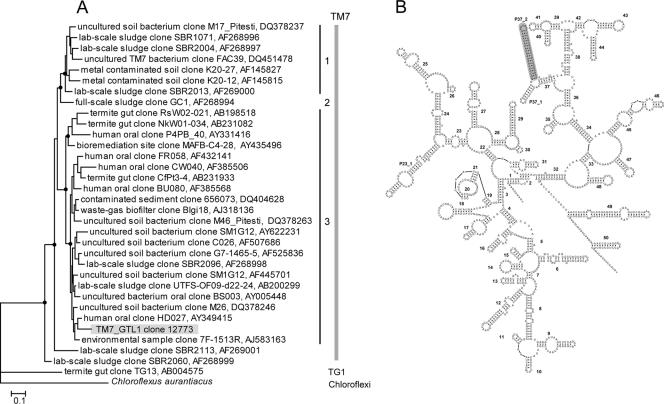
A comparison of the TM7_GTL1 SSU rRNA sequence with the >100 candidate division TM7 environmental sequences in the RDP-II database shows a maximum level of sequence identity of ∼90% to previously known clones, with the closest relative being from a human oral community (GenBank accession number AY349415). Phylogenetic analysis places TM7_GTL1 in TM7 subdivision 3 (27) (Fig. 2). A rather unique feature of the TM7_GTL1 rRNA sequence is an ∼30-nucleotide insertion which extends the P37_2 helix to 20 bp, significantly longer than that for most other bacteria, including all environmental TM7 sequences (Fig. 2). The role of P37_2 in ribosome assembly and function is unknown.
FIG. 2.
(A) Phylogenetic analysis (maximum likelihood) showing the relationships between the SSU rRNA sequence of TM7_GTL1 and previously known environmental clone sequences representing the three TM7 subdomains (27), with sequences from TG1 and Chloroflexi as outgroups. Filled circles represent bootstrap support values over 50% (shown only for major nodes). The bar indicates inferred 10% sequence divergence. GenBank accession numbers are given. (B) Secondary-structure model of TM7_GTL1 SSU rRNA, with the P37_2 helix shaded.
Sequencing, assembly, and annotation of the TM7 genomic library.
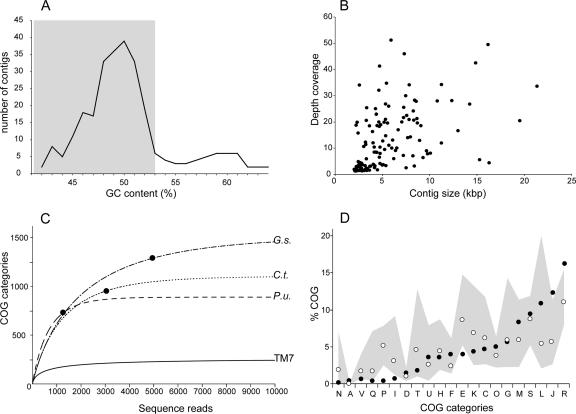
After the initial sequence assembly, in order to identify the contigs representing the Pseudomonas coisolate, we analyzed the nucleotide composition of the data set and the identity of the closest public sequence relatives. Based on sequence comparisons with completed Pseudomonas genomes, we found contigs that had multiple open reading frames with high levels of sequence identity (>90%) to known Pseudomonas genes. Those sequences also had elevated GC contents (>53%) relative to those of most of the sequences (average of <50%) (Fig. 3A). Based on these results, we separated the sequences with GC contents over 53% (42 contigs, totaling 185 kb, with an average GC content of 58.3%) from the remaining “TM7 contigs,” which had lower GC contents and closest gene homologues in more basal bacterial divisions, including Chloroflexi, Cyanobacteria, and Firmicutes. Since GC contents fluctuate within any given genome, we cannot exclude the possibilities that we removed some legitimate TM7 sequences or that the TM7 sequence pool may still contain a low number of Pseudomonas sequences. Binning approaches have been applied previously for genomic assemblies using metagenomic sequences from low-complexity communities (47, 49).
FIG. 3.
Information content analyses for the TM7_GTL1 genomic library. (A) Average GC content (percent) distribution of the assembled primary contigs. The shaded region indicates the contigs that were pooled as representing the TM7_GTL1 genome. Contigs with GC contents of >53% represent a Pseudomonas minor coisolate. (B) Average depth coverages (n-fold) for the final TM7 contigs. (C) Functional accumulation curves (COG categories) for TM7_GTL1 and representative finished bacterial genomes as a function of sequence depth (sequencing reads). The filled circle indicates the sequence depth corresponding to a 1× coverage for each of the finished genomes. P.u., Pelagibacter ubique; G.s., Geobacter sulfurreducens; C.t., Chlorobium tepidum. (D) Percentages of distribution of major COG categories in the TM7_GTL1 data (filled circles). For comparison, the median frequency values based on 502 bacterial genomes (not including obligatory parasites and symbionts) are indicated by the open circles. The gray area spans the observed distribution of frequencies in those genomes, between the minimum and maximum values.
Current limitations of libraries constructed from MDA-amplified DNA from one cell or a few cells include DNA amplification bias and the generation of chimeric sequences (51). Some of the chimeras are indicated by the presence of fragmented genes or duplicated regions across separate contigs. To address this, we analyzed the predicted open reading frames in an initially annotated data set and searched for such signs of chimerism. When detected, we eliminated the sequence reads containing chimeric junctions and reassembled the contigs. Even after this, some contigs contained chimeric regions that could not be resolved but were retained since the genes they included are valuable for the metabolic reconstruction. The final TM7 data set contained 132 contigs and 679,515 nucleotides, with an average GC content of 48.5%, and encoded 670 predicted proteins, six tRNAs, a full-length SSU rRNA gene, and a partial large-subunit rRNA gene.
To estimate the level of bias and sequence coverage across the sequenced portions of the genome, we calculated the average coverage for every contig, based on the number of sequence reads that contributed to the consensus. As shown in Fig. 3B, the coverage depths varied extensively and for some contigs exceeded 50-fold. Such variations in coverage have been observed previously (51).
An independent measure of the degree of bias was obtained by studying the accumulation functions of the gene categories. To do this, we used COG category assignments for each of the genes predicted during the annotation process and mapped the sequence reads as they were generated to the COG category list. We compared the TM7 accumulation curve to those generated by analyzing several other completed bacterial genomes of different sizes (Pelagibacter ubique, 1.3 Mbp; Geobacter sulfurreducens, 3.8 Mbp; and Chlorobium tepidum, 2.15 Mbp). Among the three completed genomes, we observed a steady accumulation of novel COG categories for approximately the first 1,000 sequence reads, after which the slopes for the individual genomes changed and eventually flattened at levels reflecting differences in genome size and total COG abundance, which is nearly twofold between Pelagibacter and Geobacter (889 versus 1,511 COG categories) (Fig. 3C). The initial accumulation slope for Pelagibacter is steeper, likely reflecting its more compact genome and its higher fraction of COG genes. For TM7, the COG accumulation curve departs from those of the other genomes and plateaus early in the sequencing progress, reaching 277 COGs, a value of about a third of that of Pelagibacter, which has the smallest genome for a free-living bacterium (20). Since it is unlikely that the TM7 genome is so much smaller than that of Pelagibacter, the rapid plateau likely reflects the amplification bias and indicates that the genomic library contains little additional information beyond what has been recovered. Overall, the frequency distributions of the different COG categories match those observed for complete genomes (Fig. 3D) and it does not appear that the bias is concentrated to specific types of genes. We did note a potential underrepresentation of ion and amino acid transport and metabolism genes (COG categories P and E) as well as a slight overrepresentation of translation/ribosomal genes and genes with a predicted general function (COG categories J and R). These may, however, represent natural characteristics of the organism rather than experimentally induced bias.
Place of TM7 within the Bacteria.
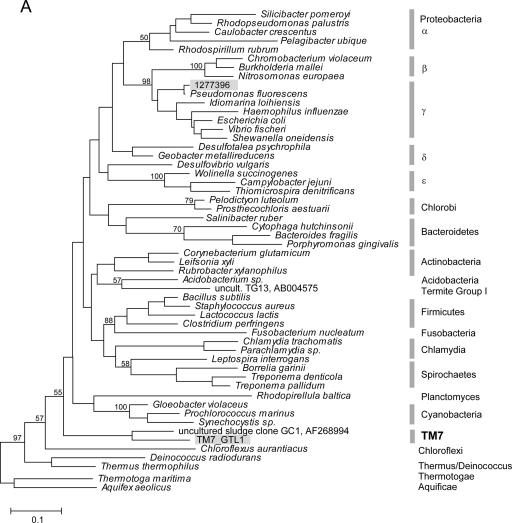
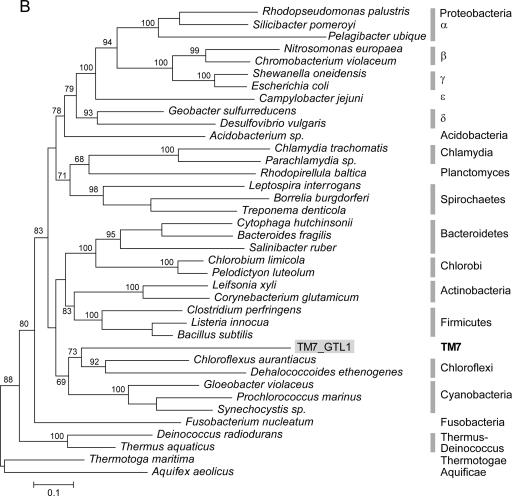
Due to limited phylogenetic information encoded within any single gene, including rRNA, the evolutionary relationships among the several dozen bacterial phyla (most without cultured representatives) are still unclear. Multiple gene phylogenies may increase resolution and in some cases have resulted in reassessments of interphyletic relationships (9). Previous analyses of environmental RNA sequences suggest a relatively basal position for TM7 in the bacterial domain, possibly related to Chloroflexi, OP10, and the Thermus-Deinococcus group (26). We used both rRNA and concatenated ribosomal protein data sets to investigate the placing of TM7 within the bacterial domain. While the rRNA tree is less resolved and suggests a close relationship to the green nonsulfur bacteria Chloroflexi, protein analysis supports a sister-like relationship with that group (Fig. 4). Several significant differences occur between the protein and the rRNA phylogenies, most notably, the close relationship between Acidobacteria and Proteobacteria as well as a deep position of Fusobacterium. These unsettled relationships have been reported previously and illustrate the difficulties in resolving the topology of the bacterial tree even when genomic data are available (9, 34).
FIG. 4.
Phylogenetic analysis (maximum likelihood) of the relationships among bacterial phyla, determined using SSU rRNA (A) and concatenated ribosomal protein sequences (B). Bootstrap support values over 50 are indicated (for major nodes only). The placement of TM7_GTL1 is indicated by shaded labels. rRNA sequence 1277396 represents the Pseudomonas coisolate of the amplified TM7_GTL1 genomic DNA. The bar indicates inferred 10% sequence divergence.
Insights into TM7 metabolism.
Although a comprehensive metabolic reconstruction was not possible using the incomplete TM7_GTL1 genomic data, we used functional gene annotations (COG, KEGG, and Pfam) to identify key steps in various metabolic pathways in order to uncover some of the physiological potentials of the organism. We identified genes for eight enzymes involved in glucose utilization, indicating the presence of glycolysis and pentose phosphate pathways. Ribulose-phosphate-3-epimerase is present and represents an important entry enzyme in the pentose interconversion pathway, which generates a variety of other sugars for downstream metabolic processes, including cell wall biosynthesis. Other represented pathways include lipid and nucleotide biosynthesis, amino acid transformations, and enzymes from the general nitrogen metabolism. We did not find evidence to suggest that TM7_GTL1 can grow autotrophically or that it can degrade certain polysaccharides, so the nature of the carbon source for this organism is unknown.
Based on sequence similarity to known transporters, we identified at least five transporters that belong to the drug H+ antiporter 1 family, involved in multidrug drug resistance. From the ABC transporter superfamily, we identified potential lipopolysaccharide exporters for lipid A (with a role in outer membrane synthesis), heavy metals, and macrolides. TM7_GTL1 also has a cytochrome P450 gene as part of the resistance mechanisms to toxic compounds. Considering that soil microbial communities are complex and involve constant competition, the presence of diverse transporters and multiple drug resistance mechanisms is expected. Additional transporters include a putative autoinducer 2 exporter (AI-2E) with a role in signaling/interspecific communication and a large conductance mechanosensitive channel which protects against osmotic cell lysis.
Among the genes involved in energy metabolism, we identified the conserved operon that encodes subunits of the FoF1 ATP synthase (contig 28). The gene order in this operon is highly conserved in bacteria, including TM7: A, C, B, delta, alpha, gamma, beta, and epsilon. One difference in TM7 is the insertion of a hypothetical gene between the alpha and gamma subunit genes. The only possible homologue of that gene is a weak hit in Desulfitobacterium hafniense, where the gene belongs to a cluster of genes of phage origin. This suggests that the gene in TM7_GTL1 has also been acquired from a viral genome. Interestingly, we also identified a second operon containing the ATP synthase subunit genes A through alpha (contig 642). Phylogenetic analysis places the second partial operon close to the ATPases from Chlorobi (not shown) and may indicate horizontal gene transfer.
Signal transduction, environmental interactions, and the cell wall.
Bacteria have evolved a variety of mechanisms to sense the environment, respond to changes, and adapt to new conditions. One of the conserved mechanisms in bacteria involves the use of the hyperphosphorylated guanosine nucleotide (p)ppGpp as a global regulator in stress response, adaptation, and interaction with other bacteria (7). We found the gene encoding the key enzyme that modulates the levels of this small effector molecule, RelA-SpoT (contig 547), potentially used during periods of nutrient starvation and growth arrest.
We identified a number of genes from two-component systems, including two-histidine kinases, a hybrid histidine kinase, a two-component winged-helix transcriptional regulator, and four CheY-like response regulators. The architecture of these proteins contains the same variation seen in other bacteria. The kinases contain either or both integral membrane and PAS/PAC sensing domains. Several pairs of kinase response regulators appear to be involved in sensing and responding to variations in available phosphate, copper/heavy metals, and osmoregulation. We did not identify any chemotaxis genes or flagellar components.
While there is no evidence to suggest that TM7_GTL1 has flagella, we identified multiple genes that make up the type IV pili, responsible for twitching motility. This represents an alternative for cells that live in nonfluid environments to move and colonize wet surfaces (32). Aside from motility, type IV pili also mediate DNA uptake and conjugation and can serve as docking sites for bacteriophages. There is a common evolutionary history between type IV pili and the type II secretion apparatus, involved in pathogenicity and environmental adaptation (37). We identified components of both systems, sometimes in multiple copies, suggesting that they play an important role in the biology of this organism. Among the type IV pilus genes, we identified PilA (encoding a putative pilin monomer), PilB assembly ATPase, PilC, PilM, PilN, and the PilT disassembly protein. Three distinct GspE genes, relatives of PilB ATPase with a role in the formation of the type II secretion apparatus, are also present, as is a putative competence factor (ComF) gene.
Supporting the view that TM7_GTL1 is engaged in active exchanges with the environment and the microbial community, we also identified gene products for the type IV secretion system, VirB4 and VirB6. Type IV secretion plays a major role in the translocation of macromolecules across the membrane and is particularly important for the exchanges of plasmids that can confer resistance to antibiotics as well as other DNA fragments and proteins. Type IV secretion also plays an important role in biofilm formation and in interactions with other bacteria and with eukaryotes (3). Since TM7_GTL1 was obtained from soil, which has high microbial and phage diversity, the cells would benefit from restriction modification systems to control the incoming of foreign DNA. We identified genes and gene fragments for a type I system (the R, S, and M subunit genes) as well as putative type IIS and type III restriction enzyme genes. We also found two different transposases, an indication that mobile genetic elements have been integrated into the TM7_GTL1 genome.
Information processing in TM7_GTL1.
The genes encoding proteins involved in TM7 chromosome replication, recombination, and DNA repair are well represented in the genomic data. Among those involved in the replication initiation complex, we identified DnaA, DnaB, and DnaG. Genes involved in replication include DNA polymerase III (beta and gamma/tau subunits), DNA gyrase (A and B subunits), and an NAD+-dependent DNA ligase. The genes involved in DNA repair, recombination, and modification are represented by those encoding uracil DNA glycosylase, adenine-specific methylase, the holiday junction helicase (RuvAB), resolvase (RuvX), and endonuclease (RuvC); recombination proteins RecA and RecF; excinuclease complex UvrABC; the RadA repair and MutT mutator proteins; and several other exo- and endonucleases. A functional domain present in over a dozen genes involved in repair is the Nudix hydrolase domain, which has been associated with controlling the levels of damaged mutagenic nucleotides. High numbers of such genes in genomes are suggested to indicate metabolic complexity and high adaptability potential (33). Similarly to what we have observed for genes encoding restriction endonucleases, genes involved in replication, recombination, and repair also appear to be overrepresented in chimeric reads and contigs.
Several transcription factors that belong to the ArsR, HxlR, TetR, MarR, and AraC families were identified; however, none of the basal transcriptional machinery genes are present in the data. Among the genes involved in translation, 10 of the 24 bacterial aminoacyl tRNA synthetase genes are present. We also recovered a large fraction of the conserved ribosomal protein superoperon, spanning 16 genes, from L3 through L6 (as part of contig 5). The order of the genes is highly conserved across the bacterial domain. Two additional ribosomal protein genes (L25 and L35) were also found elsewhere in the genome, as were other genes involved in translation and in RNA processing [elongation factor G, polynucleotide phosphorylase, RNase PH, and tRNA (uracil-5-)-methyltransferase]. The TM7_GTL1 gene for the SSU rRNA, with a sequence identical to that determined using PCR, was identified on contig 599. We also recovered most of the gene for the large-subunit rRNA (∼2.4 kb) as part of contig 19.
TM7_GTL1 genome coverage.
Due to the bias in the amplified genome library, it is difficult to estimate the genome size for TM7_GTL1. An independent measure can be attempted based on single-copy genes that are present in all genomes. Using a list of 182 bacterial core genes (31), we identified 35 genes (data not shown), suggesting a genomic coverage of ∼20% and a genome size of 3 to 3.5 Mbp. Since some of the genes are clustered, however, the genome size could be underestimated.
DISCUSSION
There is considerable interest for a more in-depth understanding of the impacts microbial communities have on many aspects that range from health and economics to small-scale environmental processes and global cycles (36). The magnitude of microbial diversity has been recognized for some time, and it might still be underestimated (41). rRNA sequences have enabled the study of that diversity and allowed the in situ visualization of some of the low-abundance microbes. However, they do not provide an understanding of the role that these organisms might play in the community. Environmental shotgun sequencing can provide a snapshot into the genomic milieu of complex communities but is unlikely to lead to metabolic reconstruction for species present at low levels. Such organisms, often members of phyla with few or no cultured representatives, may encode novel biochemical strategies and play key physiological and ecological roles in the community. While it may be desirable to study some of these organisms in culture, a targeted approach to rapidly access their genomes can offer valuable initial information about their biology.
We have shown here that it is possible to obtain a significant fraction of the genome of an uncultured bacterium, starting with several cells selectively isolated from a complex environmental sample. To achieve this, we linked two powerful methods used in microbial ecology (FISH and cell separation by flow cytometry) to whole-genome amplification and sequencing. This approach combines the high specificity derived from the stringent hybridization of oligonucleotide probes to target rRNA in a taxonomically predefined cellular population with the high sensitivity and throughput from the detection and separation of labeled cells from complex mixtures of organisms derived from flow cytometry. Because environmental bacterial populations representing a “species” are not clonal and their genomes may contain sequence and genetic map polymorphisms not reflected at the rRNA level (48), the computational burden in resolving the polymorphisms and assembling a “pan-genome” increases with the number of pooled cells. We therefore kept the number of cells at a minimum while providing an input template sufficient for whole-genome amplification.
Since MDA has been used successfully for recovering and sequencing genomes from cultured single bacterial cells, the method should allow for genomic characterization of uncultured organisms. The current technique, however, has limitations when applied to a single cell or a few cells. There is a significant sequence bias during amplification, causing variations in genome coverage. As observed previously (51), the distribution of bias appears to be random and is probably due to the limited number of initial replication initiation events and the relatively short amplified DNA fragments. When the starting amount of DNA is relatively high (ng range), the bias is not as severe (38). The amplification bias can be reduced partially by pooling multiple separate reactions; however, in the case of environmental bacteria, this will result in an increased chance of heterogeneity, due to the nonclonal nature of the population. Another limitation of MDA is the formation of chimeric structures which result in fragmented genes and difficulties in assembling large genomic contigs. We determined empirically that by using five cells and performing a modified MDA reaction protocol we could achieve the lowest level of nontarget cells while providing a template sufficient to result in a lower fraction of chimeric fragments, as judged by assembly artifacts and fragmented genes. The low level of sequences from a coisolate, a common problem in cell separation done using flow cytometry, was mitigated by GC content and taxonomic sequence binning.
We applied this approach to a member of the TM7 division for multiple reasons. TM7 bacteria are known to inhabit a wide range of environments, from soils, water, and activated sludge to termite guts (27, 35, 40). They have also been found in the human oral cavity and may be associated with periodontitis (8). There has been a sustained effort to obtain a glimpse into the physiology of TM7 bacteria in the absence of a cultivation method and to bring some representatives into pure laboratory cultures (44). In a recent study (19), a soil TM7 bacterium formed microcolonies on a membrane support, although formal species description and continuous cultivation have not yet been reported for any member of this division. We decided, therefore, that obtaining genomic information from a TM7 species would complement previous studies and may aid future cultivation efforts.
Microbiological characterization of soil communities is notoriously difficult compared to characterizations of most other environments. Besides the presence of components that interfere with efficient cell separation, FISH, and DNA isolation, soil has a heterogeneous architecture and is the home for some of the most complex microbial communities, with diversities that can reach thousands of species per gram (14). Partial genomic sequencing of a minor uncultured TM7 member from the soil community renders the approach applicable to organisms from a broad range of other environments.
Several interesting biological inferences emerged for TM7_GTL1. Phylogenetic analysis suggests that TM7 is a deep lineage most closely related to the green nonsulfur bacteria (Chloroflexi). As expected for a soil bacterium living in close proximity to so many other species, TM7_GTL1 has abundant protection mechanisms against toxins and foreign DNA, including various transporters, cytochrome P450, and several restriction-modification systems. It is a highly adaptable organism with genes for plasmid acquisition and effective DNA repair and genes linked to environmental stress response and resistance to starvation. Like many other soil bacteria, TM7_GTL1 does not appear to use flagella for motility but has genes for the type IV pilus, which may allow limited movement by twitching. This is an important mechanism used by bacteria to colonize new niches in the soil interstitial space and is also involved in biofilm formation. The available information does not indicate whether TM7_GTL1 has a specialized type of metabolism or specify the nature of its major nutrient sources. The presence of two operons for F-type ATP synthase may indicate intense metabolic activity. The available sequence data do not provide evidence sufficient to conclude whether this organism is gram negative or gram positive. Prior studies of filamentous TM7 bacteria suggest that these organisms are gram positive (27), although variations in cell wall structure are known to occur across taxonomic distances lower than division/phylum (6).
The strategy that we described can be used to obtain rapidly a much larger fraction of the genome of a target uncultured microbe than that obtainable by any other existing approach. Obvious improvements that can be made at the molecular level include reducing the amplification bias and the formation of chimeric structures. Such improvements should allow sequencing of a larger fraction of the genome, starting with a single cell. One could imagine environmental-genomics projects in which the focus is a specific group of organisms within a highly diverse community. Rather than using a whole-library shotgun approach, it would be more effective to target a significant fraction of that diversity for “species by species” characterization by flow cytometry and sequencing. Developing sorting approaches for target cells that have not been fixed could also open the way to cultivation and physiological studies of some of these rare organisms.
Acknowledgments
This work was supported by the Office of Science (BER), U.S. Department of Energy GTL program, grant no. DE-FG02-04ER63771, and the Virtual Institute of Microbial Stress and Survival (http://VIMSS.lbl.gov), supported by the U.S. Department of Energy, Office of Science, Office of Biological and Environmental Research, GTL Genomics program, through contract DE-AC0205CH11231 between the Lawrence Berkeley National Laboratory and the U.S. Department of Energy.
We thank Cheryl Kuske and Sue Barns at Los Alamos National Laboratory for collaboration and suggestions, the sequencing group at Diversa for technical support, and Melvin Simon, Natalia Ivanova, and Phil Hugenholtz for suggestions and critical reading of the manuscript.
Footnotes
Published ahead of print on 16 March 2007.
REFERENCES
- 1.Abulencia, C. B., D. L. Wyborski, J. A. Garcia, M. Podar, W. Chen, S. H. Chang, H. W. Chang, D. Watson, E. L. Brodie, T. C. Hazen, and M. Keller. 2006. Environmental whole-genome amplification to access microbial populations in contaminated sediments. Appl. Environ. Microbiol. 72:3291-3301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Amann, R. I., B. J. Binder, R. J. Olson, S. W. Chisholm, R. Devereux, and D. A. Stahl. 1990. Combination of 16S rRNA-targeted oligonucleotide probes with flow cytometry for analyzing mixed microbial populations. Appl. Environ. Microbiol. 56:1919-1925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Baron, C. 2005. From bioremediation to biowarfare: on the impact and mechanism of type IV secretion systems. FEMS Microbiol. Lett. 253:163-170. [DOI] [PubMed] [Google Scholar]
- 4.Beja, O., M. T. Suzuki, E. V. Koonin, L. Aravind, A. Hadd, L. P. Nguyen, R. Villacorta, M. Amjadi, C. Garrigues, S. B. Jovanovich, R. A. Feldman, and E. F. Delong. 2000. Construction and analysis of bacterial artificial chromosome libraries from a marine microbial assemblage. Environ. Microbiol. 2:516-529. [DOI] [PubMed] [Google Scholar]
- 5.Berry, A. E., C. Chiocchini, T. Selby, M. Sosio, and E. M. Wellington. 2003. Isolation of high molecular weight DNA from soil for cloning into BAC vectors. FEMS Microbiol. Lett. 223:15-20. [DOI] [PubMed] [Google Scholar]
- 6.Beveridge, T. J. 2001. Use of the gram stain in microbiology. Biotech. Histochem. 76:111-118. [PubMed] [Google Scholar]
- 7.Braeken, K., M. Moris, R. Daniels, J. Vanderleyden, and J. Michiels. 2006. New horizons for (p) ppGpp in bacterial and plant physiology. Trends Microbiol. 14:45-54. [DOI] [PubMed] [Google Scholar]
- 8.Brinig, M. M., P. W. Lepp, C. C. Ouverney, G. C. Armitage, and D. A. Relman. 2003. Prevalence of bacteria of division TM7 in human subgingival plaque and their association with disease. Appl. Environ. Microbiol. 69:1687-1694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ciccarelli, F. D., T. Doerks, C. von Mering, C. J. Creevey, B. Snel, and P. Bork. 2006. Toward automatic reconstruction of a highly resolved tree of life. Science 311:1283-1287. [DOI] [PubMed] [Google Scholar]
- 10.Cole, J. R., B. Chai, R. J. Farris, Q. Wang, A. S. Kulam-Syed-Mohideen, D. M. McGarrell, A. M. Bandela, E. Cardenas, G. M. Garrity, and J. M. Tiedje. 2006. The ribosomal database project (RDP-II): introducing myRDP space and quality controlled public data. Nucleic Acids Res. 35:D169-D172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Colwell, R. K. 2006. EstimateS: statistical estimation of species richness and shared species from samples. University of Connecticut, Storrs. http://viceroy.eeb.uconn.edu/EstimateS.
- 12.Connon, S. A., and S. J. Giovannoni. 2002. High-throughput methods for culturing microorganisms in very-low-nutrient media yield diverse new marine isolates. Appl. Environ. Microbiol. 68:3878-3885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Coroler, L., M. Elomari, B. Hoste, M. Gillis, D. Izard, and H. Leclerc. 1996. Pseudomonas rhodesiae sp. nov., a new species isolated from natural mineral waters. Syst. Appl. Microbiol. 19:600-607. [Google Scholar]
- 14.Crawford, J. W., J. A. Harris, K. Ritz, and I. M. Young. 2005. Towards and evolutionary ecology of life in soil. Trends Ecol. Evol. 20:81-87. [DOI] [PubMed] [Google Scholar]
- 15.Delong, E. F., G. S. Wickham, and N. R. Pace. 1989. Phylogenetic stains: ribosomal RNA-based probes for the identification of single cells. Science 243:1360-1363. [DOI] [PubMed] [Google Scholar]
- 16.De Rijk, P., J. Wuyts, and R. De Wachter. 2003. RnaViz 2: an improved representation of RNA secondary structure. Bioinformatics 19:299-300. [DOI] [PubMed] [Google Scholar]
- 17.DeSantis, T. Z., P. Hugenholtz, N. Larsen, M. Rojas, E. L. Brodie, K. Keller, T. Huber, D. Dalevi, P. Hu, and G. L. Andersen. 2006. Greengenes, a chimera-checked 16S rRNA gene database and workbench compatible with ARB. Appl. Environ. Microbiol. 72:5069-5072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Felsenstein, J. 2005. PHYLIP (phylogeny inference package, v. 3.6). University of Washington, Seattle. http://evolution.genetics.washington.edu/phylip.html.
- 19.Ferrari, B. C., S. J. Binnerup, and M. Gillings. 2005. Microcolony cultivation on a soil substrate membrane system selects for previously uncultured soil bacteria. Appl. Environ. Microbiol. 71:8714-8720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Giovannoni, S. J., H. J. Tripp, S. Givan, M. Podar, K. L. Vergin, D. Baptista, L. Bibbs, J. Eads, T. H. Richardson, M. Noordewier, M. S. Rappe, J. M. Short, J. C. Carrington, and E. J. Mathur. 2005. Genome streamlining in a cosmopolitan oceanic bacterium. Science 309:1242-1245. [DOI] [PubMed] [Google Scholar]
- 21.Gordon, D., C. Abajian, and P. Green. 1998. Consed: a graphical tool for sequence finishing. Genome Res. 8:195-202. [DOI] [PubMed] [Google Scholar]
- 22.Guindon, S., and O. Gascuel. 2003. A simple, fast, and accurate algorithm to estimate large phylogenies by maximum likelihood. Syst. Biol. 52:696-704. [DOI] [PubMed] [Google Scholar]
- 23.Hallam, S. J., N. Putnam, C. M. Preston, J. C. Detter, D. Rokhsar, P. M. Richardson, and E. F. Delong. 2004. Reverse methanogenesis: testing the hypothesis with environmental genomics. Science 305:1457-1462. [DOI] [PubMed] [Google Scholar]
- 24.Handelsman, J. 2004. Metagenomics: application of genomics to uncultured microorganisms. Microbiol. Mol. Biol. Rev. 68:669-685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hosono, S., A. F. Faruqi, F. B. Dean, Y. Du, Z. Sun, X. Wu, J. Du, S. F. Kingsmore, M. Egholm, and R. S. Lasken. 2003. Unbiased whole-genome amplification directly from clinical samples. Genome Res. 13:954-964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hugenholtz, P. 2002. Exploring prokaryotic diversity in the genomic era. Genome Biol. 3:0003.1-0003.8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hugenholtz, P., G. W. Tyson, R. I. Webb, A. M. Wagner, and L. L. Blackall. 2001. Investigation of candidate division TM7, a recently recognized major lineage of the domain Bacteria with no known pure-culture representatives. Appl. Environ. Microbiol. 67:411-419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kalyuzhnaya, M. G., R. Zabinsky, S. Bowerman, D. R. Baker, M. E. Lidstrom, and L. Chistoserdova. 2006. Fluorescence in situ hybridization-flow cytometry-cell sorting based method for separation and enrichment of type I and type II methanotroph populations. Appl. Environ. Microbiol. 72:4293-4301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lorenz, P., and J. Eck. 2005. Metagenomics and industrial applications. Nat. Rev. Microbiol. 3:510-516. [DOI] [PubMed] [Google Scholar]
- 30.Manz, W., R. Amann, W. Ludwig, M. Vancanneyt, and K. H. Schleifer. 1996. Application of a suite of 16S rRNA-specific oligonucleotide probes designed to investigate bacteria of the phylum cytophaga-flavobacter-bacteroides in the natural environment. Microbiology 142:1097-1106. [DOI] [PubMed] [Google Scholar]
- 31.Martin, H. G., N. Ivanova, V. Kunin, F. Warnecke, K. W. Barry, A. C. McHardy, C. Yeates, S. He, A. A. Salamov, E. Szeto, E. Dalin, N. H. Putnam, H. J. Shapiro, J. L. Pangilinan, I. Rigoutsos, N. C. Kyrpides, L. L. Blackall, K. D. McMahon, and P. Hugenholtz. 2006. Metagenomic analysis of two enhanced biological phosphorus removal (EBPR) sludge communities. Nat. Biotechnol. 24:1263-1269. [DOI] [PubMed] [Google Scholar]
- 32.Mattick, J. S. 2002. Type IV pili and twitching motility. Annu. Rev. Microbiol. 56:289-314. [DOI] [PubMed] [Google Scholar]
- 33.McLennan, A. G. 2006. The Nudix hydrolase superfamily. Cell. Mol. Life Sci. 63:123-143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Mira, A., R. Pushker, B. A. Legault, D. Moreira, and F. Rodriguez-Valera. 2004. Evolutionary relationships of Fusobacterium nucleatum based on phylogenetic analysis and comparative genomics. BMC Evol. Biol. 4:50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Nakajima, H., Y. Hongoh, R. Usami, T. Kudo, and M. Ohkuma. 2005. Spatial distribution of bacterial phylotypes in the gut of the termite Reticulitermes speratus and the bacterial community colonizing the gut epithelium. FEMS Microbiol. Ecol. 54:247-255. [DOI] [PubMed] [Google Scholar]
- 36.Pace, N. R. 1997. A molecular view of microbial diversity and the biosphere. Science 276:734-740. [DOI] [PubMed] [Google Scholar]
- 37.Peabody, C. R., Y. J. Chung, M. R. Yen, D. Vidal-Ingigliardi, A. P. Pugsley, and M. H. Saier, Jr. 2003. Type II protein secretion and its relationship to bacterial type IV pili and archaeal flagella. Microbiology 149:3051-3072. [DOI] [PubMed] [Google Scholar]
- 38.Pinard, R., A. de Winter, G. J. Sarkis, M. B. Gerstein, K. R. Tartaro, R. N. Plant, M. Egholm, J. M. Rothberg, and J. H. Leamon. 2006. Assessment of whole genome amplification-induced bias through high-throughput, massively parallel whole genome sequencing. BMC Genomics 7:216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Raghunathan, A., H. R. Ferguson, Jr., C. J. Bornarth, W. Song, M. Driscoll, and R. S. Lasken. 2005. Genomic DNA amplification from a single bacterium. Appl. Environ. Microbiol. 71:3342-3347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Rheims, H., F. A. Rainey, and E. Stackebrandt. 1996. A molecular approach to search for diversity among bacteria in the environment. J. Ind. Microbiol. 17:159-169. [Google Scholar]
- 41.Sogin, M. L., H. G. Morrison, J. A. Huber, D. M. Welch, S. M. Huse, P. R. Neal, J. M. Arrieta, and G. J. Herndl. 2006. Microbial diversity in the deep sea and the underexplored “rare biosphere.” Proc. Natl. Acad. Sci. USA 103:12115-12120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Stein, J. L., T. L. Marsh, K. Y. Wu, H. Shizuya, and E. F. Delong. 1996. Characterization of uncultivated prokaryotes: isolation and analysis of a 40-kilobase-pair genome fragment from a planktonic marine archaeon. J. Bacteriol. 178:591-599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Teeling, H., and F. O. Gloeckner. 2006. RibAlign: a software tool and database for eubacterial phylogeny based on concatenated ribosomal protein subunits. BMC Bioinformatics 7:66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Thomsen, T. R., B. V. Kjellerup, J. L. Nielsen, P. Hugenholtz, and P. H. Nielsen. 2002. In situ studies of the phylogeny and physiology of filamentous bacteria with attached growth. Environ. Microbiol. 4:383-391. [DOI] [PubMed] [Google Scholar]
- 45.Tringe, S. G., and E. M. Rubin. 2005. Metagenomics: DNA sequencing of environmental samples. Nat. Rev. Genet. 6:805-814. [DOI] [PubMed] [Google Scholar]
- 46.Tringe, S. G., C. von Mering, A. Kobayashi, A. A. Salamov, K. Chen, H. W. Chang, M. Podar, J. M. Short, E. J. Mathur, J. C. Detter, P. Bork, P. Hugenholtz, and E. M. Rubin. 2005. Comparative metagenomics of microbial communities. Science 308:554-557. [DOI] [PubMed] [Google Scholar]
- 47.Tyson, G. W., J. Chapman, P. Hugenholtz, E. E. Allen, R. J. Ram, P. M. Richardson, V. V. Solovyev, E. M. Rubin, D. Rokhsar, and J. F. Banfield. 2004. Community structure and metabolism through reconstruction of microbial genomes from the environment. Nature 428:37-43. [DOI] [PubMed] [Google Scholar]
- 48.Whitaker, R. J., and J. F. Banfield. 2006. Population genomics in natural microbial communities. Trends Ecol. Evol. 21:508-516. [DOI] [PubMed] [Google Scholar]
- 49.Woyke, T., H. Teeling, N. N. Ivanova, M. Huntemann, M. Richter, F. O. Gloeckner, D. Boffelli, I. J. Anderson, K. W. Barry, H. J. Shapiro, E. Szeto, N. C. Kyrpides, M. Mussmann, R. Amann, C. Bergin, C. Ruehland, E. M. Rubin, and N. Dubilier. 2006. Symbiosis insights through metagenomic analysis of a microbial consortium. Nature 443:950-955. [DOI] [PubMed] [Google Scholar]
- 50.Zengler, K., G. Toledo, M. Rappe, J. Elkins, E. J. Mathur, J. M. Short, and M. Keller. 2002. Cultivating the uncultured. Proc. Natl. Acad. Sci. USA 99:15681-15686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Zhang, K., A. C. Martiny, N. B. Reppas, K. W. Barry, J. Malek, S. W. Chisholm, and G. M. Church. 2006. Sequencing genomes from single cells by polymerase cloning. Nat. Biotechnol. 24:680-686. [DOI] [PubMed] [Google Scholar]